Abstract
miRNA-mediated repression in animals is dependent on the GW182 protein family. GW182 proteins are recruited to the miRNA repression complex through direct interaction with Argonaute proteins, and they function downstream to repress target mRNA. Here we demonstrate that in human and Drosophila melanogaster cells, the critical repressive features of both the N-terminal and C-terminal effector domains of GW182 proteins are Gly/Ser/Thr-Trp (G/S/TW) or Trp-Gly/Ser/Thr (WG/S/T) motifs. These motifs, which are dispersed across both domains and act in an additive manner, function by recruiting components of the CCR 4–NOT deadenylation complex. A heterologous yeast polypeptide with engineered WG/S/T motifs acquired the ability to repress tethered mRNA and to interact with the CCR 4–NOT complex. These results identify previously unknown effector motifs functioning as important mediators of miRNA-induced silencing in both species, and they reveal that recruitment of the CCR 4–NOT complex by tryptophan-containing motifs acts downstream of GW182 to repress mRNA s, including inhibiting translation independently of deadenylation.
MicroRNAs (miRNAs) are small, ~21-nt–long RNAs that post-transcriptionally regulate gene expression in eukaryotes. In animals, miRNAs bind to partially complementary sites in mRNAs, leading to translational repression and mRNA deadenylation and degradation1–4. miRNAs function as part of ribonucleoprotein complexes, miRNPs, with Argonaute (AGO) and GW182 family proteins being the crucial components. GW182s interact directly with AGO proteins and function downstream as effectors mediating mRNA repression. Hence, understanding the function of GW182 proteins is critical for understanding miRNA-mediated repression.
GW182 functional regions have been mapped in D. melanogaster and mammalian proteins. In D. melanogaster, three regions were found to repress tethered mRNA to a similar extent5: the N-terminal effector domain (NED) having multiple GW-repeats, the middle Q-rich region, and the C-terminal effector domain (CED) containing the poly(A) binding protein (PABP)-interacting motif 2 (PAM2) and the RNA-recognition motif (RRM). The role of the CED in repression was also previously established by others6–8. In mammals, tethering of the three regions mentioned above also represses reporter mRNA, with the major contribution being provided by the CED9–11. The mechanism by which GW182 domains repress mRNA function appears to be evolutionarily conserved, as dGW182 can repress mRNA function in mammalian cells, and human TNRC6 proteins (mammals express three counterparts of dGW182: TNRC6A, B and C) act as repressors in D. melanogaster cells5,8,9.
The CED of both human and fly GW182s interacts with PABP, and this interaction, possibly by interfering with the PABP-eIF4G association, promotes target mRNA deadenylation by recruiting, through PABP, the components of the CCR4–NOT deadenylation complex7,8,12,13. In addition, others14–16 have demonstrated the role of CCR4–NOT and PAN2–PAN3 deadenylation complexes in the deadenylation of miRNA targets. It is unclear how GW182 proteins recruit these deadenylase complexes and how translation repression is modulated. One possible model is that the interaction of CED with PABP interferes with the PABP-eIF4G association and reduces translation7,12,13. However, interfering with eIF4G-PABP interaction and binding of the CCR4–NOT complex through PABP cannot explain the repression of mRNAs bearing no poly(A) tails (reviewed in refs. 2,3), nor can it explain the repression by GW182 domains other than CED.
Previous work on the fly GW182 and human NED indicated a role for glycine-tryptophan (GW) repeats as effector motifs contributing to miRNA-mediated silencing17,18. Here we set out to investigate how the GW182 CED and NED regions bring about mRNA repression. We found that motifs bearing tryptophan residues also in contexts other than GW or WG function as important repressive sequences in the CED, both in human and D. melanogaster cells. The effector G/S/TW and WG/S/T motifs in the NED and CED recruit the components of CCR4–NOT and PAN2–PAN3 complexes in a PABP-independent manner to repress function of both poly(A)+ and poly(A)− mRNAs. These results identify the recruitment of the CCR4–NOT complex as a critical event for miRNA-mediated mRNA degradation and translation repression.
RESULTS
The CED of TNRC 6C interacts with the CCR 4–NOT complex
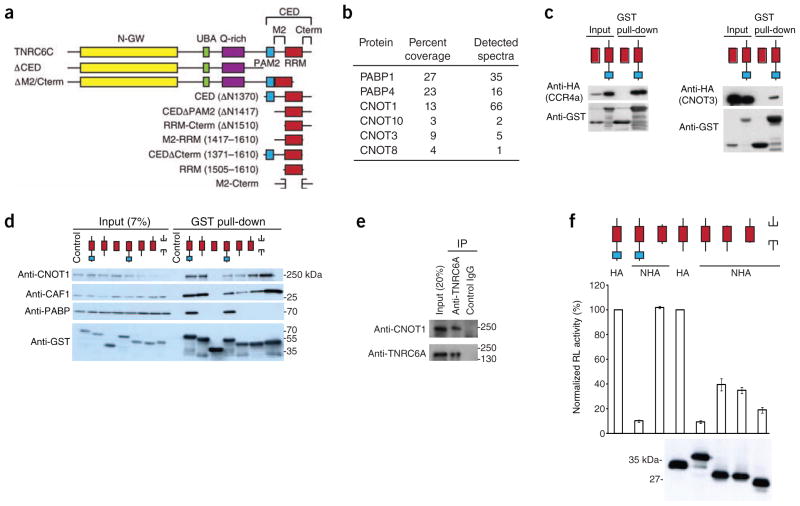
The CED of human TNRC6C (ΔN1370 fragment; Fig. 1a) functions as an autonomous repressive domain, inducing both translational inhibition and mRNA degradation9. To elucidate how the CED induces the repression of target mRNAs, it was expressed as a glutathione S-transferase (GST) fusion in HEK293T cells and used for pull-down experiments. Among the pulled-down proteins, MS identified several components of the CCR4–NOT complex, including CNOT1, its scaffolding component and CNOT8, a paralog of the deadenylase CNOT7/CAF1 (Fig. 1b). PABP was also among the interacting proteins, consistent with previous findings8,12,13. The interaction of the CED with different components of CCR4–NOT, either endogenous or ectopically expressed, was confirmed by western blotting (Fig. 1c,d). Notably, endogenous TNRC6A could also co-immunoprecipitate CNOT1 (Fig. 1e).
Figure 1.
The TNRC6C CED interacts with components of the CCR4–NOT complex. (a) Schematic representation of human TNRC6C and fragments analyzed in the study. Individual domains and regions of TNRC6C are indicated: N-GW, GW-repeat–rich region; UBA, ubiquitin associated–like domain; RRM, RNA-recognition motif; M2 and Cterm2, regions flanking RRM, constituting—together with PAM2 and RRM—the CED region. (b) MS analysis of proteins interacting with the CED. Relevant proteins are listed along with peptide coverage and amount of assigned spectra. For full list of proteins, see Supplementary Table 1. (c) Validation of the CED interaction with selected CCR4–NOT components by GST pull-down assays and western blotting. GST-RRM was used as a control. (d) M2 and Cterm regions of the CED interact with components of the CCR4–NOT complex but not with PABP. TNRC6C CED and its subfragments were used for GST pull-down assays. Inputs (7%) and pull-down assays were analyzed by western blotting. Extracts from nontransfected cells were used as controls. (e) CNOT1 co-immunoprecipitates with endogenous TNRC6A. (f) M2 and Cterm regions of TNRC6C mediate repression of tethered mRNA. HEK293T cells were co-transfected with plasmids encoding NHA-CED or indicated fragments, and RL-5BoxB and firefly luciferase–transfection control (FL-Con) reporters. As negative controls, untethered hemagglutinin-CED (HA-CED) and tethered NHA-RRM (where ‘N’ stands for tethering λ peptide; see Supplementary Fig. 2a) were expressed. Values represent percentage of Renilla luciferase activity (normalized to firefly luciferase activity) in the presence of nontethered HA-CED or HA-CEDΔPAM2. In all luciferase assays presented in this work, values represent means ± s.e.m. from three to six experiments. Expression levels of HA- or NHA-fusion proteins were estimated by western blotting.
CAF1 was reported to interact with PABP through the TOB1 protein19, raising the possibility that the CED recruits CCR4–NOT through PABP. The PAM2 motif (Fig. 1a and Supplementary Fig. 1) represents the main region in the CED responsible for its interaction with PABP in human cells8,13. Deletion of PAM2 (CEDΔPAM2) abrogated the association with PABP without affecting the interaction with CNOT1 and CAF1, suggesting that the CED interaction with CCR4–NOT is PABP-independent (Fig. 1d). Moreover, the observed interactions were not mediated by RNA, as they were resistant to micrococcal nuclease treatment (Fig. 1d and Online Methods).
To identify sequences in CEDΔPAM2 responsible for the CCR4–NOT interaction, we did pull-down assays with CEDΔPAM2 subfragments (see Fig. 1a). Deleting either M2 or C-terminal (Cterm) regions reduced the interaction with CNOT1 and CAF1. The RRM alone did not pull down CNOT1 or CAF1, whereas a fusion of M2 and Cterm regions pulled them down with an efficiency similar to that of CEDΔPAM2 (Fig. 1d).
Repression by the CED correlates with CCR 4–NOT interaction
The CED domain and its subfragments were tested for activity in repressing protein synthesis in an mRNA-tethering assay (Supplementary Fig. 2a). Tethering of the CED or CEDΔPAM2 repressed Renilla luciferase expression by approximately ten times, when compared to proteins lacking the N-peptide (Fig. 1f). Constructs lacking either M2 or Cterm regions showed reduced repression, whereas the M2-Cterm fusion repressed almost as well as CEDΔPAM2 (Fig. 1f). Hence, similarly to their requirement for the interaction with the CCR4–NOT complex, the combined M2 and Cterm regions are sufficient for effective mRNA repression8.
When analyzed in the context of full-length TNRC6C, deletion of M2 and Cterm regions alleviated mRNA repression to a level comparable to that seen when the entire CED is deleted (Supplementary Fig. 2b). Similarly, both TNRC6C deletion mutants interacted less strongly with CAF1 and CNOT1 (Supplementary Fig. 2c). The ability of both mutants to still partially repress mRNA function and associate with CCR4–NOT is readily explained by observations that, in addition to the CED, N-proximal regions of GW182s have the potential to repress mRNAs5,9,17,18 and associate with CCR4–NOT components (see below).
To determine the features of M2 and Cterm regions that repress mRNA function, we identified conserved regions of two to six amino acids by alignment of different GW182 proteins (Supplementary Fig. 1). Because their mutagenesis in the context of CEDΔPAM2 had a very limited effect (data not shown), we tested the mutations in the context of CEDΔPAM2 subfragments, M2-RRM or RRM-Cterm (Fig. 1a and Supplementary Fig. 2d–h). This analysis revealed considerable redundancy of the CED sequences responsible for mediating both the interaction with CCR4–NOT and repression of mRNA function. Unexpectedly, our results also showed that all mutations appreciably affecting both activities were in elements containing tryptophan residues, and those tryptophan residues were important for the repressive activity, in a manner that involved recruitment of CCR4–NOT (Supplementary Figs. 2d–h and 3a,b and Supplementary Results).
W-motifs represent signals recruiting deadenylase complexes
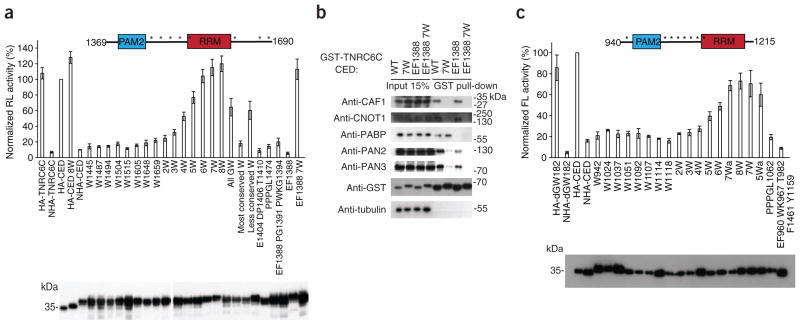
When inspecting the alignment of the CED across different species, we noted that GW or WG repeats in one GW182 homolog often align with the S/TW or WS/T repeats in other homologs (Supplementary Fig. 1). We hypothesized that reiterated G/S/TW or WG/S/T repeats (referred to as W-motifs), rather than only GW or WG repeats, must have a role in repression. The TNRC6C CED contains eight W-motifs (Fig. 2a and Supplementary Fig. 1). We analyzed the effect of Trp→Ala mutations in W-motifs on expression of the tethered mRNA (Fig. 2a). Notably, although single Trp→Ala mutations had no marked effect on repression by the CED, their combinations had a progressive additive effect. Notably, when all eight tryptophans were mutated (W8), repression by the CED was fully alleviated. We observed no alleviation when other conserved amino acid stretches were mutated in either PAM2 or M2 regions. Western blot analysis showed that the differences in repressive potential could not be explained by differences in expression levels (Fig. 2a). The most conserved tryptophan residue, Trp1515, did not contribute to repression (8W and 7W mutants differ only in the Trp1515 mutation). Trp1515 participates in the RRM structure6, whereas other W-motifs reside in regions predicted as disordered (http://dis.embl.de). Otherwise, W-motifs seem to contribute to repression independently of the degree of conservation and the context; that is, whether they are located next to glycine, serine or threonine residues (Fig. 2a).
Figure 2.
W-motifs in GW182 proteins mediate mRNA repression by recruiting CCR4–NOT and PAN2–PAN3 deadenylation complexes. (a) Mutations of tryptophan residues in W-motifs alleviate repression by the TNRC6C CED. Schematic representation of the TNRC6C CED with positions of W-motifs marked with asterisks is shown above the graph. Plasmids encoding either wild-type NHA-CED or its mutants (mutations always to alanine; when several consecutive amino acids are mutated, the number corresponds to the first residue in the mutated stretch) were co-transfected to HEK293T cells, together with RL-5BoxB and FL-Con. As negative controls, plasmids encoding untethered HA-TNRC6C or HA-CED were used. Mutants 2W through 8W contain Trp→Ala mutations in W-motifs (for details, see Online Methods). All GW, W1487 W1494 W1648 W1659; most conserved tryptophan, W1504 W1515; less conserved tryptophan, W1487 W1605 W1648 W1659. Values represent percentages of Renilla luciferase produced in the presence of untethered HA-CED control. Expression of HA- or NHA-fusion proteins was estimated by western blotting (lower panel). (b) Proteins identified as interacting with the CED in a tryptophan-dependent manner by MS (Supplementary Fig. 4a) were validated by GST pull-down assays and western blotting. Positions of protein size markers are indicated. (c) W-motifs are required for repression by the D. melanogaster GW182 CED. NHA-dGW182 CED, either wild-type or with mutations, were co-transfected with FL-5BoxB and RL-Con in S2 cells. As negative controls, plasmids encoding HA-dGW182 and HA-dGW182 CED were used. Mutants 2W through 8W contain mutations in W-motifs, with some (5Wa and 7Wa) having different combinations of mutated tryptophans (positions of W-motifs are marked with asterisks in the scheme above; for details, see Online Methods). Expression of firefly luciferase was normalized to Renilla luciferase. Values represent percentages of firefly luciferase produced in the presence of HA-CED. Expression of HA-fusions was estimated by western blotting.
Because the CED Trp→Ala mutants relieve repression activity, we determined, by MS, how these mutations affect the interaction of proteins with the CED (Supplementary Fig. 4a). As expected, the wild-type CED associated with different components of the CCR4–NOT complex. However, none of them associated with the 7W mutant, indicating that the CED interacts with CCR4–NOT in a W-dependent manner. As both wild-type and 7W mutant CEDs contain the PAM2 region, each associated with PABP. We also observed that the PAN2–PAN3 deadenylase complex components were present among proteins bound by wild-type but not 7W mutant fusions, though PAN2 and PAN3 were found in smaller amounts than CCR4–NOT proteins.
We also analyzed the pull-down assays by western blotting (Fig. 2b). Both CNOT1 and CAF1 interacted with wild-type CED but not with its 7W mutant. Mutations of W-motifs also strongly affected association with PAN2 and PAN3 but had no major effect on interaction with PABP. In two out of four experiments, however, PABP binding was slightly affected in the 7W mutant (1.5-fold to two-fold; not shown). This could be explained by the secondary weak PABP binding site located in the M2 or Cterm regions7,13. Interactions with PABP through this site seemed to be indirect8, suggesting that they occur through components of the CCR4–NOT or PAN2–PAN3 complexes.
We have mapped regions in the CED required for PABP and CCR4–NOT interactions, so we were able to determine the interdependence of these interactions. Mutations in PAM2 that disrupted the CED-PABP interaction (mutant EF1388; mutations are always to alanine; when several consecutive amino acids are mutated, the number corresponds to the first residue in the mutated stretch) did not affect the association of CED with CCR4–NOT, whereas the 7W mutant that did not interact with CCR4–NOT still interacted with PABP (Fig. 2b). Hence, the CED interactions with CCR4–NOT and PABP are independent. The PAN2–PAN3 interactions were more complex: mutation of PAM2 somewhat reduced binding of PAN2 and PAN3, though not as strongly as mutations of W-motifs, and the double EF1388 7W mutant showed no PAN2–PAN3 binding (Fig. 2b). These results suggest that PAN2–PAN3 is primarily recruited through the function of W-motifs but that it can also weakly interact with the CED through PABP, which is consistent with the direct PAN3-PABP interaction previously described20.
To investigate whether the role of W-motifs in repression is conserved across the species, we also introduced Trp→Ala mutations into the eight W-motifs in the dGW182 CED (Fig. 2c and Supplementary Fig. 1). The mutant proteins were tethered to the firefly luciferase reporter FL-5BoxB, expressed in fly S2 cells. As in the case of the TNRC6C CED, mutations alleviated repression in an additive manner, leading to almost no repression when all tryptophans were mutated. In contrast, mutation of other conserved sequences had no appreciable effect (Fig. 2c).
Taken together, our data indicate that the role of W-motifs in mRNA repression is evolutionarily conserved and that W-motifs function by recruiting CCR4–NOT and PAN2–PAN3 complexes independently of PABP.
Repression by NED and CED follows a similar mechanism
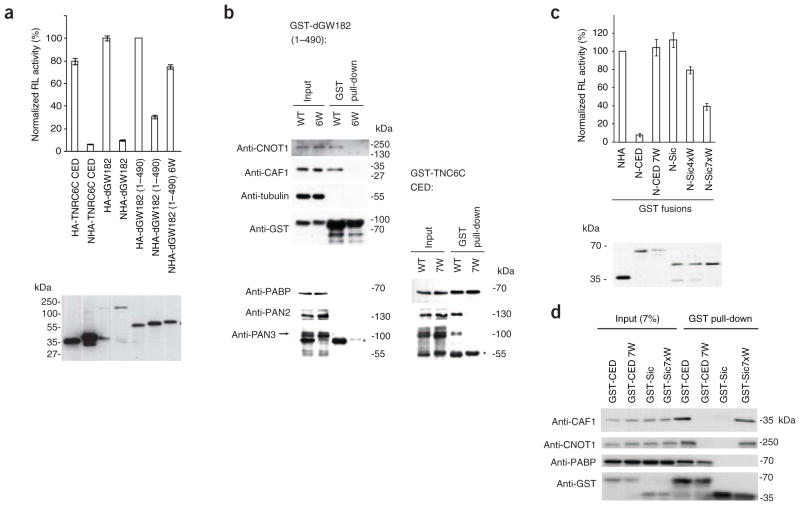
To test if the recruitment of the CCR4–NOT complex represents a mechanism conserved across different effector domains and across species, we analyzed the function of the dGW182 NED in human HEK293T cells. Our previous work demonstrated that the dGW182 NED is able to repress the tethered mRNA in human cells9, and we investigated whether mutations in W-motifs in that region would affect its repressive potential. Because the 205–490 dGW182 fragment, studied previously in S2 cells, was less effective in human cells (data not shown), a longer 1–490 fragment was used instead. We observed that six Trp→Ala mutations in GW repeats in the 205–490 region (mutant NHA-dGW182(1–490)6W) led to a marked alleviation of repression (Fig. 3a), similar to that observed in D. melanogaster S2 cells17.
Figure 3.
W-motifs present in the dGW182 NED and the engineered yeast protein fragment repress tethered mRNA and recruit components of CCR4–NOT. (a) The dGW182 NED W-motifs function in mRNA repression. HEK293T cells were transfected with RL-5BoxB, FL-Con and plasmids expressing either full-length NHA-dGW182 or its NED (1–490) (WT or 6W mutant; for description of the mutant, see ref. 17). As negative controls, HA-dGW182 and HA-dGW182(1–490) were used. As positive controls, TNRC6C NHA-CED and full-length NHA-dGW182 were tethered. Values represent percentages of Renilla luciferase produced in the presence of HA-dGW182(1–490). Western blot analysis of HA- or NHA-fusion proteins is presented below. (b) GST fusions of the dGW182(1–490), WT and 6W mutant, expressed in HEK293T cells, were used for GST pull-down assays. Inputs (7% for anti-CNOT1, anti-CAF1, anti-tubulin and anti-GST; 15% for anti-PABP, anti-PAN2 and anti-PAN3) and the pulled-down material were analyzed by western blotting, using indicated antibodies. Additional western blots (on the right) for PABP, PAN2 and PAN3 represent pull-down assays done with the TNRC6C GST-CED analyzed in parallel on the same gel. Anti-PAN3 antibody cross-reacts with GST (asterisk). (c) W-motifs are sufficient to induce repression of tethered mRNA. HEK293T cells were transfected with RL-5BoxB, FL-Con and plasmids encoding engineered N-Sic-GST protein fusions having either four (N-Sic4xW-GST) or seven (N-Sic7xW-GST) W-motifs. N-Sic-GST containing no tryptophan residues, and NHA-GST, served as controls; plasmids encoding TNRC6C N-CED-GST, WT and 7W mutant were transfected for comparison. (d) GST pull-down assays with GST-Sic7xW, GST-CED (positive control), and GST-CED 7W and GST-Sic (negative controls), were done as in Figure 1d. The pulled-down material was analyzed by western blotting, using indicated antibodies.
Analysis of interaction partners of the dGW182 NED(1–490) in HEK293T cells revealed that it interacts with CNOT1 and CAF1 in a W-dependent manner (Fig. 3b), suggesting that the mechanism of mRNA repression by different GW182 domains is similar and involves the recruitment of CCR4–NOT through W-motifs. Neither PABP nor PAN2–PAN3 was detected in the NED GST pull-down assays, whereas they were pulled down with the TNRC6C CED (Fig. 3b, lower panels). Thus, interaction with PABP and PAN2–PAN3 may not be required for repression by the NED.
Engineered W-motifs are sufficient to induce repression
We investigated whether W-motifs are not only required but also sufficient to induce mRNA repression. We introduced X→Trp mutations (with X corresponding to any amino acid) to the unstructured fragment of the yeast protein Sic1p21. The resulting engineered proteins, having either four (Sic4xW) or seven (Sic7xW, Supplementary Fig. 4b and Supplementary Methods) sequences resembling the W-motifs, were fused to N- and GST polypeptides and their activity tested in the tethering assay. Notably, the proteins containing W-motifs were able to repress Renilla luciferase-5BoxB (RL-5BoxB) mRNA, with the degree of repression being dependent on the number of motifs (Fig. 3c). Moreover, GST pull-down experiments revealed that both CAF1 and CNOT1, but not PABP, were bound by Sic7xW but not the control tryptophan-free fragment (Fig. 3d). Hence, W-motifs are not only necessary but also sufficient to induce mRNA repression by recruiting CCR4–NOT.
W-motifs function in a genuine miRNA-mediated repression
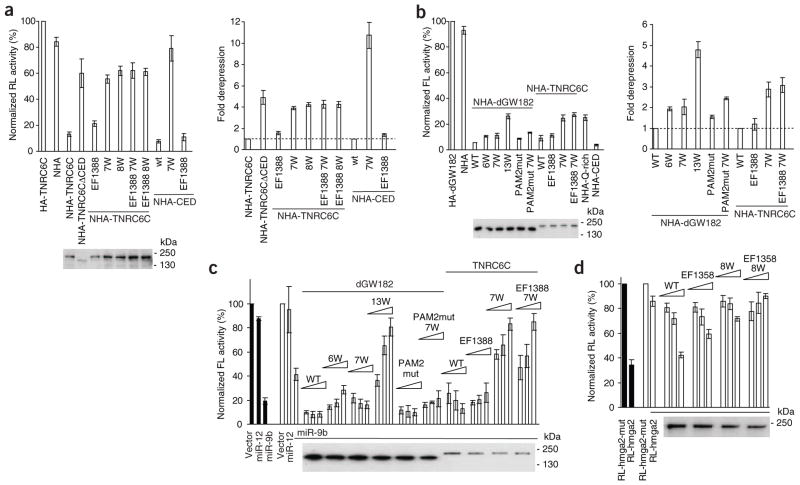
We next investigated whether W-motifs also function in the context of full-length GW182 proteins. Mutation of tryptophan residues in W-motifs of the CED strongly compromised the repressive potential of TNRC6C in HEK293T cells (Fig. 4a, mutants 7W and 8W, ~four-fold effect; for clarity, the data are also shown as fold derepression in the right panels of Fig. 4a,b). A more marked effect (~ten-fold) of tryptophan mutations on activity of the CED alone (Fig. 4a; see also Fig. 2a) is readily explained by the potential of the TNRC6 N-proximal sequences to partially repress the tethered mRNA9,18. In the context of the full-length TNRC6C, the PAM2 mutation EF1388 led to moderate alleviation of repression, consistent with previous data8.
Figure 4.
W-motifs are necessary for repression by full-length GW182 and function in bona fide miRNA repression. (a) W-motifs are required for repression by tethered full-length TNRC6C. The experiment was done as in Figure 2a but included the full-length TNRC6C. The right panel shows fold derepression relative to repression induced by WT NHA-TNRC6C or NHA-CED taken to be a value of 1 (broken line). Western analysis of expression levels of relevant mutants in a and other panels, with anti-HA antibody, is shown below the graphs. (b) Mutations in W-motifs lead to partial derepression of tethered mRNAs in D. melanogaster S2 cells. The assay was done as in Figure 2c but with the full-length dGW182 and TNRC6C. 6W, 7W and EF1388 mutations were described in Figures 2 and 3a but are here introduced into the full-length proteins. 13W mutant combines 6W and 7W; PAM2mut has EF960 WK967 Thr982 mutated. NHA-Q–rich (1080–1245) and NHA-CED represent TNRC6C fragments. In the right panel, data are presented as in a. (c) W-motifs are required to rescue depletion of endogenous dGW182. Endogenous dGW182 was depleted in D. melanogaster S2 cells with dsRNA (open bars); a batch of cells was treated with GFP-specific dsRNA as a control (black bars). Cells were transfected with RL-Con, FL-nerfin, and plasmids encoding miR-9b or miR-12, or the empty vector. To rescue depletion of dGW182, increasing amounts of plasmids encoding NHA-dGW182, NHA-TNRC6C or their mutants were co-transfected. In panels c and d, extracts from cells transfected with highest plasmid concentrations were used for western blotting. (d) W-motifs are necessary to complement the knockdown of endogenous TNRC6 proteins. HeLa cells were transfected with siRNAs targeting three endogenous TNRC6 proteins (open bars) or AllStars siRNA (negative control, black bars), RL-hmga2 reporter containing let-7 sites or its mutant version (RL-hmga2 mut), and increasing amounts of plasmids expressing NHA-TNRC6A or its mutants: 8W has Trp→Ala mutations in W-motifs within the CED region (Supplementary Fig. 1 and Online Methods); EF1358 has PAM2 mutated.
In D. melanogaster S2 cells, mutating W-motifs also led to alleviation of repression induced by either dGW182 or TNRC6C, though the effects were less pronounced than in human cells (Fig. 4b). This can be explained by a marked contribution of the Q-rich domains of these proteins to the repression in S2 cells (Fig. 4b, NHA-Q-rich and ref. 5). For dGW182, mutating W-motifs in either NED (mutant 6W) or CED (7W) alone had only a mild effect (~two-fold), but combining these mutations (13W) led to more than four-fold alleviation of repression. Mutating seven tryptophans within the CED of TNRC6C alleviated repression ~three-fold, with mutations in PAM2 having no effect (Fig. 4b).
Having demonstrated that W-motifs function in the context of full-length GW182 proteins, we analyzed their importance in a bona fide miRNA repression assay. We depleted S2 cells of the endogenous dGW182 and tested tryptophan mutants of dGW182 for activity to rescue miRNA repression. To assess miRNA-mediated silencing, cells were co-transfected with the firefly luciferase–nerfin (FL-nerfin) reporter and the plasmid expressing miR-9b, which targets the FL-nerfin 3′ UTR. miR-9b efficiently repressed FL-nerfin mRNA in control cells (Fig. 4c, black bars), and depletion of dGW182 (open bars) partially alleviated miR-9b–induced repression; as expected, transfection of a plasmid encoding wild-type dGW182 resistant to RNAi rescued the repression. Mutations of tryptophans in either NED (6W) or CED (7W) had only a minor effect on the functionality of dGW182 in the rescue, consistent with independent repression by NED and CED domains5. However, combining the tryptophan mutations in both regions led to a strong alleviation of repression, demonstrating the role of W-motifs in miRNA-mediated silencing. Mutation of the PAM2 motif had no appreciable effect.
Because GW repeats present in the N-terminal part of dGW182 contribute to dAGO1 binding22, we tested if mutations of tryptophans introduced into dGW182 affect its interaction with dAGO1. We found that whereas the 7W mutant interacted with dAGO1 as efficiently as wild-type dGW182, the 6W and 13W mutants showed lower levels of binding (Supplementary Fig. 4c). Consequently, it is possible that tryptophan residues in the NED contribute to the rescue not only by enhancing the CCR4–NOT interaction (Fig. 3b) but also by increasing the affinity of dGW182 for dAGO1. However, as 6W and 13W mutants have similar dAGO1-binding properties (Supplementary Fig. 4c), we can conclude that W-motifs in the CED are required for the dGW182 function in miRNA repression (Fig. 4c).
Because human TNRC6C is able to complement the knockdown of dGW182 in S2 cells8 (Fig. 4c), we tested the effect of tryptophan mutations on its function in rescue experiments. Notably, mutations of the W-motifs within the CED region (7W) strongly alleviated repression by TNRC6C. This is consistent with findings that the CED represents the major repressive region of human GW182 proteins6,9,11. To test the requirement of W-motifs for miRNA repression in human cells, we used a reporter having the 3′ UTR of the HMGA2 gene (RL-hmga2), which is targeted by let-7 miRNA23,24. This miRNA is expressed endogenously in HeLa cells, and it represses RL-hmga2 by about three times when compared with its mutant version that has disabled let-7 sites (Fig. 4d, black bars). Depletion of all three TNRC6 proteins by RNAi led to almost full alleviation of the repression (Fig. 4d, open bars), which could be rescued with the wild-type TNRC6A (we used a TNRC6A paralog, as it functions most efficiently in the complementation assay8). Mutation of PAM2 (EF1358) partially interfered with the rescue, consistent with the previous report8. Notably, mutations of W-motifs either alone (8W) or in combination with PAM2 mutation (EF1358 8W) led to a nearly complete loss of TNRC6A function in miRNA repression. We conclude that W-motifs of both D. melanogaster and human GW182s are important for bona fide miRNA-mediated silencing.
Role of W-motifs and CCR 4–NOT in poly(A)− mRNA repression
Recruitment of the CCR4–NOT deadenylase explains how miRNAs and tethered GW182 silencing domains induce deadenylation and mRNA decay2,3. Indeed, we observed that tethering of the dGW182 CED induces deadenylation of the FL-5BoxB reporter and that this effect is dependent on W-motifs (Supplementary Fig. 5). Do the CED and CCR4–NOT also mediate the translational repression known to be induced by miRNA machinery2–4? To address this question we first tested whether the dGW182 CED can repress, in a W-motif–dependent manner, tethered mRNAs in which the polyadenylation signal is substituted by either a histone stem loop (HSL) or a hammerhead ribozyme (HhR). These mRNAs, FL-5BoxB-HSL and FL-5BoxB-HhR, were previously shown to have no poly(A) and to undergo translational repression in S2 cells in response to tethered dGW182, without changes in mRNA levels25. Tethering of dGW182 to FL-5BoxB-HSL and FL-5BoxB-HhR repressed their activity by four and two times, respectively (Fig. 5a), as reported25. Tethering of the dGW182 CED or its longer version extending to the dGW182 C terminus (CED*) was slightly less inhibitory, but, notably, the inhibition was nearly fully relieved by mutating W-motifs. Similarly to the effect of CED domains, direct tethering of the fly Caf1 (dCAF1) and human CNOT1 (the D. melanogaster clone is not available) reduced, by 55% to 75%, activity of both poly(A)+ and poly(A)− reporters in S2 cells (Fig. 5b). Although the inhibition of poly(A)+ RNA by either the dGW182 CED domain or CCR4–NOT components was associated with a decrease of approximately two times in mRNA levels, repression of poly(A)− mRNAs was not accompanied by pronounced mRNA degradation (Fig. 5b).
Figure 5.
The CED W-motifs and CCR4–NOT complex contribute to repression of poly(A)− mRNAs in fly cells. (a) The CED W-motifs contribute to repression of poly(A)− mRNA in fly cells. S2 cells were co-transfected with plasmids encoding NHA fusions of the WT dGW182 CEDs (NHA-CED or NHA-CED*) or its indicated mutants, together with plasmids encoding the indicated reporters (FL-5BoxB, FL-5BoxB-HSL or FL-5BoxB-HhR) and RL-Con. Normalized firefly luciferase activity is indicated as the percentage of activity in cells expressing NHA-lacZ set as 100%. Expression of relevant HA- and NHA-fusion proteins was estimated by western blotting and is shown in the panel on the right. (b) Tethering dCAF1 or human CNOT1 represses poly(A)+ and poly(A)− mRNAs in fly cells. Cells were co-transfected with plasmids expressing HA or NHA fusions of dCAF1 or human CNOT1 and plasmids encoding indicated reporters. Normalized firefly luciferase activity is indicated as the percentage of activity in cells expressing HA fusions of dCAF1 or human CNOT1 set as 100%. Expression levels of HA- and NHA-fusion proteins were estimated by western blotting (shown above the graph). HA- and NHA-CNOT1 were only detectable after enrichment by anti-HA antibody immunoprecipitation. Lower signal of the NHA-tagged, compared to HA-tagged protein, may be partially due to the lower reactivity of anti-HA antibody with the internally located epitope. Analysis of mRNA levels by northern blotting is shown below the graph. Identity of analyzed reporters (including Renilla luciferase mRNA as a reference) is shown on the left, and the co-transfected CCR4–NOT complex components are indicated at the bottom.
We also investigated whether human TNRC6C CED and human CCR4–NOT proteins can repress tethered mRNA independently of poly(A) in HEK293T cells. We found that both classes of proteins repress activity of the poly(A)− reporter that was either expressed from plasmids or transfected as in vitro transcribed mRNA, the latter bearing the cordycepin residue at the 3′ end to prevent its potential adenylation in the cell. Inhibition of the poly(A)− mRNAs was not accompanied by their degradation (Supplementary Figs. 6a–e and 7a–d and Supplementary Results).
Collectively, these results show that recruitment of the GW182 CED or components of CCR4–NOT also induces silencing of poly(A)− mRNAs, without any accompanying RNA degradation, suggesting that the CCR4–NOT complex mediates not only mRNA deadenylation but also translational repression.
Repression of poly(A)− RNA by GW182 depends on CCR 4–NOT
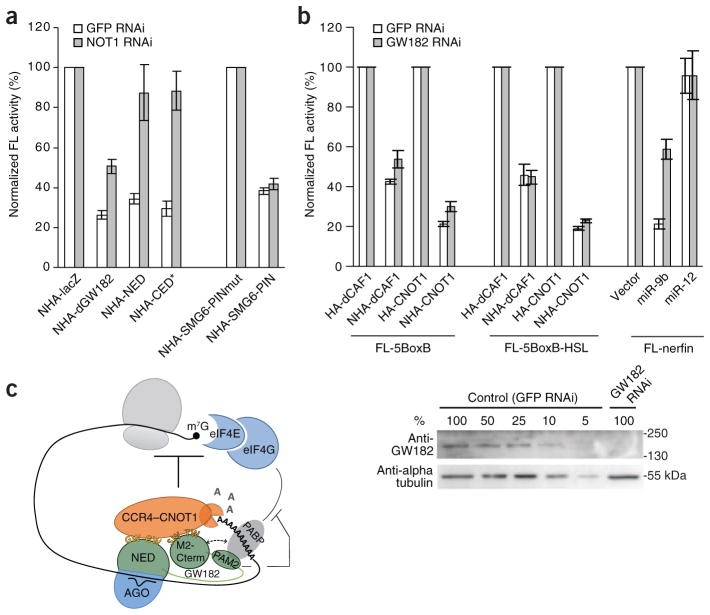
If the CCR4–NOT complex functions downstream of GW182 during repression of poly(A)− mRNAs, the inhibitory effect of GW182 should be dependent on CCR4–NOT. To address this assumption, dGW182 and its fragments were tested for their ability to repress the poly(A)− mRNA in S2 cells depleted of NOT1, a large CCR4–NOT complex scaffolding protein26. Depletion of NOT1 resulted in a marked alleviation of repression, more pronounced for the fragments of dGW182 (2.5-fold to three-fold) than the full-length dGW182 (two-fold) (Fig. 6a). This is probably due to dGW182 also containing domains (for example, Q-rich5) that may repress mRNA by a CCR4–NOT–independent mechanism.
Figure 6.
Repression of poly(A)− RNA by tethering dGW182 or its fragments depends on NOT1, but repression by tethered CCR4–NOT components is dGW182-independent. (a) Repression of FL-5BoxB-HSL reporter by tethering dGW182 or its fragments is alleviated in S2 cells depleted of NOT1. S2 cells treated with dsRNA targeting GFP or NOT1 were co-transfected with plasmids expressing either NHA fusions of dGW182 and its fragments or the PIN domain (either WT or a catalytic mutant thereof) of the endonuclease SMG6, and also reporter plasmids FL-5BoxB-HSL and RL-Con. Normalized firefly luciferase activity is indicated as percentage of the activity in cells expressing NHA-lacZ or SMG6-PINmut, set as 100%. The NOT1 depletion affected the repression by dGW182 and its fragments but had no effect on repression by SMG6-PIN that targets mRNA for endonucleolytic degradation35, supporting the specificity of the effect. (b) Repression of FL-5BoxB and FL-5BoxB-HSL reporters by tethered dCAF1 and human CNOT1 is unaffected in S2 cells depleted of dGW182. Normalized firefly luciferase activity is indicated as the percentage of activity in cells expressing HA-dCAF1 or HA-CNOT1, or cells transfected with pAC5.1 (empty vector), each set as 100%. The efficiency of GW182 depletion was analyzed by western blotting (lower panel). Lanes 1–5, dilutions of the extract from S2 cells treated with GFP-specific (control) dsRNA. (c) Scheme illustrating a possible mode of action of GW182 proteins in miRNA-mediated repression. GW182 proteins are recruited to mRNA through direct interaction with the miRNA–AGO complex. The GW182 NED and CED regions both recruit, through the W-motifs, the CCR4–NOT complex that represses translation and leads to mRNA deadenylation. Interaction of the GW182 PAM2 motif with PABP may interfere with the PABP-eIF4G association, thus contributing to translational inhibition and mRNA deadenylation. The PABP interaction with the CED M2/C-term regions (broken line) may be mediated by the CCR4–NOT complex (see text).
The observation that repression of poly(A)− RNA by tethering dGW182 and its fragments depends on NOT1 suggested that the CCR4–NOT complex also acts downstream of GW182 in translational repression. Consistently, repression caused by tethering of the CCR4–NOT proteins dCAF1 and CNOT1 to FL-5BoxB-HSL RNA was not affected by depletion of endogenous dGW182 (Fig. 6b). Of note, the dGW182 depletion resulted in partial (30–40%) alleviation of the repression of the poly(A)+ FL-5BoxB reporter (Fig. 6b). This is consistent with results indicating that GW182 affects repression not only through the recruitment of CCR4–NOT but also through the association with PABP, and the latter interaction has been shown to be important for miRNA-induced deadenylation7,12,13. We conclude that the CCR4–NOT complex also functions downstream of GW182 during repression of poly(A)− mRNAs, consistent with its role in mediating inhibition of translation.
DISCUSION
We here provide evidence that human and D. melanogaster GW182 proteins repress mRNAs by recruiting the CCR4–NOT complex to the mRNA, in a PABP-independent manner. This recruitment specificity comes from W-motifs that are dispersed throughout the N- and C-terminal regions of the proteins and that act in an additive manner. Moreover, we found that recruitment of CCR4–NOT represses both poly(A)+ and poly(A)− mRNAs, arguing that this complex, in addition to catalyzing mRNA deadenylation, also mediates miRNA-induced translational repression.
The following evidence supports the conclusion that W-motifs represent critical signals for recruiting CCR4–NOT and inducing mRNA repression. (i) Exhaustive mutagenesis of the CED identified redundant W-containing elements in the CED M2 and Cterm regions and demonstrated a strong correlation between repression and interaction with CCR4–NOT. (ii) Introduction of an increasing number of Trp→Ala mutations, in both GW (or WG) and S/TW (or WS/T) contexts, across the CED regions of either TNRC6C or dGW182, had an additive effect on alleviating repression, regardless of whether these substitutions were tested in the CED or full-length proteins. (iii) W-motifs present in the NED and CED regions functioned in an additive manner and by similar mechanisms that involved the recruitment of the CCR4–NOT complex. (iv) In the assay measuring bona fide miRNA repression, the activity of dGW182, TNRC6C and TNRC6A to rescue miRNA-mediated silencing in GW182-depleted cells was strongly compromised upon mutation of W-motifs. (v) Finally, fragments of the yeast protein Sic1p having engineered W-motifs acquired the ability to repress mRNA and to interact with the CCR4–NOT components. Hence, W-motifs are not only required but also sufficient to induce repression by recruitment of the CCR4–NOT complex. Notably, two motifs in TNRC6C, identified in an accompanying paper27 as important for mediating deadenylation and CCR4–NOT interaction in vitro, also contain tryptophan residues.
It is unlikely that alleviation of mRNA repression by Trp→Ala substitutions is due to perturbation by the higher-order structure of the polypeptides or by their folding upon binding to target proteins. First, the mutated W-motifs are located in the NED and CED regions that are predicted to be disordered (http://dis.embl.de). Indeed, NMR analysis of the TNRC6C NED confirmed its disordered character (F. Laughlin, M. Chekulaeva, W.F. and F. Allain, unpublished data). Second, in the case of the CED ‘half ’ regions—that is, the M2-RRM and RRM-Cterm regions—mutating even one or two tryptophan residues had an appreciable effect on repression. Third, the Sic1p protein fragment used for the gain-of-repression experiments is known to be unstructured21 and, apart from engineered W-motifs, shows no sequence similarity to repressive GW182 fragments.
We also observed that the CED domain interacts with the PAN2–PAN3 complex in a manner dependent on W-motifs. Others15 have previously shown that PAN2 contributes to miRNA-mediated deadenylation, most probably at its initial stage. Our data indicate that PAN2–PAN3 is primarily recruited through the function of the W-motifs in the CED, but it can also weakly interact with the CED through PABP (Fig. 2c), consistent with the previously described direct PAN3-PABP interaction20.
The additive contribution of W-motifs, distributed in disordered protein regions, raises the question of how these motifs promote the interaction of GW182 and CCR4–NOT. Does the sheer quantity of the motifs just increase the probability of initial productive interactions? Do the tryptophan-containing regions recruit more than one CCR4–NOT complex at a time? One model of GW182 function is reminiscent of protein-protein interactions reported for the U2AF homology motif (UHM) of the U2 snRNP factor U2AF65 (ref. 28). In that case, the spliceosome component SF3b155 binds to the U2AF65 UHM through motifs having an essential tryptophan and consensus RWD/E. Similarly to GW182 proteins, SF3b155 contains an unstructured region with seven RWD/E repeats28.
The CCR4–NOT components CAF1 and CNOT1 were previously identified as important for miRNA-mediated deadenylation in both flies and mammals, and it has been suggested that the interaction of GW182 with PABP might lead to the recruitment of CCR4–NOT to mRNA7,12,14–16. Our data indicate that recruitment of CCR4–NOT by W-motifs present in CED and NED regions is independent of PABP and represents either a complementary or alternative mechanism for repression. The critical observation in our study was that deletion of PAM2 or its mutation that disrupts CED-PABP interaction did not affect the CED association with CCR4–NOT and mRNA repression, whereas the CED 7W mutant, which still interacted with PABP but not with CCR4–NOT, was inactive in repression (Fig. 2). Moreover, the dGW182 NED region, which is repressive in both S2 and HEK293T cells, interacted with the CCR4–NOT complex components but not with PABP (Fig. 3b). Similarly, the repressive yeast Sic1p fragment associated with the CCR4–NOT proteins but not PABP (Fig. 3d). The association between the TNRC6C CED and CCR4–NOT most probably occurs through the CNOT1 subunit of the complex, because human CNOT1, but not CNOT6 or CNOT7/CAF1, interacted with the CED in the yeast two-hybrid system (Supplementary Fig. 8). CNOT1 was also by far the most effectively pulled down CCR4–NOT complex component identified by MS (Fig. 1b and Supplementary Fig. 4a).
One of the most important findings of our work is that components of the CCR4–NOT complex are able to repress not only polyadenylated but also poly(A)-free mRNAs. The observation that repression of poly(A)− RNA by dGW182 and its fragments depends on CCR4–NOT, whereas repression by tethering of CCR4–NOT proteins is dGW182-independent, indicates that the CCR4–NOT complex acts downstream of GW182 proteins also during repression of poly(A)− mRNAs. Together with the finding that the CCR4–NOT repression of poly(A)− RNAs is not associated with a decrease in mRNA levels, these data strongly implicate the CCR4–NOT proteins in mediating translational repression induced by miRNAs. These results are consistent with recent work29 showing that tethering of CAF1 to the microinjected reporter mRNA can repress translation at the initiation step in Xenopus laevis oocytes. Our experiments extend these results by demonstrating that the CCR4–NOT complex may be responsible for translational repression induced by miRNAs. We also found that in HEK293T and S2 cells, the tethering of CAF1 and, notably, other subunits of the CCR4–NOT complex, repressed mRNA activity (Supplementary Fig. 7c), without affecting the levels of poly(A)− mRNA (Fig. 5b and Supplementary Fig. 7b). Jointly, these observations indicate that W-motif–mediated recruitment of the CCR4–NOT complex causes both translational repression and deadenylation of target mRNAs (see model in Fig. 6c). We find it interesting that in yeast and in fly, the CCR4–NOT complex is known to interact with the translational repressor Dhh1/Me31b30,31, whose orthologs in other organisms are known to be required for miRNA-mediated repression32–34, suggesting a possible mechanism by which the CCR4–NOT complex could repress translation.
ONLINE METHODS
Cell culture, transfections, RNAi and luciferase assays
Human HEK293T cells were grown in DMEM (GIBCO BRL) supplemented with 2 mM l-glutamine and 10% (v/v) FCS buffer. Transfections were done in 6-, 12-, 24- and 96-well plates with nanofectin (PAA Laboratories), according to manufacturer’s instructions. In tethering experiments, cells were transfected with 1 ng RL-5BoxB, 20 ng FL-Con and 20–30 ng HA- or NHA-fusion constructs per well in a 96-well plate. For other formats, the amount of plasmids was adjusted proportionally. Cells were lysed 24 h after transfection. For TNRC6 rescue experiments, HeLa cells stably expressing Tet-On machinery36 were transfected using attractene reagent (Qiagen). Per well of the 96-well plate, transfection mixtures contained 10 ng of the let-7 reporter plasmid, increasing amounts of NHA-TNRC6A or its point mutants (20, 60 and 180 ng), and either siRNAs specific to TNRC6A, B and C (5′-GCCUAAUCUCCGUGCUCAATT-3′, 5′-GGCCUUGUAUUGCCAGCAATT-3′ and 5′-GCAUUAAGUGCUAAACAAATT-3′ (Microsynth; sequences represent sense strands), 0.53 pmol each; or 1.6 pmol AllStars siRNA negative control (Qiagen). TNRC6A plasmids were made resistant to siRNA by introducing silent point mutations. Let-7 reporter plasmids (kindly provided by J. Béthune) encoded Renilla luciferase fused to the human HMGA2 3′ UTR, either WT with sites recognized by let-7 (RL-hmga2), or mutant in which let-7 sites were mutated (RL-hmga2 mut)37,38, as well as FL-Con, both under control of the tetracycline-responsive element. Expression of reporters was induced with 1 μg ml−1 doxycycline 2 d after transfection and cells were lysed 4 h after induction. D. melanogaster S2 cells were transfected in 96-well plates with Cellfectin II and PLUS reagents (Invitrogen). In tethering experiments, we transfected 5 ng FL-5BoxB plasmid, 30 ng RL-Con, and 20–30 ng plasmid encoding HA- or NHA-fusion protein per well. Cells were lysed 3 d after transfection. In rescue experiments, transfection mixtures contained 5 ng FL-nerfin reporter plasmid, 30 ng RL-Con and 5 ng of either an empty vector or a plasmid encoding miR-9b or miR-12 per well of a 96-well plate; plasmids encoding dGW182, TNRC6C and their mutants were added in increasing amounts from 3–30 ng. RNAi experiments were conducted as described39 using dsRNA targeting the dGW182 3′ UTR or the coding region of NOT1. S2 cells were treated with dsRNA twice, on days 1 and 4, transfected on day 6 and lysed on day 9.
Luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega). In all luciferase assays, values represent means ± s.e.m. from three to six independent experiments.
CED mutants containing mutations in W-motifs
Positions of single tryptophan mutations are as indicated in Figure 2a,c. Other mutants in the TNRC6C CED are designated as follows: 2W stands for W1445 W1487; 3W, W1445 W1487 W1494; 4W, W1445 W1487 W1494 W1659; 5W, W1445 W1487 W1494 W1648 W1659; 6W, W1445 W1487 W1494 W1605 W1648 W1659; 7W, W1445 W1487 W1494 W1504 W1605 W1648 W1659; 8W, W1445 W1487 W1494 W1504 W1515 W1605 W1648 W1659; all GW, W1487 W1494 W1648 W1659; most conserved, W1504 W1515; and less conserved, W1487 W1605 W1648 W1659.
For selecting most conserved and less conserved W-motifs mutated in the last two mutants, the protein alignment included sequences of more GW182 proteins than the one shown in Supplementary Figure 1 (data not shown).
The mutants in the dGW182 CED are designated as follows: 2W stands for W1107 W1114; 3W, W1107 W1114 W1118; 4W, W1092 W1107 W1114 W1118; 5W, W1051 W1092 W1107 W1114 W1118; 6W, W1037 W1051 W1092 W1107 W1114 W1118; 7Wa, W1024 W1037 W1051 W1092 W1107 W1114 W1118; 8W, W942 W1024 W1037 W1051 W1092 W1107 W1114 W1118A; 8Wa, W942 W1024 W1037 W1051 W1092 W1107 W1114 W1350; 7W, W942 W1024 W1037 W1051 W1092 W1107 W1114; and 5Wa, W942 W1024 W1037 W1051 W1092.
The 8W mutant of the TNRC6A contains the following mutations: W1420A W1450A W1494A W1505A W1518A W1619A W1666A W1676A (see Supplementary Fig. 1).
Pull-down assays and western blotting
For GST pull-down assays, HEK293T cells grown in a 10-cm dish were transfected with 5 μg plasmid expressing GST-TNRC6C CED, GST-dGW182(1–490) (or mutants thereof), GST-Sic or GST-Sic7xW. Cells were lysed 24 h after transfection and GST-fusions were pulled down as described40. In short, cells were lysed in buffer A (50 mM Tris-HCl, pH 7.5, 150 mM KCl, 0.5% (v/v) Triton X-100, 1× complete EDTA-free protease inhibitor mix (Roche)), and cleared lysates were treated with micrococcal nuclease (10 ng μl−1) for 25 min at 20 °C. We have verified that this treatment eliminates RNA-dependent interactions (see, for example, Fig. 6c in ref. 40). The lysates were incubated with glutathione (GSH)-Sepharose beads (GE Healthcare) for 2 h at 4 °C; beads were washed 3× with buffer A containing 0.1% (v/v) Triton X-100, and GST-fusions were eluted with 50 mM GSH. For anti-TNRC6A immunoprecipitations, HeLa cells were lysed in buffer B (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM DTT, 0.5% (v/v) NP40, 1× complete EDTA-free protease inhibitor (Roche)), treated with micrococcal nuclease as described above and incubated with anti-TNRC6A antibody (Bethyl A302-330A) or, as a negative control, with rabbit IgG (Sigma) bound to Dynabeads Protein G (Invitrogen) overnight at 4 °C. Beads were washed 3× with buffer B containing 0.1% (v/v) NP-40 and boiled in Laemmli SDS-PAGE buffer.
The following primary antibodies were used for western blotting: anti-TNRC6A, 1:5000 (Bethyl A302-329A); anti-CNOT1, 1:250 dilution (provided by M. Collart); anti-CAF1 (Abnova), 1:1,000; anti-PABP (Cell Signaling Technology), 1:5,000; anti-PAN2, 1:1,000 and anti-PAN3, 1:500 (both provided by A.-B. Shyu); anti-dGW182, 1:2,000 (provided by E. Izaurralde); anti-GST (GE Healthcare), 1:10,000; anti-α-tubulin (Sigma T5168), 1:10,000; anti-HA tag (Roche 3F10), 1:5,000; anti-HA tag (Santa Cruz sc-7392), 1:2,000; and anti-LexA (Santa Cruz sc-7544), 1:2,000.
Supplementary Material
Acknowledgments
We thank M. Collart (University of Geneva), A.-B. Shyu (University of Texas Medical School), H.T.M. Timmers (University of Utrecht), E. Izaurralde (Max Planck Institute for Developmental Biology), N. Sonenberg (McGill University), K. Schönig (Zentralinstitut für Seelische Gesundheit), J. Bethune (Friedrich Miescher Institute (FMI)), E. Wahle (Martin Luther University, Halle) and T. Yamamoto (University of Tokyo) for reagents; R. Sack, D. Klei, and the FMI Protein Analysis Facility for MS analysis; M. Tsai for help with the Y2H assay; and H. Gut, I. Loedige, J. Krol, J. Bethune, M. de la Mata, N. Thoma, F. Allain, E. Izaurralde and N. Sonenberg for stimulating discussions. M. Chekulaeva is the recipient of long-term postdoctoral fellowships from the Human Frontiers Science Program and Engelhorn Stiftung. J.A. is funded by the German National Academic Foundation. R.P. is supported by funds from the Howard Hughes Medical Institute. This work was supported by the European Community FP6 Program ‘Sirocco’. The FMI is supported by the Novartis Research Foundation.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
M. Chekulaeva, H.M., J.T.Z., J.A., M. Colic, R.P. and W.F. designed the experiments.
M. Chekulaeva, H.M., J.T.Z., J.A. and M. Colic conducted the experiments.
M. Chekulaeva, H.M., R.P. and W.F. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 4.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chekulaeva M, Filipowicz W, Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eulalio A, et al. The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing. Nucleic Acids Res. 2009;37:2974–2983. doi: 10.1093/nar/gkp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of miRNA targets and is required for target release. Mol Cell Biol. 2009;23:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010;29:4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol Cell Biol. 2009;29:4144–4155. doi: 10.1128/MCB.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chekulaeva M, Parker R, Filipowicz W. The GW/WG repeats of Drosophila GW182 function as effector motifs for miRNA-mediated repression. Nucleic Acids Res. 2010;38:6673–6683. doi: 10.1093/nar/gkq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao B, et al. Divergent GW182 functional domains in the regulation of translational silencing. Nucleic Acids Res. 2011;39:2534–2547. doi: 10.1093/nar/gkq1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ezzeddine N, et al. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqui N, et al. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J Biol Chem. 2007;282:25067–25075. doi: 10.1074/jbc.M701256200. [DOI] [PubMed] [Google Scholar]
- 21.Mittag T, et al. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höck J, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 27.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nat Struct Mol Biol. doi: 10.1038/nsmb.2149. (published online 7 Oct 2011) [DOI] [PubMed] [Google Scholar]
- 28.Thickman KR, Swenson MC, Kabogo JM, Gryczynski Z, Kielkopf CL. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J Mol Biol. 2006;356:664–683. doi: 10.1016/j.jmb.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Temme C, et al. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbee SA, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eulalio A, et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weidenfeld I, et al. Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 2009;37:e50. doi: 10.1093/nar/gkp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Höck J, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chekulaeva M, Filipowicz W, Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.