Abstract
Recent genome-wide association studies have identified many promising schizophrenia candidate genes and demonstrated that common polygenic variation contributes to schizophrenia risk. However, whether these genes represent perturbations to a common but limited set of underlying molecular processes (pathways) that modulate risk to schizophrenia remains elusive, and it is not known whether these genes converge on common biological pathways (networks) or represent different pathways. In addition, the theoretical and genetic mechanisms underlying the strong genetic heterogeneity of schizophrenia remain largely unknown. Using 4 well-defined data sets that contain top schizophrenia susceptibility genes and applying protein-protein interaction (PPI) network analysis, we investigated the interactions among proteins encoded by top schizophrenia susceptibility genes. We found proteins encoded by top schizophrenia susceptibility genes formed a highly significant interconnected network, and, compared with random networks, these PPI networks are statistically highly significant for both direct connectivity and indirect connectivity. We further validated these results using empirical functional data (transcriptome data from a clinical sample). These highly significant findings indicate that top schizophrenia susceptibility genes encode proteins that significantly directly interacted and formed a densely interconnected network, suggesting perturbations of common underlying molecular processes or pathways that modulate risk to schizophrenia. Our findings that schizophrenia susceptibility genes encode a highly interconnected protein network may also provide a novel explanation for the observed genetic heterogeneity of schizophrenia, ie, mutation in any member of this molecular network will lead to same functional consequences that eventually contribute to risk of schizophrenia.
Key words: genome-wide association study, schizophrenia susceptibility genes, protein-protein interaction, common molecular networks, genetic heterogeneity, enrichment
Introduction
Schizophrenia is a severe mental disorder that affects about 1% of the world’s population.1 Family, twin, and adoption studies have demonstrated a strong genetic component of schizophrenia, with heritability estimates of about 80%.2,3 Despite the successful identification of multiple promising candidate genes, the underlying molecular mechanisms of schizophrenia remain largely unknown, and much of the genetic heterogeneity of schizophrenia remains undocumented.
In the past several decades, investigators have been fixating on the high heritability and strong genetic heterogeneity of schizophrenia.4,5 First, investigators found that different genetic variants, including single-nucleotide polymorphisms (SNPs),6,7 copy number variations (CNVs),8–10 and large-scale structural abnor malities such as deletions and insertions could contribute to schizophrenia risk.11,12 Second, although traditional molecular genetic studies (including linkage and association studies) have successfully identified multiple schizophrenia susceptibility genes, it is frequently observed that different genetic variants are identified in different ethnic populations. Furthermore, few identified schizophrenia genes are consistently replicated in genetic independent populations. These convergent lines of evidence strongly support the large genetic heterogeneity of schizophrenia. Nevertheless, the theoretical and genetic mechanisms underlying the strong genetic heterogeneity of schizophrenia remain largely unknown.
The advent of genome-wide association studies (GWAS) provides an opportunity to study the genetic heterogeneity of schizophrenia. Compared with traditional genetic association studies, the power of GWAS significantly increases as the sample size dramatically increases, and the throughput of GWAS is quite high (it can assess millions of genetic variants [SNPs] unbiased, in one test).13,14 Recently, the Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC) reported five new loci that reached the genome-wide significance level through the analysis of a large data set that consists of 17 836 schizophrenia cases and 33 859 controls.7 PGC identified 81 top SNPs from different linkage disequilibrium (LD) regions where at least 1 SNP in each LD region had surpassed P < 2×10−5 in stage 1. They followed up on these SNPs in stage 2 and identified 7 loci that were significantly associated with schizophrenia in meta-analysis of stages 1 and 2. To evaluate whether these 81 top SNPs represent true associations, a sign test for consistency between stages 1 and 2 was performed, and a highly significant result was observed (P < 10−6), with the same direction of effect observed in stage 1 also being observed in stage 2 for 49 of 59 SNPs when excluding the major histocompatibility complex (MHC) region. Hamshere et al further replicated 78 of the 81 SNPs in a large sample (2652 cases and 4539 controls), and they found 98% (confidence interval: 78%–100%) of the original set of 78 SNPs represent true associations.15 These highly significant and convergent evidences strongly suggest that the 81 top SNPs identified by PGC are authentic schizophrenia susceptibility variants. Though these findings are crucial and represent true associations, whether these identified associations represent perturbations to a common but limited set of underlying molecular processes (pathways) that modulate risk to schizophrenia (ie, whether the interaction networks between the proteins encoded by genes near these 81 top SNPs are statistically significant compared with random networks) remains elusive, and it is not known whether these SNPs converge on common biological pathways or represent different pathways. Therefore, it is important to explore the biological meaning behind these identified SNPs. To this aim, using high-confidence pairwise protein interaction resources, we first investigated the protein-protein interactions (PPIs) among proteins that were encoded by genes around these 81 top SNPs identified in Schizophrenia PGC. We then validated our results using another independent data set that consists of genome-wide significant schizophrenia susceptibility genes. Finally, using empirical functional data (transcriptome data) that derived from clinical samples, we tested our hypothesis generated from bioinformatic analyses of GWAS data.
Methods
Data Sets Used in This Study
The selection of high-quality schizophrenia susceptibility genes is essential to PPI network analysis. In this study, we selected 4 well-defined data sets that contain top schizophrenia genes. The first data set is from a recent report of Schizophrenia PGC.7 Ripke et al performed a large-scale GWAS and identified 81 top SNPs. They evaluated these SNPs in a large independent sample and found most of them are authentic schizophrenia susceptibility variants.7 Hamshere et al further validated that most of the 81 top SNPs are true risk variants for schizophrenia in a large sample.15 These high-confidence SNPs represent the most promising genetic variants for schizophrenia so far. We further translated these SNPs into genes using the method developed by Rossin et al.16 In brief, the region around an associated SNP was defined using LD and recombination hot spot information from HapMap (http://www.hapmap.org). For a given SNP, we defined the wingspan of the SNP as the region containing SNPs with r 2>.5 to the associated SNP. We then extended this region to the nearest hot spots. A gene’s residence in a locus is defined by whether 50kb upstream and 50kb downstream (to include regulatory DNA) of the coding region of the gene’s longest isoform overlaps the SNP wingspan. (For more details, please refer to Disease Association Protein-Protein Link Evaluator [http://www.broadinstitute.org/mpg/dapple/dapple.php] and the article by Rossin et al.16)
The second data set consists of carefully curated top schizophrenia candidate genes. Genes that are significantly associated with schizophrenia in recent GWAS represent the most promising candidate genes for schizophrenia.6,7,15,17–24 Therefore, investigating the potential PPI between these top schizophrenia candidate genes will provide the most useful information.
The third data set is from recent published work by Ayalew et al.25 Ayalew et al identified 42 top schizo phrenia candidate genes using translational convergent func tional genomics (CFG). The CFG method used multiple independent lines of evidence to identify and prioritize schizophrenia susceptibility genes. It integrated data from published GWAS data sets for schizophrenia with other pivotal data, including gene expression data from human postmortem brain samples and human-induced pluripotent stem cell–derived neuronal cells, as well as human blood gene expression data and relevant animal model brain and blood gene expression data. In addition, the CFG integrated other human genetic data (ie, linkage, copy number variant, or association) for schizophrenia and relevant mouse model genetic evidence. Because CFG integrated many pivotal data sets from schizophrenia studies, the genes identified by Ayalew et al represent high-confidence candidate genes for schizophrenia.
The fourth data set is from the recent work of Fillman et al.26 They quantified gene expression levels in the dorsolateral prefrontal cortex (DLPFC) of 20 individuals with schizophrenia and their matched controls using RNA sequencing (RNA-Seq). They detected 798 differentially regulated transcripts present in individuals with schizophrenia compared with the controls. These 798 differentially expressed transcripts represent 548 unique protein-coding genes.
PPI Network Analysis
To investigate the physical interactions between proteins encoded by top schizophrenia candidate genes, we used the method developed by Rossin et al.16 Protein products of the schizophrenia susceptibility genes were defined as schizophrenia-associated proteins, which were further used to construct PPI networks. The PPI networks (including direct and indirect) among schizophrenia genes were extracted from InWeb, a well-characterized PPI database developed by Lage et al.27 InWeb contains 169801 high-confidence pairwise interactions that are defined by a rigorously tested signal-to-noise threshold compared with well-established interactions from MINT, BIND, IntAct, and KEGG. If there is in vitro evidence of interactions between 2 schizophrenia-associated proteins, these 2 proteins are connected by 1 edge. In the PPI network, the nodes represent proteins, while the edges represent physical interactions. The PPI networks can be classified into 2 types, direct and indirect networks. In direct networks, any 2 associated proteins are connected by exactly 1 edge. In indirect networks, associated proteins connect through common interactor proteins (not known to be associated with disease) with which the associated proteins each share an edge.
Edge metrics and node metrics were used to assess the network properties. The edge metric is the direct network connectivity parameter defined as the number of edges in the direct network. The direct network’s connectivity equals 1 if 2 different associated proteins directly bind to each other. Therefore, the direct network’s connectivity represents the total number of direct edges in the direct network. Further information about the network properties can be found in the work completed by Rossin et al.16
Assessment of the Significance of the PPI Network
To evaluate whether schizophrenia susceptibility genes are significantly connected via PPIs, we used a permutation test to assess the significance of networks built from PPI data. Briefly, a within-degree node-label permutation approach was performed. For a given number of proteins, we first constructed the direct and indirect networks, which are built from interactions among schizophrenia-associated proteins according to InWeb.27 Various network parameters (such as direct network connectivity) were obtained according to the constructed PPI network. These network parameters derived from our real network (ie, network built from schizophrenia susceptibility genes) were further used to generate the random networks. We first generated a random network that has nearly the exact same structure (ie, structurally equivalent random networks) as the original one that is extracted from the InWeb database. The node labels (ie, the protein names) were then randomly reassigned to nodes of equal binding degree. This approach assumes a null distribution of connectivity that is entirely a function of the binding degree of individual proteins. We built 10 000 random networks, and each of them had the same size (number of proteins), number of edges (connectivity), and per-protein binding degree as InWeb. The significance of our real PPI network was then assessed through permutation. With this method, we are able to test the nonrandomness of our network conditional on the exact binding degree distribution of schizophrenia proteins. For more details, please refer to the article of Rossin et al.16
Gene Set Enrichment Analysis
We examined whether genes near the top 81 SNPs from PGC were enriched for specific functional categories. In addition, Fillman et al quantified gene expression levels in the DLPFC of 20 individuals with schizophrenia and their matched controls in a recent study.26 They detected 798 differentially regulated transcripts present in individuals with schizophrenia compared with the controls. These differentially regulated transcripts represent 548 protein-coding genes. We also tested whether specific gene ontology (GO) terms were enriched among 548 genes with significantly altered expression levels in DLPFC of schizophrenia patients. The primary analysis was performed using the “DAVID Bioinformatics Resources 6.7” Web site (http://david.abcc.ncifcrf.gov/).28,29 GO terms—biological processes (GO_BP), cellular components (GO_CC), and molecular functions (GO_MF)—were used. The Benjamini-Hochberg procedure was used to correct the P values of the enriched GO terms.
KEGG Pathway Analysis of Genes That Participate in the Highly Interconnected PPI Network and Gene Expression Profiling Analysis
Detailed information on KEGG pathway and gene expression profiling analyses can be found in the online supplementary methods.
Results
Proteins Encoded by Genes Defined by the Top 81 SNPs From Schizophrenia PGC Form a Highly Significant Interconnected Network
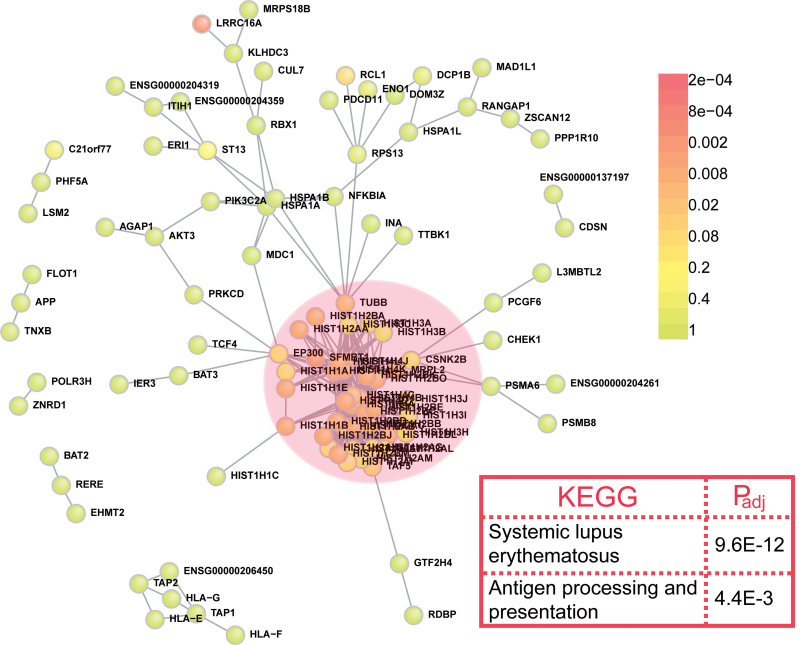
To evaluate the interaction between proteins encoded by genes near the 81 top SNPs from the Schizophrenia PGC, we constructed and assessed the PPI network. These 81 top SNPs were first translated into protein-coding genes using the wingspan method developed by Rossin et al.16 The PPI network was then constructed using the translated genes. We found there were 104 schizophrenia-associated proteins participating in the direct network (figure 1) among the 265 genes (see online supplementary tables 1 and 2) defined by these 81 top SNPs. We tested this degree of interconnectivity through permutation (n = 10 000 permutations) and found the direct PPI network of genes from those near the 81 top SNPs had significantly more edges than expected by chance (P = 9.9 × 10−4, corrected) (figure 1 and online supplementary figure 1 and table 2). The PPI network is thus highly significant compared with 10 000 random networks and is also significant for indirect connectivity (P = .004, corrected). These results indicate that genes near the 81 top SNPs encode directly interacting proteins beyond the level expected by chance, suggesting these genes perturb common molecular networks that modulate schizophrenia risk.
Fig. 1.
Proteins encoded by genes that were defined by the top 81 single-nucleotide polymorphisms (SNPs) from Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC) form a highly significant interconnected network. Protein-protein interaction (PPI) network constructed by genes that defined by top 81 SNPs from Schizophrenia PGC. There were 104 disease proteins participating in the direct network and 343 direct interactions in total. This degree of interconnectivity is statistically highly significant (P = 9.9 × 10− 4, corrected) compared with 10 000 random networks, which only have the 206 direct edges count expected by chance. The core of this highly interconnected network is composed of genes that are involved in nucleosome assembly (pink circle), suggesting an enrichment of nucleosome assembly genes in schizophrenia susceptibility loci. KEGG pathway analysis of the genes that participate in the direct network is shown in the red box. P values were corrected by the Benjamini-Hochberg procedure in DAVID.
Because candidate genes in the PPI network were defined by the wingspan method, we tested whether those candidate genes in the PPI network are located more closely to the associated SNPs than those still in the wingspan regions yet not in the PPI network (detailed methods can be found in online supplementary material). Overall, we did not find a pattern of significance (P > .05; see online supplementary figure 2). This observation is likely due to the fact that the genes in the PPI network were from a few major associated SNPs: among the 81 seed SNPs, only 23 of them have genes included in our PPI network. Nevertheless, we indeed observed a larger proportion of genes (13/23 = 56.5%) in the network that are most closely located (rank = 1) to the associated SNPs.
We further explored whether genes defined by these 81 top SNPs were enriched for specific functional categories. Gene set enrichment analyses were performed using “DAVID Bioinformatics Resources 6.7” (http://david.abcc.ncifcrf.gov/).28 KEGG, BioCarta, BBID, and PANTHER pathway databases were included, and GO terms—biological processes (GO_BP), cellular components (GO_CC), and molecular functions (GO_MF)—were used. GO analysis using biological process as a key word revealed that nucleosome (chromatin) assembly genes were significantly enriched in these gene sets (P = 5.72×10−15, corrected; table 1). In addition, other GO terms such as antigen processing and presentation (P = 1.1×10−10, corrected), response to unfolded protein (P = 4.3×10–8, corrected), response to protein stimulus (P = 3.64×10−7, corrected), immune response (P = 9.7×10−4, corrected), and response to nutrient (P = 1.1×10−2, corrected) were also found to be overrepresented in these gene sets. GO analysis using cellular components (CC) as a key word revealed similar results, and DAVID functional clustering further confirmed these findings.
Table 1.
Gene Set Enrichment Analysis of Genes Defined by 81 Top Single-Nucleotide Polymorphisms (SNPs) From the Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC)
| Category | Database ID | Term | Padj |
|---|---|---|---|
| GOTERM_BP_FAT | GO:0006334 | Nucleosome assembly | 5.72E-15 |
| GOTERM_BP_FAT | GO:0019882 | Antigen processing and presentation | 1.08E-10 |
| GOTERM_BP_FAT | GO:0006986 | Response to unfolded protein | 4.32E-08 |
| GOTERM_BP_FAT | GO:0002474 | Antigen processing and presentation of peptide antigen via MHC class I | 2.23E-07 |
| GOTERM_BP_FAT | GO:0048002 | Antigen processing and presentation of peptide antigen | 3.93E-07 |
| GOTERM_BP_FAT | GO:0051789 | Response to protein stimulus | 3.64E-07 |
| GOTERM_BP_FAT | GO:0006952 | Defense response | 4.46E-05 |
| GOTERM_BP_FAT | GO:0002504 | Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 4.40E-04 |
| GOTERM_BP_FAT | GO:0006955 | Immune response | 9.77E-04 |
| GOTERM_BP_FAT | GO:0007584 | Response to nutrient | .0111 |
| GOTERM_BP_FAT | GO:0048660 | Regulation of smooth muscle cell proliferation | .0290 |
| GOTERM_BP_FAT | GO:0031667 | Response to nutrient levels | .0297 |
Note: MHC, major histocompatibility complex. The table shows GO terms identified by DAVID that are enriched among 256 genes defined by 81 top SNPs from Schizophrenia PGC. P adj values in the table represent P values adjusted by the Benjamini-Hochberg procedure in DAVID. The terms in bold matched the GO terms from the analysis of 548 genes with significantly altered expression levels in dorsolateral prefrontal cortex (DLPFC) of schizophrenia patients.
To identify whether specific biological pathways (KEGG)30 are enriched among genes participating in this direct network, we conducted an analysis of those pathways. We found systemic lupus erythematosus (corrected P = 9.6×10−12) and antigen processing and presentation (corrected P = 4.4×10−3) pathways were enriched among genes forming this interconnected PPI network (figure 1 and online supplementary figures 3 and 4). Interestingly, a recent expression study also found these 2 pathways (systemic lupus erythematosus and antigen processing and presentation pathways) are significantly enriched in the differentially expressed genes identified in schizophrenia patients compared with healthy controls,31 suggesting these 2 pathways may represent authentic dysregulated processes/pathways in schizophrenia.
To further validate common biological processes that were perturbed in schizophrenia, we investigated expression data from a recent study of Fillman et al.26 In that study, gene expression levels in the DLPFC of 20 individuals with schizophrenia and their matched controls were quantified using RNA-Seq. The analysis implicated a set of 548 genes with significantly altered expression levels in DLPFC of schizophrenia patients compared with matched controls. The functional analysis of the differentially expressed genes with DAVID identified multiple significant GO terms (see online supplementary table 3). Strikingly, we noticed many of the identified terms matched the terms from the GO analysis of 256 genes defined by 81 top SNPs from Schizophrenia PGC (table 1). These overlapping GO terms include immune response (P = 3.91×10−4, corrected), response to unfolded protein (P = .0065, corrected), defense response (P = .013, corrected), response to protein stimulus (P = .021, corrected), and response to nutrient levels (P = .034, corrected). These corroborating, consistent results strongly suggest that multiple lines of evidence converge on similar functions and processes in schizophrenia.
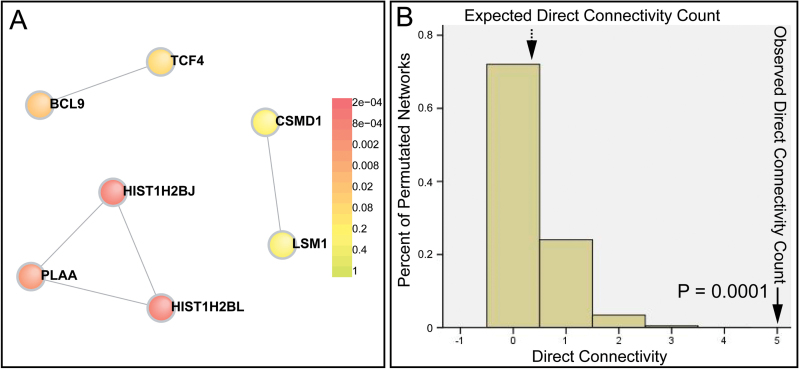
Bias may be introduced in the construction of the PPI network because there were highly linked genes from the MHC region. Therefore, we tested a curated data set containing genes that are significantly associated with schizophrenia in recent GWAS (see online supplementary table 4; many genes from MHC region were excluded). We thereby pinpointed the most promising candidate genes for schizophrenia so far. Specifically, we found there were 7 schizophrenia-associated proteins participating in the direct network (figure 2) among the 30 seed genes (see online supplementary tables 2 and 4) that reached GWAS significance level. We tested this degree of interconnectivity through permutation (n = 10000 permutations) and found these genes significantly interacted and the network is significant for both direct connectivity (P = .0001; figure 2 and online supplementary table 2) and indirect connectivity (P = .001; see online supplementary figure 5). To avoid potential bias, we generated the PPI network using another database of physical interactions (GeneMANIA),32 and similar results were obtained (see online supplementary figure 6). Interestingly, GO analysis revealed that nucleosome assembly genes were significantly enriched again (P = 6.61×10−14) in this network (see online supplementary table 5).
Fig. 2.
Protein products encoded by genome-wide significant schizophrenia susceptibility genes significantly interacted. (A) Protein-protein interaction (PPI) network constructed with genes that were significantly associated with schizophrenia in recent genome-wide association studies (GWAS) of schizophrenia. (B) Significant network (P = .0001) compared with 10 000 random networks, suggesting significant physical interactions between protein products of top schizophrenia susceptibility genes. Structurally equivalent random networks were built from a within-degree node-label permutation method. An empirical distribution was constructed for a direct connectivity count and used to assess the significance of networks. Numbers on the x-axis represent the direct network connectivity (the number of edges in the direct network), which were enumerated for the disease networks and 10 000 random networks. The plotted histogram represents random expectation (the dashed arrowhead), and the solid arrowheads indicate the schizophrenia network (observed). The y-axis represents percent of permutated networks (eg, the percentage of networks with 5 edges in the direct network is 0.0001, which is statistically significant compared with 10 000 random networks).
Top 42 Schizophrenia Candidate Genes Identified by CFG Encode a Significantly Interconnected PPI Network
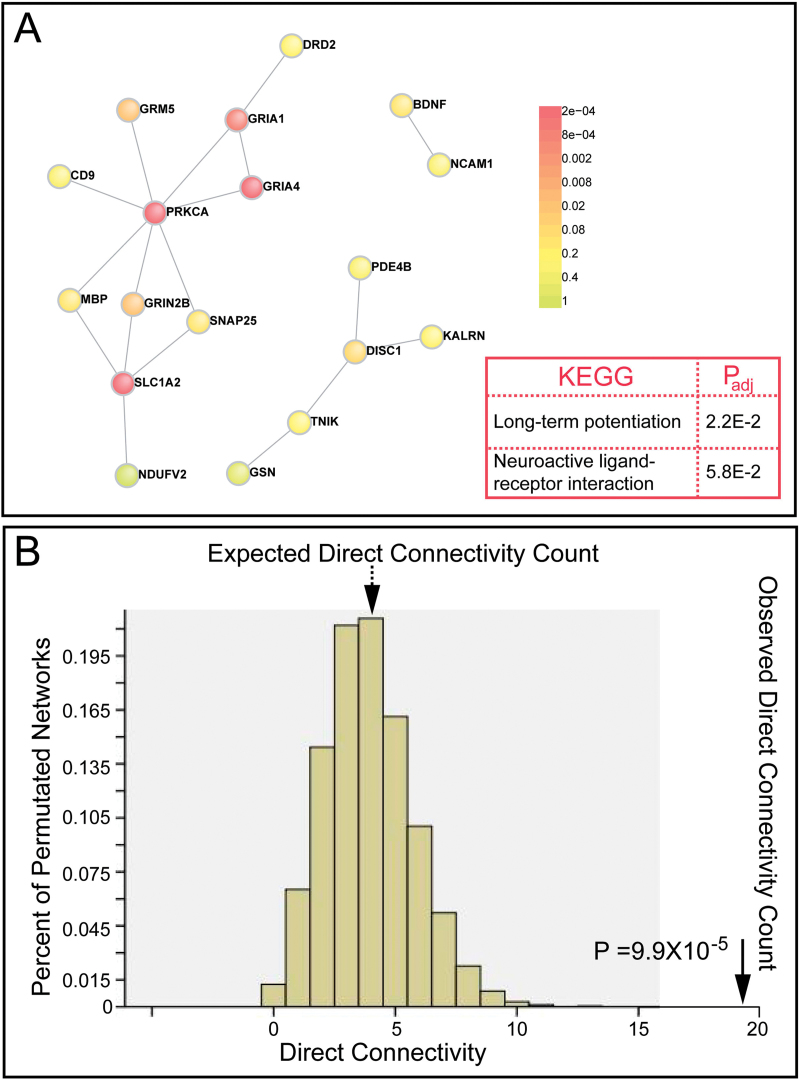
To further validate that schizophrenia susceptibility genes significantly physically interacted and may perturb common biological networks, we tested a third independent data set from recent work by Ayalew et al.25 Ayalew et al identified 42 top schizophrenia candidate genes (see online supplementary table 6) using translational CFG, which integrated GWAS with gene expression studies in both human and animal models. Among the 42 schizophrenia susceptibility genes (see online supplementary table 6) that were identified by Ayalew et al, we found there were 18 schizophrenia-associated proteins participating in the direct network (figure 3). Again, we noticed these top genes from CFG encode a densely interconnected PPI network (figure 3). We further tested this degree of interconnectivity through permutation (n = 10000 permutations) and found the PPI network of genes from CFG is statistically significant compared with 10000 random networks (corrected P = 9.9 × 10−5; figure 3 and online supplementary table 2). The PPI network is also significant for indirect connectivity (P = .008, corrected; see online supplementary figure 7). We noticed that the main inclusion criteria of these tops genes identified by CFG are genetic association studies, expression data from humans and animals, and animal models. That is, they are not identified through PPIs. Therefore, our observation that top genes from CFG formed a highly interconnected PPI network provides robust evidence that top schizophrenia genes form a highly interconnected network. Once again, these significant results support that top schizophrenia genes encode directly interacting proteins beyond the level expected by chance, suggesting these genes perturb common molecular networks that modulate schizophrenia risk. This result also indicates CFG is a useful and promising tool in delineating the genetic mechanisms of schizophrenia. We also performed KEGG biological pathway analysis and found long-term potentiation (corrected P = 2.2 × 10−2) is significantly overrepresented among genes forming the direct network (figure 3 and online supplementary figure 8).
Fig. 3.
Top 42 schizophrenia candidate genes identified by convergent functional genomics (CFG) encode a significantly interconnected protein-protein interaction (PPI) network. (A) PPI network constructed with top schizophrenia genes that were identified using translational CFG of schizophrenia. (B) The direct connectivity network is statistically highly significant (has more edges) compared with 10 000 random networks (P = .000099, corrected), suggesting top schizophrenia susceptibility genes are preferential toward physical interaction. KEGG pathways that were significantly overrepresented among genes forming the direct network are shown in the red box. P values were corrected by the Benjamini-Hochberg procedure in DAVID.
Differentially Expressed Genes Detected in DLPFC of Individuals With Schizophrenia Encode Directly Interacting Proteins Beyond Results Expected by Chance
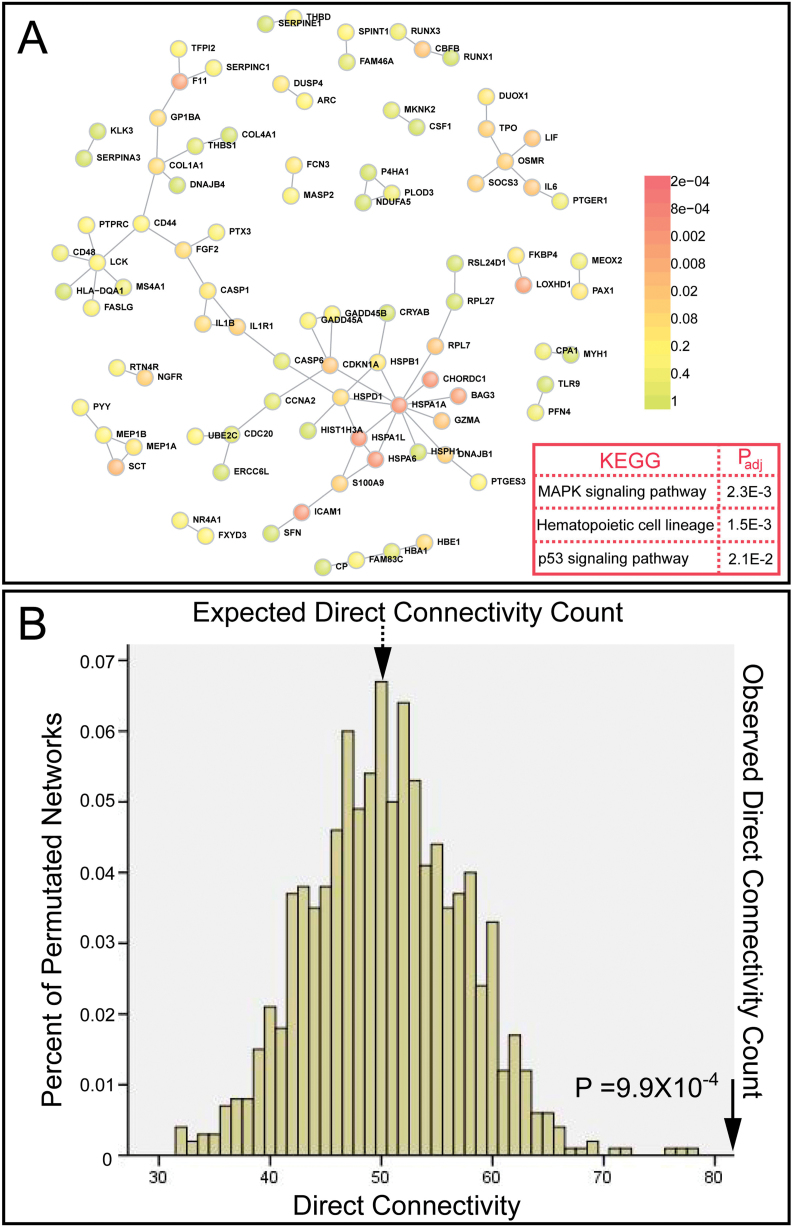
To test our hypothesis generated by analyzing top genes from GWAS and CFG, we performed PPI analysis using transcriptome data from clinical samples. Recently, Fillman et al used next-generation sequencing (RNA-Seq) to quantify gene expression levels in the DLPFC of 20 individuals with schizophrenia and their matched controls.26 They identified 798 differentially expressed transcripts present in people with schizophrenia compared with controls. They further confirmed their results using quantitative polymerase chain reaction and Western blot in an expanded cohort (n = 74). We tested the PPI of the differentially expressed genes from this expression data set. Among the 548 differentially expressed seed genes, 92 seed proteins participated in the direct network (figure 4 and online supplementary table 2). A permutation test further supported that these identified dysregulated genes formed a statistically significant interconnected network (corrected P = 9.9×10−4; figure 4 and online supplementary table 2). This result provides additional evidence that supports our original finding that schizophrenia susceptibility genes form a densely interconnected network. Finally, we performed a KEGG biological pathway analysis and found the mitogen-activated protein kinase signaling pathway (corrected P = 2.3×10−3), hematopoietic cell lineage (corrected P = 1.5×10−3), and p53 signaling biological pathways (corrected P = 2.1×10−2) were significantly enriched among genes that form the direct network (figure 4 and online supplementary figures 9–11). In summary, these consistent results suggest that multiple lines of evidence from different schizophrenia studies converge on common molecular networks and biological processes. This functional convergence also indicates that common molecular networks and biological processes modulate schizophrenia risk.
Fig. 4.
Differentially expressed genes detected in dorsolateral prefrontal cortex (DLPFC) of individuals with schizophrenia encode a significantly interconnected protein-protein interaction (PPI) network. (A) PPI network constructed using differentially expressed genes identified in DLPFC of individuals with schizophrenia. (B) The direct connectivity network is statistically significant (has more edges) compared with 10 000 random networks (P = .000099, corrected), implying significant physical interaction among proteins that are encoded by dysregulated genes identified in individuals with schizophrenia. KEGG pathways that were significantly enriched among genes participating in the direct network are shown in the red box. P values were corrected by the Benjamini-Hochberg procedure in DAVID.
Genes Identified in PPI Networks Are Expressed in the Brain and Immune Tissues
Because many of the candidate genes were from GWAS and CFG analyses of schizophrenia, we further explored their expression profile in human tissues. We found a large proportion of the genes identified in the PPI network are expressed in brain tissues (see online supplementary figures 12–14). In addition, we found genes identified in the PPI network are preferentially expressed in brain and immune tissues (see online supplementary figure 15). Of note, our GO analysis also revealed an enrichment of immune genes among genes defined by the top 81 SNPs from Schizophrenia PGC (table 1). In fact, accumulating evidence strongly suggest that immune-related genes may play pivotal roles in schizophrenia. First, multiple genetic linkage and association studies have revealed that many immune genes are significantly associated with schizophrenia.7,17,19,33–35 Second, dysregulation of immune-associated genes were frequently observed in schizophrenia patients.26,31,36–38 Taken together, these results indicate genes identified in the PPI network are expressed in brain tissues, suggesting these genes may play important roles in brain function. In addition, our results provide further evidence of the dysregulation of immune-associated genes in schizophrenia.
Discussion
Schizophrenia is a complex mental disorder with high heritability and strong genetic heterogeneity. To elucidate the genetic and molecular mechanisms underlying the substantial genetic heterogeneity of schizophrenia, many efforts have been made in past decades. However, little progress has been made because traditional genetic studies have intrinsic limitations (eg, lower throughput and power), which make it hard to evaluate all of the genetic variants in an unbiased manner in one test. Because only limited genes or variants can be identified, it is difficult to study the overall interaction patterns between schizophrenia susceptibility genes. In addition, although many PPI databases have been established, most of them focus on PPI between 2 specific proteins and do not provide a statistical framework to evaluate the significance of our interesting networks. Therefore, despite the successful identification of multiple promising candidate genes, the underlying interactions among schizophrenia susceptibility genes remain largely unknown and much of the genetic heterogeneity of schizophrenia remains undocumented.
Fortunately, the advent of GWAS provides an opportunity to investigate the strong genetic heterogeneity of schizophrenia. Compared with traditional genetic studies, GWAS have higher throughput and power. Because GWAS could identify numerous high-confidence candidate genes (variants) in one test, we can assess the PPI between these identified genes.39 To date, multiple GWAS of schizophrenia have been conducted and many pivotal schizophrenia susceptibility genes have been identified. In addition, accumulating evidence clearly indicates common polygenic variation contributes to schizophrenia risk. More importantly, Rossin et al developed a reliable and efficient method to evaluate whether genes in loci associated with complex traits are statistically significantly connected via PPI.16 In this study, we used a PPI network analysis to study the PPIs among top schizophrenia susceptibility genes. We present convergent and statistically significant evidence that top schizophrenia candidate genes encode a highly interconnected network. Our study may provide useful methodological guidelines to investigate the complex genetic heterogeneity of schizophrenia. Considering at least 4 independent data sets were used in this study and these data sets were from different sources (eg, GWAS of schizophrenia, CFG of schizophrenia, and expression study of individuals with schizophrenia), these convergent and consistent results suggest that top schizophrenia genes interact significantly and form a densely interconnected network.
It is worth noting that gene set enrichment analysis of 2 independent data sets (ie, 256 genes defined by 81 top SNPs from Schizophrenia PGC and 548 genes with significantly altered expression levels in DLPFC of schizophrenia patients) identified many overlapping GO terms. Considering the data type of these 2 data sets were different (ie, one is from a GWAS of schizophrenia, while the other is from an expression study of schizophrenia patients), these overlapping terms may represent authentic biological processes that were perturbed in schizophrenia. Furthermore, these consistent results provide robust evidence that common molecular networks or biological processes were perturbed in schizophrenia.
Taken together, these converging lines of evidence indicate schizophrenia susceptibility genes encode proteins that significantly directly interact and form a substantially interconnected network, suggesting perturbations to common underlying molecular networks or processes that modulate schizophrenia risk. Our findings that schizophrenia susceptibility genes encode a highly interconnected protein network may also provide novel explanation for the observed genetic heterogeneity of schizophrenia, ie, mutations in any member of this molecular network or pathway will lead to same functional consequences that eventually contribute to schizophrenia risk.
Supplementary Material
Supplementary material is available at http://schizophre niabulletin.oxfordjournals.org.
Funding
National Natural Science Foundation of China grants (81271006 to L.G., 81060081 to L.H.); Hangzhou City Health Science Foundation grant (20120633B25 to L.G.); Jiangxi Provincial Natural Science Foundation (2010GZY0089 to L.H.); National Institutes of Health grant (R01LM011177 to Z.Z.); 2010 National Alliance for Research in Schizophrenia and Affective Disorders Young Investigator Award (to P.J.).
Supplementary Material
Acknowledgments
We thank Drs Chris Cotsapas and Elizabeth J. Rossin for their help in PPI analysis. We are grateful to Rebecca H Posey for her help in language editing. The authors report no financial relationships with commercial interests. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. Jablensky A, Sartorius N, Korten A, et al. Incidence worldwide of schizophrenia. Br J Psychiatry. 1987; 151: 408–409 [DOI] [PubMed] [Google Scholar]
- 2. Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17 [PubMed] [Google Scholar]
- 3. Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003; 60: 1187–1192 [DOI] [PubMed] [Google Scholar]
- 4. Pulver AE, Mulle J, Nestadt G, et al. Genetic heterogeneity in schizophrenia: stratification of genome scan data using co-segregating related phenotypes. Mol Psychiatry. 2000; 5: 650–653 [DOI] [PubMed] [Google Scholar]
- 5. Beckmann H, Franzek E. The genetic heterogeneity of “schizophrenia.”. World J Biol Psychiatry. 2000; 1: 35–41 [DOI] [PubMed] [Google Scholar]
- 6. O’Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055 [DOI] [PubMed] [Google Scholar]
- 7. Ripke S, Sanders AR, Kendler KS, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011; 43:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. St Clair D. Copy number variation and schizophrenia. Schizophr Bull. 2009;35:9–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levinson DF, Duan J, Oh S, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011; 168: 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010; 167: 899–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543 [DOI] [PubMed] [Google Scholar]
- 12. Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008; 455:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009; 5: e1000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008; 9: 356–369 [DOI] [PubMed] [Google Scholar]
- 15. Hamshere ML, Walters JT, Smith R, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry.101038/mp201267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rossin EJ, Lage K, Raychaudhuri S, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J, Zhou G, Ji W, et al. Common variants in the BCL9 gene conferring risk of schizophrenia. Arch Gen Psychiatry. 2011; 68: 232–240 [DOI] [PubMed] [Google Scholar]
- 19. Steinberg S, de Jong S, Andreassen OA, et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011;20:4076–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rietschel M, Mattheisen M, Degenhardt F, et al. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiatry. 2012;17:906–917 [DOI] [PubMed] [Google Scholar]
- 21. Ikeda M, Aleksic B, Yamada K, et al. Genetic evidence for association between NOTCH4 and schizophrenia supported by a GWAS follow-up study in a Japanese population. Mol Psychiatry. 2012.2010.1038/mp.2012.2074 [DOI] [PubMed] [Google Scholar]
- 22. Shi Y, Li Z, Xu Q, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 2011;43:1224–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yue WH, Wang HF, Sun LD, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011; 43: 1228–1231 [DOI] [PubMed] [Google Scholar]
- 24. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009; 460:748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ayalew M, Le-Niculescu H, Levey DF, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012; 17: 887–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fillman SG, Cloonan N, Catts VS, et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2012.10.1038/mp.2012.110 [DOI] [PubMed] [Google Scholar]
- 27. Lage K, Karlberg EO, Størling ZM, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007; 25: 309–316 [DOI] [PubMed] [Google Scholar]
- 28. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4: 44–57 [DOI] [PubMed] [Google Scholar]
- 29. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu J, Sun J, Chen J, et al. RNA-Seq analysis implicates dysregulation of the immune system in schizophrenia. BMC Genomics. 2012; 13(suppl 8):S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shirts BH, Wood J, Yolken RH, Nimgaonkar VL. Association study of IL10, IL1beta, and IL1RN and schizophrenia using tag SNPs from a comprehensive database: suggestive association with rs16944 at IL1beta. Schizophr Res. 2006; 88: 235–244 [DOI] [PubMed] [Google Scholar]
- 34. Xu M, He L. Convergent evidence shows a positive association of interleukin-1 gene complex locus with susceptibility to schizophrenia in the Caucasian population. Schizophr Res. 2010; 120: 131–142 [DOI] [PubMed] [Google Scholar]
- 35. Yoshida M, Shiroiwa K, Mouri K, et al. Haplotypes in the expression quantitative trait locus of interleukin-1β gene are associated with schizophrenia. Schizophr Res. 2012;140:185–191 [DOI] [PubMed] [Google Scholar]
- 36. Gardiner EJ, Cairns MJ, Liu B, et al. Gene expression analysis reveals schizophrenia-associated dysregulation of immune pathways in peripheral blood mononuclear cells. J Psychiatr Res. 2013; 47: 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Michel M, Schmidt MJ, Mirnics K. Immune system gene dysregulation in autism and schizophrenia. Dev Neurobiol. 2012; 72: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fineberg AM, Ellman LM. Inflammatory cytokines and neurological and neurocognitive alterations in the course of schizophrenia. [published online ahead of print]Biol Psychiatry. 2013; doi: 10.1016/j.biopsych.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011; 27: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.