Abstract
Purpose
To provide an evidenced-based review of the genetic basis of the corneal endothelial dystrophies.
Methods
A review of the English-language peer-reviewed literature describing the molecular genetic basis of posterior polymorphous corneal dystrophy (PPCD), congenital hereditary endothelial dystrophy (CHED), Fuchs endothelial corneal dystrophy (FECD) and X-linked endothelial corneal dystrophy (XECD) was performed.
Results
Mutations in several genes have been implicated as playing a pathogenic role in the corneal endothelial dystrophies: VSX1 mutations in PPCD1; COL8A2 mutations in PPCD2 and FECD; ZEB1 mutations in PPCD3 and FECD; and SLC4A11 mutations in CHED2 and FECD. However, linkage, association and familial segregation analyses support a role of only one gene in each corneal endothelial dystrophy: ZEB1 in PPCD3, SLC4A11 in CHED2 and COL8A2 in FECD (early onset). In addition, insufficient evidence exists to consider the autosomal dominant form of CHED (CHED1) as distinct from PPCD.
Conclusions
An accurate classification of the corneal endothelial dystrophies requires a critical review of the evidence to support the role of each suggested chromosomal locus, gene and genetic mutation associated with a corneal endothelial dystrophy. Only after the separation of evidence from opinion is performed can a critical examination of the molecular pathways that lead to endothelial dysfunction in each of these disorders be accurately performed.
Keywords: Corneal Dystrophy, Posterior Polymorphous,1; Corneal Dystrophy, Posterior Polymorphous, 2; Corneal Dystrophy, Posterior Polymorphous, 3; Corneal Endothelial Dystrophy 1; Corneal Endothelial Dystrophy 2; Corneal Dystrophy, Fuchs Endothelial, 1; Corneal Dystrophy, Fuchs Endothelial, 2; Corneal Dystrophy, Fuchs Endothelial, Early-Onset; Corneal Dystrophy, Fuchs Endothelial, Late-Onset
INTRODUCTION
The corneal dystrophies are a group of bilateral inherited disorders associated with the development of either opacities in the normally clear cornea or endothelial dysfunction that may result in corneal edema and loss of corneal clarity. Several dystrophies that primarily affect the corneal endothelium have been described: posterior polymorphous corneal dystrophy (PPCD; MIM #122000) congenital hereditary endothelial dystrophy (CHED; MIM # 217700), Fuchs endothelial corneal dystrophy (FECD; MIM #613267), and X-linked endothelial dystrophy (XECD; MIM # 300779). The prevalence of each of the corneal endothelial dystrophies varies significantly from one population to another, although they constitute common indications for corneal transplantation in published series from around the world.1, 2 In the United States, FECD affects as much as 5% of the population over 40 years of age, and visually significant corneal edema secondary to FECD is the most common indication for corneal transplantation, accounting for 47% of endothelial keratoplasty procedures performed in 2012.3, 4 While autosomal recessive CHED is relatively uncommon in the United States, it is one of the most frequently encountered corneal dystrophies in countries where consanguineous marriages are common, and is a well-recognized cause of congenital corneal edema,. Less common than FECD but more common than CHED in the United States, PPCD is a dominantly inherited corneal endothelial dystrophy that is associated with a varied phenotype, ranging from asymptomatic corneal endothelial changes to congenital corneal edema and glaucoma, the latter of which develops in 15–40% of affected patients.(Figure 1)5, 6 X-linked endothelial dystrophy remains the least common of the corneal endothelial dystrophies, reported in only a single family to date.7 Seven consecutive generations of an Austrian family demonstrated a wide range of phenotypic features, ranging from asymptomatic endothelial changes to congenital corneal edema.7 Given apparent x-linked inheritance pattern, linkage analysis was performed for the X chromosome, revealing evidence of significant linkage to a 14.79 Mb region on Xq25, although the genetic basis remains unknown.7
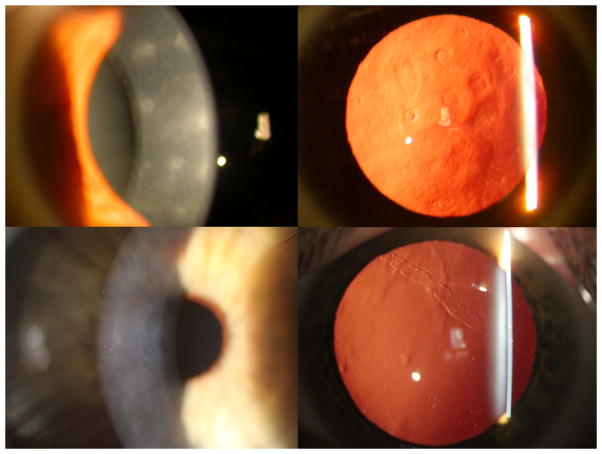
Figure 1.
Slit lamp photomicrographs of individuals with PPCD demonstrating the classic endothelial abnormalities of geographic gray opacities (top left and right), clustered endothelial “vesicles” surrounded by gray opacity (bottom left) and endothelial band, seen in retroillumination against the red reflex (bottom right).
Significant advances have been made in the last decade in elucidating the genetic basis of the corneal endothelial dystrophies.8 Mutations in the zinc finger E-box binding homeobox 1 gene (ZEB1), the solute carrier family 4 member 11 gene (SLC4A11) and the collagen, type VIII, alpha-2 gene (COL8A2) have been convincingly demonstrated to play a role in PPCD, CHED and an early onset form of FECD, respectively. However, some evidence exists to suggest that mutations in each of these genes also cause another of the corneal endothelial dystrophies (SLC4A11 mutations in FECD, COL8A2 mutations in PPCD and ZEB1 mutations in FECD).9–13 This review will critically examine what is known about the genetic basis of the corneal endothelial dystrophies, based on an analysis of the supporting evidence.
Posterior polymorphous corneal dystrophy
Locus heterogeneity has been demonstrated for PPCD, which has been mapped to the pericentromeric region of chromosome 20 (the PPCD1 locus), associated with mutations in the collagen, type VIII, alpha 2 gene (COL8A2) on chromosome 1 (the PPCD2 locus) and associated with nonsense mutations in the zinc finger E-box binding homeobox 1 gene (ZEB1), also known as the transcription factor 8 gene (TCF8), on chromosome 10 (the PPCD3 locus).14–18 While compelling evidence exists to support the involvement of a gene in the PPCD1 locus and ZEB1 in the pathogenesis of PPCD, insufficient evidence exists to support a role for COL8A2, or any other gene in the PPCD2 locus, in the pathogenesis of PPCD.
Posterior polymorphous corneal dystrophy 1
A locus for PPCD (PPCD1) has been identified in the pericentromeric region of chromosome 20 through linkage analysis by our group and two others.5, 19, 20 The region of chromosome 20 that is contained within the overlapping support intervals from all three studies, the PPCD1 common support interval, is a 1.8 Mb region containing 30 mapped genes (Human Annotation Release 104). We screened the exonic regions of these genes and identified the minor alleles of 7 SNPs that segregated with the affected phenotype, although each was also identified in control individuals, indicating that none is a candidate for the causal variant for PPCD1. However, the demonstration of association of each of these SNPs with the disease allele provides compelling evidence that the pathogenic mutation lies within the PPCD1 locus.20 In addition, a mouse model of PPCD1 has been mapped to a region of chromosome 2 that is syntenic with the PPCD1 locus on human chromosome 20.21
Posterior polymorphous corneal dystrophy 2
Although a locus containing the collagen, type VIII, alpha 2 gene (COL8A2) on chromosome 1 has been proposed for PPCD (termed PPCD2), linkage to this locus has never been demonstrated in a family with PPCD, and the only identified presumed pathogenic sequence variant identified in COL8A2 was not shown to segregate with the affected phenotype in the pedigree in which it was identified.13 After associating mutations in COL8A2 with an early onset variant of FECD, investigators screened COL8A2 in PPCD families, given the clinical and histopathologic similarities between PPCD and FECD.13 After identifying a missense amino acid change in two individuals affected with PPCD from one family (no unaffected family members were screened), the investigators proposed COL8A2 as the PPCD2 locus, although no coding region mutations were identified in 13 other affected patients. Subsequently, our group and others screened COL8A2 in PPCD families and failed to identify any pathogenic coding region mutations, supporting our belief that the originally reported missense variant in COL8A2 is actually a rare polymorphism.22, 23
Posterior polymorphous corneal dystrophy 3
ZEB1, a zinc finger transcription factor that binds to DNA at a conserved sequence (CACCTG) that is known as an E2 box, plays a critically important role both in development (e.g. embryonic gastrulation) and disease (e.g. tumor metastasis) through the repression of transcription of genes important for maintaining the epithelial phenotype.24 ZEB1 is involved in epithelial-to-mesenchymal transition (EMT) through its regulation by TGF-β, FGF and the miR200 family of microRNAs and repression of transcription of E-cadherin (CDH1), an epithelial cell marker.24 While FGF-2 is known to mediate the endothelial-to-mesynchymal transformation (EnMT) that results in retrocorneal fibrous membrane formation by HCEnC, the role of ZEB1 in EnMT is unknown and the actual mechanism by which ZEB1 mutations lead to the similar HCEnC dysregulation and loss of normal HCEnC function that characterize PPCD3 is currently unclear.25 ZEB1 is expressed in the human corneal endothelium and is known to repress the transcription of a variety of genes, including several involved in EMT, and is suggested to negatively regulate the transcription of other genes, including collagen, type IV, alpha 3 (COL4A3).15, 26 While COL4A3 has been shown to be expressed in the infant and adult DM, it has been reported as absent in the adult human corneal endothelium.15, 27 However, in the presence of a truncating mutation in ZEB1, COL4A3 expression has been demonstrated in the corneal endothelium of an individual affected with PPCD3.15 These findings, and the identification of six E2 boxes in the 5 kb upstream of the COL4A3 transcription initiation site, suggest that ZEB1 participates in the negative regulation of COL4A3 transcription. Therefore, ZEB1 haploinsufficiency in the cornea secondary to one of the 24 truncating mutations identified to date is hypothesized to lead to COL4A3 expression, resulting in the abnormal endothelial proliferation, corneal thickening and iridocorneal adhesions observed in PPCD.(Table 1) However, the demonstration of COL4A3 expression in the normal adult human corneal endothelium and inversely related expression levels of ZEB1 and COL4A3 mRNA in normal and PPCD3 human corneal endothelium challenges this hypothesis and suggests that it is the relative expression of COL4A3, not the simple presence or absence of COL4A3 expression, that differentiates normal from affected corneal endothelium.28
Table 1.
ZEB1 mutations associated with PPCD3 and SLC4A11 mutations associated with CHED.
| Dystrophy | Gene (Locus) | Exon/Intron | Nucleotide change | AA change | Ref |
|---|---|---|---|---|---|
| PPCD3 | ZEB1 (10p11.2) | E1 | c.1A>G | p.(Met1?) | 1 |
| c.2T>G | p.(Met1?) | 2 | |||
| c.34C>T | p.(Gln12*) | 2 | |||
| E4 | c.449delG | p(Gly150Alafs*36) | 3 | ||
| E5 | c.640C>T | p.(Gln214*) | 2 | ||
| c.672delA | p.(Gly225Glufs*7) | 4 | |||
| c.689_690delAT | p.(His230Argfs*7) | 3 | |||
| E7 | c.929dupA | p.(Cys311Valfs*25) | 2 | ||
| c.973C>T | p.(Arg325*) | 2 | |||
| c.1124delT | p.(Phe375Serfs*31) | 5 | |||
| c.1332_1335delCAAT | p.(Ile444Metfs*48) | 6 | |||
| c.1348C>T | p.(Gln450*) | 6 | |||
| c.1387_1390delCCTT | p.(Pro463Trpfs*29) | 5 | |||
| c.1482dupA | p.(Glu495Argfs*10) | 2 | |||
| c.1569delA | p.(Val526*) | 2 | |||
| c.1576dupG | p.(Val526Glyfs*3) | 6 | |||
| c.1913_1914delCA | p.(Ser638Cysfs*5) | 3 | |||
| c.2157C>G | p.(Tyr719*) | 5 | |||
| c.2182G>T | p.(Glu728*) | 6 | |||
| c.2324dupA | p.(Glu776Glyfs*44) | 5 | |||
| E8 | c.2650delC | p.(Gln884Argfs*37) | 3 | ||
| E9 | c.2916_2917delTG | p.(Gly973Valfs*14) | 6 | ||
| c.2990_2991delAG | p.(Glu997Alafs*7) | 2 | |||
| c.3116_3117delAG | p.(Glu1039Glyfs*6) | 3 | |||
| CHED | SLC4A11 (20p13–p12) | E2 | c.140delA | p.(Tyr47Serfs*69) | 7 |
| c.246_247delTTinsA | p.(Phe84Leufs*32) | 8 | |||
| E3 | c.306delC | p.(Gly103Valfs*13) | 7 | ||
| c.334C>T | p.(Arg112*) | 7 | |||
| E4 | c.353_356delAGAA | p.(Lys118Thrfs*12) | 9 | ||
| c.374G>A | p.(Arg125His) | 10 | |||
| c.427G>A | p.(Glu143Lys) | 7 | |||
| c.473_480delGCTTCGCC | p.(Arg158Glnfs*4) |
11 7 10 12 |
|||
| c.478G>A | p.(Ala160Thr) | 10 | |||
| E5 | c.618_619delAG | p.(Val208Alafs*38) | 7 | ||
| c.625C>T | p.(Arg209Trp) | 7 | |||
| c.637T>C | p.(Ser213Pro) | 11 | |||
| c.638C>T | p.(Ser213Leu) | 7 | |||
| E6 | c.654 (−97)_778 (−1488)del698 | p.(Cys219Lysfs*49) | 10 | ||
| c.695G>A | p.(Ser232Asn) | 13 | |||
| c.697C>T | p.(Arg233Cys) | 7 | |||
| c.720G>A | p.(Trp240*) | 14 | |||
| E7 | c.806C>T | p.(Ala269Val) | 10 | ||
| c.812C>T | p.(Thr271Met) | 15 | |||
| c.859_862delGAGAinsCCT | p.(Glu287Profs*21) | 16 | |||
| c.878_889del12 | p.(Glu293_Glu296d el) | 7 | |||
| c.985A>T | p.(Arg329*) | 13 | |||
| I7 | c.996+25del19 | N/A |
7 12 |
||
| I8 | c.1091-1G>C | splice site inactivation | 7 | ||
| E9 | c.1156T>C | p.(Cys386Arg) |
14 10 17 |
||
| c.1179G>C | p.(Gly394Arg) | 12 | |||
| c.1202C>A | p.(Thr401Lys) | 7 | |||
| E10 | c.1253G>A | p.(Gly418Asp) | 7, 12 | ||
| E11 | c.1317_1322del6ins8 | p.(Leu440Valfs*6) | 7 | ||
| c.1378_1381delTACGins A | p.(Tyr460_Ala461d elinsThr) | 11 | |||
| c.1391G>A | p.(Gly464Asp) | 9 | |||
| c.1418T>G | p.(Leu473Arg) | 7 | |||
| c.1463G>A | p.(Arg488Lys) | 11 | |||
| E12 | c.1466C>T | p.(Ser489Leu) | 7, 9 | ||
| E13 | c.1704_1705delCT | p.(His568Hisfs*177) | 8 | ||
| c.1751C>A | p.(Thr584Lys) | 7 | |||
| E14 | c.1813C>T | p.(Arg605*) |
8 7,9 |
||
| c.1894G>T | p.(Glu632*) | 8 7 | |||
| E15 | c.2017_2019delTTC | p.(Phe673del) | 16 | ||
| I15 | c.2067 −6_−16 delinsGGCCGGCCGG | N/A | 9 | ||
| c.2067+1G>A | splice site inactivation | 12 | |||
| E16 | c.2233_2240dupTATGAC AC | p.(Ile748Metfs*5) | 11 | ||
| c. 2236C>T | p.(Arg757*) | 12 | |||
| I16 | c.2240+1G>A | splice site inactivation |
14 17 |
||
| E17 | c.2263C>T | p.(Arg755Trp) |
14 7, 10 |
||
| c.2264G>A | p.(Arg755Gln) |
8 9 14 7 |
|||
| c.2318C>T | p.(Pro773Leu) | 7,10 | |||
| c.2389_2391delGAT | p.(Asp797del) | 7 | |||
| c.2398C>T | p.(Gln800*) | 14 | |||
| c.2407C>T | p.(Gln803*) | 7 | |||
| c.2411G>A | p.(Arg804His) | 8 | |||
| c.2420delTinsGG | p.(Leu807Argfs*71) | 8 | |||
| c.2423_2454del32 | p.(Leu808Profs*110) | 11 | |||
| I17 | c.2437-1G>A | splice site inactivation | 14 | ||
| E18 | c.2470G>A | p.(Val824Met) | 11 7 17 | ||
| c.2498C>T | p.(Thr833Met) | 8 | |||
| c.2506C>T | p.(Glu836*) | 14 | |||
| c.2518-2520delCTG | p.(Leu840del) | 17 | |||
| c.2528T>C | p.(Leu843Pro) | 11 | |||
| c.2566A>G | p.(Met856Val) | 11 | |||
| c.2606G>A | p.(Arg869His) | 8 | |||
| c.2605C>T | p.(Arg869Cys) |
9
14 7 |
|||
| E19 | c.2618T>C | p.(Leu873Pro) | 10 | ||
| c.2623C>T | p.(Arg875*) | 7 |
Vincent AL, Niederer RL, Richards A, et al. Phenotypic characterisation and ZEB1 mutational analysis in posterior polymorphous corneal dystrophy in a New Zealand population. Mol Vis 2009;15:2544–53.
Aldave AJ, Yellore VS, Yu F, et al. Posterior polymorphous corneal dystrophy is associated with TCF8 gene mutations and abdominal hernia. Am J Med Genet A 2007;143A:2549–56.
Bakhtiari P, Frausto RF, Roldan AN, et al. Exclusion of pathogenic promoter region variants and identification of novel nonsense mutations in the zinc finger E-box binding homeobox 1 gene in posterior polymorphous corneal dystrophy. Mol Vis 2013;19:575–80.
Nguyen DQ, Hosseini M, Billingsley G, et al. Clinical phenotype of posterior polymorphous corneal dystrophy in a family with a novel ZEB1 mutation. Acta Ophthalmol 2010;88:695–9.
Liskova P, Tuft SJ, Gwilliam R, et al. Novel mutations in the ZEB1 gene identified in Czech and British patients with posterior polymorphous corneal dystrophy. Hum Mutat 2007;28:638.
Krafchak CM, Pawar H, Moroi SE, et al. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet 2005;77:694–708.
Sultana A, Garg P, Ramamurthy B, et al. Mutational spectrum of the SLC4A11 gene in autosomal recessive congenital hereditary endothelial dystrophy. Mol Vis 2007;13:1327–32.
Jiao X, Sultana A, Garg P, et al. Autosomal recessive corneal endothelial dystrophy (CHED2) is associated with mutations in SLC4A11. J Med Genet 2007;44:64–8.
Vithana EN, Morgan P, Sundaresan P, et al. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2). Nat Genet 2006;38:755–7.
Hemadevi B, Veitia RA, Srinivasan M, et al. Identification of mutations in the SLC4A11 gene in patients with recessive congenital hereditary endothelial dystrophy. Arch Ophthalmol 2008;126:700–8.
Desir J, Moya G, Reish O, et al. Borate transporter SLC4A11 mutations cause both Harboyan syndrome and non-syndromic corneal endothelial dystrophy. J Med Genet 2007;44:322–6.
Aldahmesh MA, Khan AO, Meyer BF, Alkuraya FS. Mutational spectrum of SLC4A11 in autosomal recessive CHED in Saudi Arabia. Invest Ophthalmol Vis Sci 2009;50:4142–5.
Aldave AJ, Yellore VS, Bourla N, et al. Autosomal recessive CHED associated with novel compound heterozygous mutations in SLC4A11. Cornea 2007;26:896–900.
Ramprasad VL, Ebenezer ND, Aung T, et al. Novel SLC4A11 mutations in patients with recessive congenital hereditary endothelial dystrophy (CHED2). Mutation in brief #958. Online. Hum Mutat 2007;28:522–3.
Shah SS, Al-Rajhi A, Brandt JD, et al. Mutation in the SLC4A11 gene associated with autosomal recessive congenital hereditary endothelial dystrophy in a large Saudi family. Ophthalmic Genet 2008;29:41–5.
Kumar A, Bhattacharjee S, Prakash DR, Sadanand CS. Genetic analysis of two Indian families affected with congenital hereditary endothelial dystrophy: two novel mutations in SLC4A11. Mol Vis 2007;13:39–46.
Paliwal P, Sharma A, Tandon R, et al. Congenital hereditary endothelial dystrophy - mutation analysis of SLC4A11 and genotype-phenotype correlation in a North Indian patient cohort. Mol Vis 2010;16:2955–63.
Congenital Hereditary Endothelial Dystrophy
Congenital hereditary endothelial dystrophy is unique among the corneal dystrophies in that both autosomal dominant (CHED1) and autosomal recessive (CHED2) forms have been described. and mapped to distinct loci on chromosome 20. However, a careful examination of the literature indicates that CHED1 is not sufficiently distinguishable from PPCD1 to consider it a separate corneal endothelial dystrophy. Therefore, the nomenclature should be revised to reflect that only autosomal recessive CHED is sufficiently well characterized and distinct from the other endothelial dystrophies to be considered a unique corneal endothelial dystrophy.
Congenital Hereditary Endothelial Dystrophy 1
A review of the English language literature reveals only five families reported with an apparently dominantly inherited form of CHED, several of which were reported in one or more publications: a Caucasian American pedigree published by Maumenee in 1960 and subsequently reported by Judisch and Maumenee in 1978 (Family 1)29, 30; a Norwegian pedigree published by Odland in 1968 (Family 2)31; a British family originally reported by Pearce and colleagues in 1969, and subsequently reported by Kirkness in 1987 and Toma in 1995 (Family 3)32–34; a Caucasian American pedigree published by Kanai and colleagues in 1971 and subsequently reported by Levinson and colleagues in 1973 (Family 4)35–37; and another Caucasian American pedigree reported by Levinson and colleagues in 1973 (Family 5)37. We believe that the only report that provides sufficient evidence of both an endothelial dystrophy and an autosomal dominant inheritance pattern is the family originally published by Pearce and colleagues.32 However, based on the clinical, histopathologic and electron microscopic findings presented in the original and subsequent publications of this family, the reported affected individuals most likely have PPCD.
Family 1
The original pedigree reported by Maumenee in 1960 demonstrates three consecutive generations of affected individuals without any affected parent-offspring pairs, leading to uncertainty regarding the mode of inheritance.30 When the family was reascertained by Judisch and Maumenee almost two decades later, a history of consanguinity was obtained that clearly established the autosomal recessive inheritance pattern in this family.29 As Judisch and Maumenee acknowledged that they had never seen an autosomal dominant CHED pedigree, the comparison of the clinical features of the dominant and recessive forms of CHED that they reported relied upon the clinical features reported by Pearce and colleagues and by Odland.29
Family 2
A review of article published by Odland in 1968 clearly demonstrates that the pedigree he reported had a corneal stromal dystrophy, not an endothelial dystrophy. Although several members of the family had a history of congenital corneal opacification, slit lamp examination revealed that the opacities were due to a “large number of small flakes and spots throughout all layers of the stroma.”31 The fact that a lamellar keratoplasty in one of the affected individuals successfully improved the patient’s vision to 5/7.5 (20/30) indicates that the corneal opacification was due to stromal opacification, not endothelial dysfunction. In addition, as histopathologic examination of the excised corneal specimens revealed amorphous material deposited between the collagen lamellae that stained with Masson trichrome, Odland attributed the corneal opacification to a “degeneration of the corneal lamellae”.31
Family 3
The pedigree reported by Pearce that was subsequently reported by Kirkness in 1987 and Toma in 1995 likely has PPCD based on clinical, histopathologic, electron microscopic and linkage analyses.29, 32–34 The severity and early onset of the corneal edema, requiring corneal transplantation in 10 of the 39 (25.6%) affected individuals in this family, was relied upon by the authors as a distinguishing feature between CHED1 from PPCD, although the authors acknowledged that the two share many clinical features. However, this percentage is not significantly different from the 23.3% (17/73) of individuals with PPCD that have undergone corneal transplantation (in several cases for congenital corneal edema) in the authors’ ongoing study of PPCD (unpublished data). Additionally, the description of a thickened and grey-colored DM, associated with “occasional irregular white areas” and a “beaten copper appearance, similar to that of early Fuchs’ dystrophy” in affected individuals is consistent with the grayish opacification of DM, vesicular and band-shaped endothelial opacities and endothelial guttae characteristic of PPCD.32 Electron microscopic examination of two excised corneal specimens demonstrated a normal appearing anterior banded portion of DM and a markedly thickened posterior nonbanded portion, similar to what is seen in PPCD.32 While the authors compared the EM findings to those described for FECD, they did not mention that the findings were consistent with PPCD, as it was two years later in 1971 that the electron microscopic features of PPCD were first published.38
In 1987, Kirkness and colleagues published a report of the clinical, light and electron microscopic findings in additional members of the family originally reported by Pearce.33 Based on these findings, the authors acknowledged that CHED1 and PPCD may have a similar clinical and histologic appearance, and thus that the two conditions could actually represent different manifestations of the same entity.33
In 1995, Toma and colleagues reported the results of linkage analysis in the same family, demonstrating linkage to a 2.7 cM region on chromosome 20 located within the original PPCD1 locus.34 The subsequent mapping of three other families with PPCD to this region led to the identification of a common 2.4 cM PPCD1 interval that is nearly identical to the CHED1 interval.(Figure 2) Potential explanations include the possibility that the two endothelial dystrophies are allelic variants, that two or more genes that are involved in corneal endothelial cell function are located in the common linked interval for the two endothelial dystrophies, or that the CHED1 family actually has PPCD1. Given the clinical, histopathologic, ultrastructural similarities between CHED1 and PPCD1, we believe that it is most plausible that the CHED1 family actually has PPCD1.
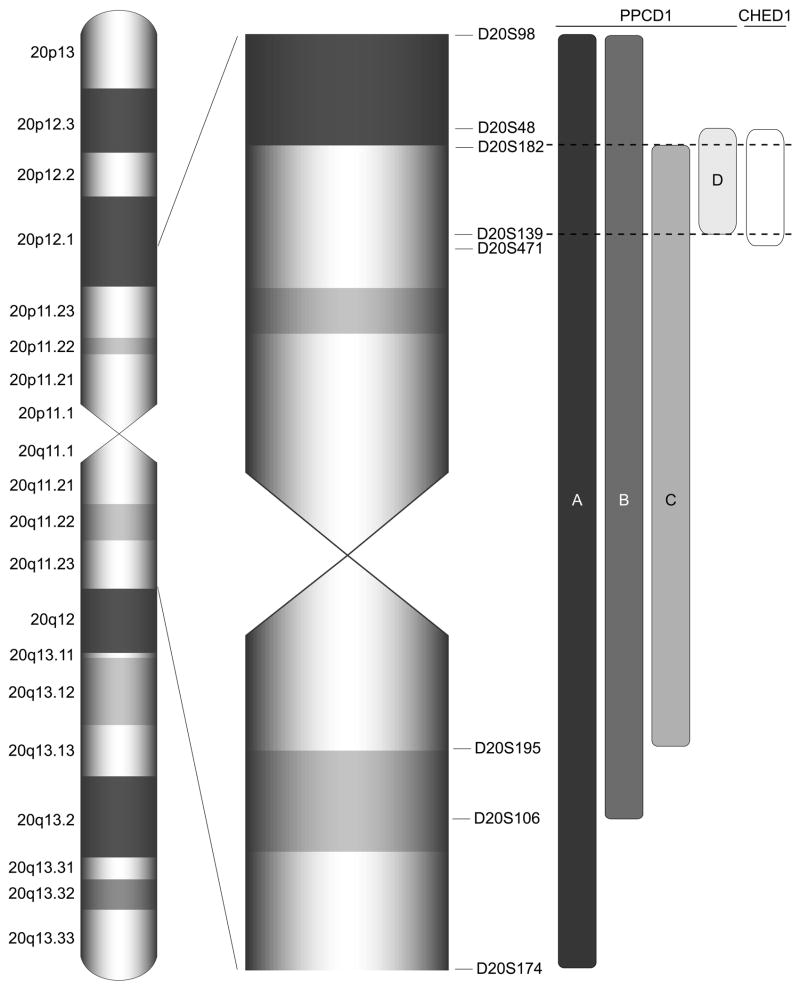
Figure 2.
Ideogram of pericentromeric portion of chromosome 20 demonstrating the relationship of the CHED1 locus, defined by the markers D20S48 and D20S471,34 to the intervals to which PPCD1 has been mapped in families A,5, 68, B19, C,20 and D19. The interval that is common to PPCD1 in each of the three studies is defined by the markers D20S182 and D20S139, which is nearly identical to the CHED1 interval. PPCD: posterior polymorphous corneal dystrophy 1; CHED1: congenital hereditary endothelial dystrophy 1.
Family 4
A pedigree reported initially by Kanai and colleagues in 1971 and again by Levenson and colleagues in 1973 also appears to have PPCD based on the clinical, light and electron microscopic findings. The authors described affected monozygotic twin sisters and three affected offspring of one of the affected twins.35, 36 Observed clinical features atypical for CHED included the presence of only peripheral corneal endothelial changes and overlying stromal edema in the affected mother, whose CDVA measured 20/25 OU and who was asymptomatic. A similar pattern of peripheral corneal edema was described in each of the affected woman’s three children as well, one of whom required corneal transplantation. Electron microscopic examination of the excised corneal button demonstrated a posterior collagenous layer interposed between DM and the corneal endothelium, and replacement of the normal endothelial monolayer with 2–3 cell layers. These findings, as well as the microvilli observed on the posterior cell surface, are now recognized, characteristic features of PPCD, which were likely not recognized as such by the authors as they were first published later that year.38
Family 5
In addition to the aforementioned family, Levenson and colleagues reported a second family with CHED affecting individuals in consecutive generations in their 1973 publication.37 Affected members of this family demonstrated endothelial findings that are now recognized as classic for PPCD, including: endothelial vesicles that the authors described as “grayish-white, circular lesions…some with clearer centers..along with circular or oval clear vacuole-like lesions. A denser whitish haze often surrounded these vacuole-like lesions, which on occasion were strung out several in a row”; and endothelial bands described as “double-contoured, nearly straight lesions that traversed the greater extent of both central corneas”.(Figure 3)37 The authors acknowledged that the endothelial changes were consistent with those associated with PPCD, but differentiated between CHED and PPCD in this pedigree due to the absence of involvement of the posterior stroma in asymptomatic affected individuals and the diagnosis of congenital corneal edema in one affected individual. However, the demonstration of regions of endothelial cell layer duplication, characteristic of PPCD, on histopathologic examination of the excised corneal button from this individual and the fact that PPCD is now a well-recognized cause of congenital corneal edema provide additional convincing evidence that both pedigrees reported by Levenson and colleagues have PPCD.39, 40
Figure 3.
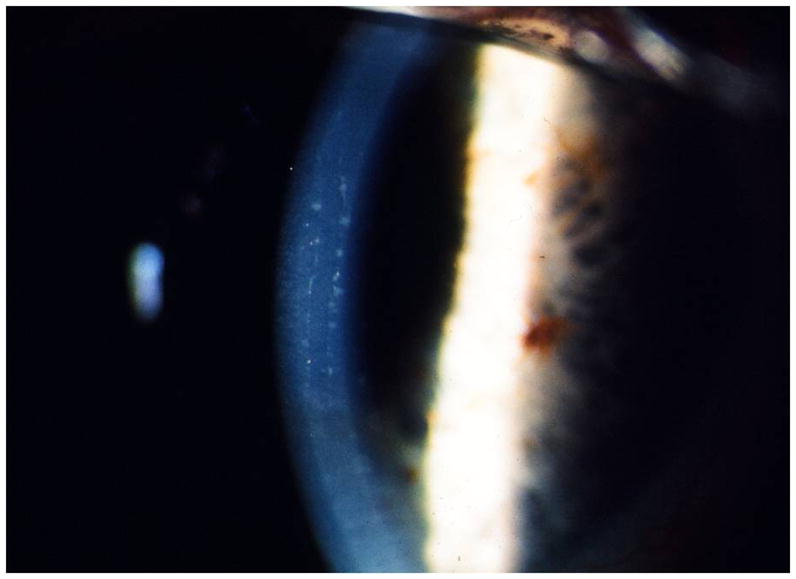
Slit lamp photomicrograph of corneal endothelial bands in affected member of a family reported with CHED1 in 1973. The appearance of the parallel bordered endothelial bands is now recognized as characteristic of PPCD. Photo courtesy of Dr. Jeremy Levenson.
Congenital Hereditary Endothelial Dystrophy 2
In 1999, two linkage studies utilizing markers located across the PPCD1 locus on chromosome 20, which contains the CHED1 locus, excluded CHED2 from this interval.41, 42 Later the same year, Hand and colleagues reported localization of CHED2 to a region of 20p13 distinct from the CHED1 locus using homozygosity mapping.43 Seven years later, two different groups reported that SLC4A11 mutations caused CHED2.44, 45 (Figure 4) Vithana and colleagues postulated that mutations in SLC4A11, which encodes bicarbonate transporter-related protein-1 (BTR1), resulted in failure of the BTR1 protein to reach the plasma membrane, where it functions as a sodium-borate cotransporter.45 Indeed, transfection of an immortalized cell line with mutant SLC4A11 cDNAs demonstrated little to no expression of the mature protein and very little of the mutant BTR1 protein localized to the cell surface.45 To date, 74 mutations in 17 of the 19 coding exons of SLC4A11 have been identified, although 32 of the 136 (23.5%) pedigrees screened to date do not demonstrate coding region mutations in SLC4A11.(Tables 1 and 2) Screening of the putative SLC4A11 promoter region in 20 of these 32 families failed to demonstrate any presumed pathogenic variants. Thus, it is possible that locus heterogeneity exists for CHED2 as it does for PPCD1, although linkage to the CHED2 locus on 20p13 has been demonstrated in each of the three families in whom genome-wide linkage analysis has been performed.43, 45, 46
Figure 4.
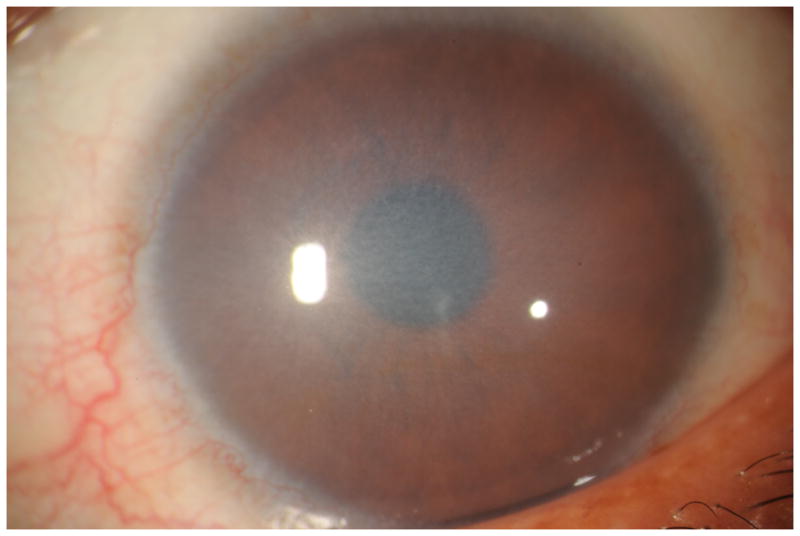
Slit lamp photomicrograph of diffuse corneal edema in a 30-year-old woman with a history of long-standing impaired vision. Screening of SLC4A11 identified a homozygous p.Thr271Met mutation previously associated with CHED2, confirming the diagnosis. Photo courtesy of Dr. Majid Moshirfar.
Table 2.
Published series of SLC4A11 screening in congenital hereditary endothelial dystrophy
| Publication | Year of Publication | Location | # of probands screened | # of probands with mutation | # of probands without a mutation | Promoter screened? |
|---|---|---|---|---|---|---|
| Jiao et al1 | 2006 | Hyderabad, India | 16 | 12 | 4 | No |
| Vithana et al2 | 2006 | Madurai, India | 10 | 10 | 0 | |
| Sultana et al3 | 2007 | Hyderabad, India | 42 | 35 | 7 | No |
| Kumar et al4 | 2007 | Bangalore, India | 2 | 2 | 0 | |
| Ramprasad et al5 | 2007 | Chennai, India | 9 | 9 | 0 | |
| Aldave et al6 | 2007 | Los Angeles, US | 1 | 1 | 0 | |
| Desir et al7 | 2007 | Brussels, Belgium | 7 | 7 | 0 | |
| Hemadevi et al8 | 2008 | Madurai, India | 20 | 11 | 9 | Yes |
| Shah et al9 | 2008 | Riyadh, Saudi Arabia | 2 | 1 | 1 | No |
| Aldahmesh et al10 | 2009 | Riyadh, Saudi Arabia | 7 | 7 | 0 | |
| Paliwal et al11 | 2010 | New Delhi, India | 20 | 9 | 11 | Yes |
| Total | 136 | 104 | 32 |
Jiao X, Sultana A, Garg P, et al. Autosomal recessive corneal endothelial dystrophy (CHED2) is associated with mutations in SLC4A11. J Med Genet 2007;44:64–8.
Vithana EN, Morgan P, Sundaresan P, et al. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2). Nat Genet 2006;38:755–7.
Sultana A, Garg P, Ramamurthy B, et al. Mutational spectrum of the SLC4A11 gene in autosomal recessive congenital hereditary endothelial dystrophy. Mol Vis 2007;13:1327–32.
Kumar A, Bhattacharjee S, Prakash DR, Sadanand CS. Genetic analysis of two Indian families affected with congenital hereditary endothelial dystrophy: two novel mutations in SLC4A11. Mol Vis 2007;13:39–46.
Ramprasad VL, Ebenezer ND, Aung T, et al. Novel SLC4A11 mutations in patients with recessive congenital hereditary endothelial dystrophy (CHED2). Mutation in brief #958. Online. Hum Mutat 2007;28:522–3.
Aldave AJ, Yellore VS, Bourla N, et al. Autosomal recessive CHED associated with novel compound heterozygous mutations in SLC4A11. Cornea 2007;26:896–900.
Desir J, Moya G, Reish O, et al. Borate transporter SLC4A11 mutations cause both Harboyan syndrome and non-syndromic corneal endothelial dystrophy. J Med Genet 2007;44:322–6.
Hemadevi B, Veitia RA, Srinivasan M, et al. Identification of mutations in the SLC4A11 gene in patients with recessive congenital hereditary endothelial dystrophy. Arch Ophthalmol 2008;126:700–8.
Shah SS, Al-Rajhi A, Brandt JD, et al. Mutation in the SLC4A11 gene associated with autosomal recessive congenital hereditary endothelial dystrophy in a large Saudi family. Ophthalmic Genet 2008;29:41–5.
Aldahmesh MA, Khan AO, Meyer BF, Alkuraya FS. Mutational spectrum of SLC4A11 in autosomal recessive CHED in Saudi Arabia. Invest Ophthalmol Vis Sci 2009;50:4142–5.
Paliwal P, Sharma A, Tandon R, et al. Congenital hereditary endothelial dystrophy - mutation analysis of SLC4A11 and genotype-phenotype correlation in a North Indian patient cohort. Mol Vis 2010;16:2955–63.
Fuchs Endothelial Corneal Dystrophy
While FECD was originally considered a sporadic disorder, several reports published between 1970 and 1980 documented multiple familial cases of FECD, establishing an autosomal dominant inheritance pattern.47–49 These reports also established a female preponderance among affected individuals, with a female-to-male ratio of 2.5:1.0 identified by Krachmer and colleagues.48 Several linkage and association studies have been performed in both a less common early onset form of FECD as well as the classic, late-onset form of FECD. While the early onset form of FECD has been linked to only a single locus, chromosome 1p, significant linkage has been demonstrated for the late onset form of FECD to chromosomes 13 (FCD1),50, 51 18 (FCD2),52 and 5 (FCD3).53 Efforts to identify the genetic basis of both the early and late onset forms of FECD have involved screening functional candidate genes within these loci, as well as genes implicated in the pathogenesis of other corneal endothelial dystrophies.
Early Onset FECD
The first linkage analysis in a family with FECD was published by Biswas and colleagues in 2001.13 Analyzing a family with an early onset form of FECD, the investigators identified a 6–7 cM interval on chromosome 1p34.3-p32. The investigators chose COL8A2 from the genes mapped to this linked interval as a functional candidate gene as it is a major component of DM.13, 54 Screening of COL8A2 revealed a missense mutation, p.Gln455Lys, that segregated with the affected phenotype in the family and was not identified in controls.13 The authors identified the same mutation in two other early-onset FECD families, but did not comment on whether segregation with the affected phenotype was demonstrated in either family (it was present in 8/8 affected individuals screened in one family, but screening of unaffected individuals was not mentioned).13 While the authors also identified another missense substitution in a family with FECD, p.Arg155Gln, and two other missense substitutions in individuals with sporadic FECD, p.Arg304Gln and p.Arg434His, the identification of p.Arg155Gln and p.Arg434His in unaffected individuals in subsequent reports indicates that these are non-pathogenic variants.22, 55, 56 Additionally, the absence of significant association between any SNPs in the COL8A2 locus and FECD in two independently performed gemone-wide association studies (GWAS) argues against a role for COL8A2 in the classic, late onset form of FECD.4, 57 However, additional evidence to support the pathogenicity of COL8A2 mutations in early onset FECD was provided by the identification of p.Leu450Trp and p.Gln455Val in two additional families with early onset FECD in which the mutations segregated with the affected phenotype.58,56 In the family in which the p.L450W mutation was identified, Gottsch and colleagues noted a 1:1 female-to-male ratio and clinical and histopathologic characteristics of the endothelial guttae that were distinct from the classic late onset form of FECD.58 Therefore, given the different demographic, morphologic and temporal features of the early and late-onset forms of FECD, it appears that COL8A2 mutations are causative of an atypical, less common variant of FECD, but do not play a role in the phenotypically distinct late onset form of FECD.58
Late Onset (Classic) FECD
SLC4A11
As with the other endothelial dystrophies, the initial efforts to identify the genetic basis of late-onset FECD consisted of linkage analyses and screening of genes implicated in other corneal endothelial dystrophies. Following the identification of SLC4A11 mutations in individuals with CHED2, Vithana and colleagues screened SLC4A11 in 89 individuals with sporadic and familial FECD, identifying 4 individuals with presumed pathogenic variants (3 missense and 1 frameshift).9 However, as 3 of the 4 cases were sporadic, and no affected family members of the fourth individual were available for testing, segregation was not demonstrated for any of the variants.9 To support their contention that the identified variants were functionally significant, the authors investigated the effects of the 3 heterozygous missense mutations on protein expression and localization. The demonstration of significantly decreased expression of 2 of the 3 mutant proteins in transfected HEK cells and reduced levels of all 3 mutant proteins localizing to the cell surface provided evidence that the identified missense mutations in SLC4A11 did adversely affect protein expression and localization.9 However, as acknowledged by the authors, the conclusion that the identified heterozygous SLC4A11 mutations are causative of FECD indicates that the parents of individuals with CHED2, heterozygous for the mutations in their affected offspring, would be expected to have FECD. The absence of the clinical features of FECD in the parents of individuals with CHED2 that they had previously reported was attributed by the authors to the fact that the parents were not yet old enough to demonstrate the clinical features of FECD.9
In a subsequent series of 192 individuals with sporadic FECD and small nuclear families with FECD, Riazuddin and colleagues identified an additional 7 heterozygous SLC4A11 missense mutations that they considered causative of FECD based on their absence in control chromosomes and biochemical studies demonstrating associated impaired intracellular localization and or posttranslational protein modification.10 However, segregation was demonstrated for only 1 of the 7 variants, in a small pedigree (2 affected, 2 unaffected and 1 indeterminate individuals screened).10
In spite of the evidence in these two publications supporting a role for SLC4A11 mutations in a minority of cases of FECD, additional evidence will need to be provided to convincingly demonstrate a pathogenic role for SLC4A11 in FECD. FECD has been mapped to multiple chromosomal loci, but never to the SLC4A11 locus on chromosome 20, arguing against a role for SLC4A11 in FECD.4, 57 Assuming that it does have a role, speculation that the identified SLC4A11 heterozygous missense mutations may lead to FECD through a dominant negative effect is inconsistent with the proposed pathogenic mechanism of haploinsufficiency caused by the identified heterozygous frameshift mutation.9 As the authors did not determine whether the frameshift mutation resulted in the formation of a truncated protein product or nonsense mediated decay of the mRNA prior to protein production, it remains unclear whether this variant does indeed lead to SLC4A11 haploinsuffiency.9 In addition, only 1 of the 11 SLC4A11 mutations identified in individuals with FECD was shown to segregate with the affected phenotype in the family in which it was identified. Given the proposed dominant negative effect of the identified missense mutations, segregation should be demonstrable in an affected pedigree given that at least 50% of individuals with FECD have a family history.48 Additionally, the clinical features of FECD should be present in the parents and/or grandparents (over 50 years of age, the average age of onset of FECD) of individuals with CHED2, leading to an increased prevalence of FECD in the population and the need for corneal transplantation for symptomatic individuals.58 However, FECD is uncommonly encountered in countries where CHED2 is most prevalent, and is an uncommon indication for corneal transplantation in these countries. Of the 136 CHED2 probands in whom SLC4A11 screening has been reported, 119 were from India and 9 were from Saudi Arabia, with the remaining 8 from Belgium and the United States.(Table 1) Two large studies examining the indications for thousands of corneal transplants performed in both Northern and Southern India found that only approximately 1% of the corneal transplants were performed for FECD.59, 60 In Saudi Arabia, two large studies each looking at thousands of corneal transplants performed in the Kingdom found that only 0.6% to 2.2% are performed for FECD.61, 62 Thus, FECD is a very uncommon indication for corneal transplantation in countries where CHED2 is most common. This draws into question whether the heterozygous SLC4A11 mutations present in all parents of individuals with CHED2 are causative of FECD, as this would be expected to result in a higher prevalence of FECD in the population, and a greater number of affected individuals requiring corneal transplantation than is observed. If one explanation for this apparent discrepancy is that particular missense variants in SLC4A11 are pathogenic in the heterozygous state (and thus cause FECD) while other missense variants are pathogenic only in the compound heterozygous or homozygous state (and thus cause CHED2), the underlying mechanisms will need to be convincingly demonstrated before such an explanation will gain general acceptance.
ZEB1 (TCF8)
Given the identification of pathogenic ZEB1 frameshift mutations in PPCD, Mehta and colleagues screened ZEB1 in 74 FECD probands (8 familial and 66 sporadic cases). As only two coding region variants were identified, one of which was a synonymous substitution (family members were unavailable to determine segregation), the investigators concluded that ZEB1 does not play a significant role in FECD.11 However, two years later Riazuddin and colleagues reported five presumed causative ZEB1 missense mutations in 7 of 384 unrelated individuals with FECD.12 As 6 of the 7 variants were sporadic, segregation analysis was not possible. However, a missense mutation was identified in a family in which affected and unaffected individuals were available for screening. As the variant was identified in only 7/12 affected individuals, the investigators performed an abbreviated GWAS that failed to reveal evidence of a linkage until it was repeated with the assumption that all affected individuals who did not demonstrate the mutation possessed a mutation elsewhere in the genome.12 Using this conditional model, the investigators identified suggestive linkage to 9p. Genotyping of markers in the 9p locus identified 2 with a LOD score of > 3, and thus the authors designated this as the fourth locus for FECD (FCD4).12
TCF4 (E2-2)
In 2010, Baratz and colleagues published the results of a GWAS for FECD in which a significant association was noted between an intronic SNP (rs613872) in TCF4 and FECD in both the discovery and replication cohorts.4 Although haplotype analysis across the TCF4 locus revealed that 4 SNPs were independently associated with FECD, the significance of the SNPs to FECD remains unclear, as no TCF4 coding region variants were associated with FECD.4 E2-2, the protein encoded by TCF4, is a helix-loop-helix transcription factor that regulates cell growth and differentiation.4, 63 Like ZEB1, E2-2 is expressed in the corneal endothelium and represses E-cadherin expression, thus playing an essential role in EMT.4, 24 As E2-2 regulates ZEB1 expression, Baratz and colleagues hypothesized that the identified TCF4 variants may alter E2-2 function, thereby affecting ZEB1 expression, similar to the proposed effect of the aforementioned ZEB1 missense mutations.4
The association of the intronic SNP (rs613872) in TCF4 and FECD has been replicated by several other investigators in both association and linkage studies.57, 64–66 However, as the minor allele of rs613872 did not segregate with the affected status in three families previously mapped to the FCD2 locus, the investigators concluded that the genetic basis of FCD2 is independent of rs613872, which may only be tagging other more rare alleles in the FCD2 locus that are involved in the pathogenesis of FECD.57, 64 Recently, Wieben and colleagues described an expansion of a non-coding trinucleotide repeat in TCF4 that was strongly associated with FECD.67 Greater than 50 trinucleotide repeats was identified in 52 of 66 (79%) FECD cases and only 2 of 63 (3%) controls, providing a sensitivity and specificity of 79% and 96%, respectively. As the identified TGC repeat was more specific for FECD than the SNP rs613872, the investigators concluded that this suggests that FECD is a trinucleotide expansion disease associated with non-coding trinucleotide repeats, similar to Friedreich’s ataxia, myotonic dystrophy type 1 and fragile X syndrome.
Supplementary Material
Acknowledgments
Support provided by National Eye Institute grant 1R01 EY022082 and an unrestricted grant from Research to Prevent Blindness (AJA)
Footnotes
Conflict of interest statement: None of the authors has a proprietary or commercial interest in any of the materials discussed in this article.
References
- 1.Dorrepaal SJ, Cao KY, Slomovic AR. Indications for penetrating keratoplasty in a tertiary referral centre in Canada, 1996–2004. Can J Ophthalmol. 2007;42:244–50. [PubMed] [Google Scholar]
- 2.Ghosheh FR, Cremona FA, Rapuano CJ, et al. Trends in penetrating keratoplasty in the United States 1980–2005. Int Ophthalmol. 2008;28:147–53. doi: 10.1007/s10792-007-9177-z. [DOI] [PubMed] [Google Scholar]
- 3.Eye Bank Association of America. Eye Banking Statistical Report 2012 [Google Scholar]
- 4.Baratz KH, Tosakulwong N, Ryu E, et al. E2-2 protein and Fuchs’s corneal dystrophy. N Engl J Med. 2010;363:1016–24. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- 5.Heon E, Mathers WD, Alward WL, et al. Linkage of posterior polymorphous corneal dystrophy to 20q11. Hum Mol Genet. 1995;4:485–8. doi: 10.1093/hmg/4.3.485. [DOI] [PubMed] [Google Scholar]
- 6.Krachmer JH. Posterior polymorphous corneal dystrophy: a disease characterized by epithelial-like endothelial cells which influence management and prognosis. Trans Am Ophthalmol Soc. 1985;83:413–75. [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid E, Lisch W, Philipp W, et al. A new, X-linked endothelial corneal dystrophy. Am J Ophthalmol. 2006;141:478–87. doi: 10.1016/j.ajo.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Weiss JS, Moller HU, Lisch W, et al. The IC3D classification of the corneal dystrophies. Cornea. 2008;27 (Suppl 2):S1–83. doi: 10.1097/ICO.0b013e31817780fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vithana EN, Morgan PE, Ramprasad V, et al. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008;17:656–66. doi: 10.1093/hmg/ddm337. [DOI] [PubMed] [Google Scholar]
- 10.Riazuddin SA, Vithana EN, Seet LF, et al. Missense mutations in the sodium borate cotransporter SLC4A11 cause late-onset Fuchs corneal dystrophy. Hum Mutat. 2010;31:1261–8. doi: 10.1002/humu.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta JS, Vithana EN, Tan DT, et al. Analysis of the posterior polymorphous corneal dystrophy 3 gene, TCF8, in late-onset Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2008;49:184–8. doi: 10.1167/iovs.07-0847. [DOI] [PubMed] [Google Scholar]
- 12.Riazuddin SA, Zaghloul NA, Al-Saif A, et al. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010;86:45–53. doi: 10.1016/j.ajhg.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas S, Munier FL, Yardley J, et al. Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet. 2001;10:2415–23. doi: 10.1093/hmg/10.21.2415. [DOI] [PubMed] [Google Scholar]
- 14.Aldave AJ, Yellore VS, Yu F, et al. Posterior polymorphous corneal dystrophy is associated with TCF8 gene mutations and abdominal hernia. Am J Med Genet A. 2007;143A:2549–56. doi: 10.1002/ajmg.a.31978. [DOI] [PubMed] [Google Scholar]
- 15.Krafchak CM, Pawar H, Moroi SE, et al. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet. 2005;77:694–708. doi: 10.1086/497348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liskova P, Tuft SJ, Gwilliam R, et al. Novel mutations in the ZEB1 gene identified in Czech and British patients with posterior polymorphous corneal dystrophy. Hum Mutat. 2007;28:638. doi: 10.1002/humu.9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen DQ, Hosseini M, Billingsley G, et al. Clinical phenotype of posterior polymorphous corneal dystrophy in a family with a novel ZEB1 mutation. Acta Ophthalmol. 2010;88:695–9. doi: 10.1111/j.1755-3768.2009.01511.x. [DOI] [PubMed] [Google Scholar]
- 18.Vincent AL, Niederer RL, Richards A, et al. Phenotypic characterisation and ZEB1 mutational analysis in posterior polymorphous corneal dystrophy in a New Zealand population. Mol Vis. 2009;15:2544–53. [PMC free article] [PubMed] [Google Scholar]
- 19.Gwilliam R, Liskova P, Filipec M, et al. Posterior polymorphous corneal dystrophy in Czech families maps to chromosome 20 and excludes the VSX1 gene. Invest Ophthalmol Vis Sci. 2005;46:4480–4. doi: 10.1167/iovs.05-0269. [DOI] [PubMed] [Google Scholar]
- 20.Yellore VS, Papp JC, Sobel E, et al. Replication and refinement of linkage of posterior polymorphous corneal dystrophy to the posterior polymorphous corneal dystrophy 1 locus on chromosome 20. Genet Med. 2007;9:228–34. doi: 10.1097/gim.0b013e31803c4dc2. [DOI] [PubMed] [Google Scholar]
- 21.Shen AL, O’Leary KA, Dubielzig RR, et al. The PPCD1 mouse: characterization of a mouse model for posterior polymorphous corneal dystrophy and identification of a candidate gene. PLoS One. 2010;5:e12213. doi: 10.1371/journal.pone.0012213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi A, Fujiki K, Murakami A, et al. Analysis of COL8A2 gene mutation in Japanese patients with Fuchs’ endothelial dystrophy and posterior polymorphous dystrophy. Jpn J Ophthalmol. 2004;48:195–8. doi: 10.1007/s10384-003-0063-6. [DOI] [PubMed] [Google Scholar]
- 23.Yellore VS, Rayner SA, Emmert-Buck L, et al. No pathogenic mutations identified in the COL8A2 gene or four positional candidate genes in patients with posterior polymorphous corneal dystrophy. Invest Ophthalmol Vis Sci. 2005;46:1599–603. doi: 10.1167/iovs.04-1321. [DOI] [PubMed] [Google Scholar]
- 24.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–87. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JG, Kay EP. NF-kappaB is the transcription factor for FGF-2 that causes endothelial mesenchymal transformation in cornea. Invest Ophthalmol Vis Sci. 2012;53:1530–8. doi: 10.1167/iovs.11-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, El-Naggar S, Darling DS, et al. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–88. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabosova A, Azar DT, Bannikov GA, et al. Compositional differences between infant and adult human corneal basement membranes. Invest Ophthalmol Vis Sci. 2007;48:4989–99. doi: 10.1167/iovs.07-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yellore VS, Rayner SA, Nguyen CK, et al. Analysis of the role of ZEB1 in the pathogenesis of posterior polymorphous corneal dystrophy. Invest Ophthalmol Vis Sci. 2012;53:273–8. doi: 10.1167/iovs.11-8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Judisch GF, Maumenee IH. Clinical differentiation of recessive congenital hereditary endothelial dystrophy and dominant hereditary endothelial dystrophy. Am J Ophthalmol. 1978;85:606–12. doi: 10.1016/s0002-9394(14)77091-6. [DOI] [PubMed] [Google Scholar]
- 30.Maumenee AE. Congenital hereditary corneal dystrophy. Am J Ophthalmol. 1960;50:1114–24. doi: 10.1016/0002-9394(60)90998-3. [DOI] [PubMed] [Google Scholar]
- 31.Odland M. Dystrophia corneae parenchymatosa congenita. A clinical, morphological and histochemical examination. Acta Ophthalmol (Copenh) 1968;46:477–85. doi: 10.1111/j.1755-3768.1968.tb02832.x. [DOI] [PubMed] [Google Scholar]
- 32.Pearce WG, Tripathi RC, Morgan G. Congenital endothelial corneal dystrophy. Clinical, pathological, and genetic study. Br J Ophthalmol. 1969;53:577–91. doi: 10.1136/bjo.53.9.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirkness CM, McCartney A, Rice NS, et al. Congenital hereditary corneal oedema of Maumenee: its clinical features, management, and pathology. Br J Ophthalmol. 1987;71:130–44. doi: 10.1136/bjo.71.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toma NM, Ebenezer ND, Inglehearn CF, et al. Linkage of congenital hereditary endothelial dystrophy to chromosome 20. Hum Mol Genet. 1995;4:2395–8. doi: 10.1093/hmg/4.12.2395. [DOI] [PubMed] [Google Scholar]
- 35.Kanai A, Kaufman HE. Further electron microscopic study of hereditary corneal edema. Invest Ophthalmol. 1971;10:545–54. [PubMed] [Google Scholar]
- 36.Kanai A, Waltman S, Polack FM, Kaufman HE. Electron microscopic study of hereditary corneal edema. Invest Ophthalmol. 1971;10:89–99. [PubMed] [Google Scholar]
- 37.Levenson JE, Chandler JW, Kaufman HE. Affected asymptomatic relatives in congenital hereditary endothelial dystrophy. Am J Ophthalmol. 1973;76:967–71. doi: 10.1016/0002-9394(73)90090-1. [DOI] [PubMed] [Google Scholar]
- 38.Boruchoff SA, Kuwabara T. Electron microscopy of posterior polymorphous degeneration. Am J Ophthalmol. 1971;72:879–87. doi: 10.1016/0002-9394(71)91683-7. [DOI] [PubMed] [Google Scholar]
- 39.Cibis GW, Krachmer JA, Phelps CD, Weingeist TA. The clinical spectrum of posterior polymorphous dystrophy. Arch Ophthalmol. 1977;95:1529–37. doi: 10.1001/archopht.1977.04450090051002. [DOI] [PubMed] [Google Scholar]
- 40.Levy SG, Moss J, Noble BA, McCartney AC. Early-onset posterior polymorphous dystrophy. Arch Ophthalmol. 1996;114:1265–8. doi: 10.1001/archopht.1996.01100140465021. [DOI] [PubMed] [Google Scholar]
- 41.Callaghan M, Hand CK, Kennedy SM, et al. Homozygosity mapping and linkage analysis demonstrate that autosomal recessive congenital hereditary endothelial dystrophy (CHED) and autosomal dominant CHED are genetically distinct. Br J Ophthalmol. 1999;83:115–9. doi: 10.1136/bjo.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanis AB, Al-Rajhi AA, Taylor CM, et al. Exclusion of AR-CHED from the chromosome 20 region containing the PPMD and AD-CHED loci. Ophthalmic Genet. 1999;20:243–9. doi: 10.1076/opge.20.4.243.2273. [DOI] [PubMed] [Google Scholar]
- 43.Hand CK, Harmon DL, Kennedy SM, et al. Localization of the gene for autosomal recessive congenital hereditary endothelial dystrophy (CHED2) to chromosome 20 by homozygosity mapping. Genomics. 1999;61:1–4. doi: 10.1006/geno.1999.5920. [DOI] [PubMed] [Google Scholar]
- 44.Jiao X, Sultana A, Garg P, et al. Autosomal recessive corneal endothelial dystrophy (CHED2) is associated with mutations in SLC4A11. J Med Genet. 2007;44:64–8. doi: 10.1136/jmg.2006.044644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vithana EN, Morgan P, Sundaresan P, et al. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2) Nat Genet. 2006;38:755–7. doi: 10.1038/ng1824. [DOI] [PubMed] [Google Scholar]
- 46.Mohamed MD, McKibbin M, Jafri H, et al. A new pedigree with recessive mapping to CHED2 locus on 20p13. Br J Ophthalmol. 2001;85:758–9. doi: 10.1136/bjo.85.6.754d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenblum P, Stark WJ, Maumenee IH, et al. Hereditary Fuchs’ Dystrophy. Am J Ophthalmol. 1980;90:455–62. doi: 10.1016/s0002-9394(14)75011-1. [DOI] [PubMed] [Google Scholar]
- 48.Krachmer JH, Purcell JJ, Jr, Young CW, Bucher KD. Corneal endothelial dystrophy. A study of 64 families. Arch Ophthalmol. 1978;96:2036–9. doi: 10.1001/archopht.1978.03910060424004. [DOI] [PubMed] [Google Scholar]
- 49.Cross HE, Maumenee AE, Cantolino SJ. Inheritance of Fuchs’ endothelial dystrophy. Arch Ophthalmol. 1971;85:268–72. doi: 10.1001/archopht.1971.00990050270002. [DOI] [PubMed] [Google Scholar]
- 50.Meadows DN, Eghrari AO, Riazuddin SA, et al. Progression of Fuchs corneal dystrophy in a family linked to the FCD1 locus. Invest Ophthalmol Vis Sci. 2009;50:5662–6. doi: 10.1167/iovs.09-3568. [DOI] [PubMed] [Google Scholar]
- 51.Sundin OH, Jun AS, Broman KW, et al. Linkage of late-onset Fuchs corneal dystrophy to a novel locus at 13pTel-13q12. 13. Invest Ophthalmol Vis Sci. 2006;47:140–5. doi: 10.1167/iovs.05-0578. [DOI] [PubMed] [Google Scholar]
- 52.Sundin OH, Broman KW, Chang HH, et al. A common locus for late-onset Fuchs corneal dystrophy maps to 18q21.2–q21. 32. Invest Ophthalmol Vis Sci. 2006;47:3919–26. doi: 10.1167/iovs.05-1619. [DOI] [PubMed] [Google Scholar]
- 53.Riazuddin SA, Eghrari AO, Al-Saif A, et al. Linkage of a mild late-onset phenotype of Fuchs corneal dystrophy to a novel locus at 5q33.1–q35. 2. Invest Ophthalmol Vis Sci. 2009;50:5667–71. doi: 10.1167/iovs.09-3764. [DOI] [PubMed] [Google Scholar]
- 54.Kapoor R, Sakai LY, Funk S, et al. Type VIII collagen has a restricted distribution in specialized extracellular matrices. J Cell Biol. 1988;107:721–30. doi: 10.1083/jcb.107.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aldave AJ, Rayner SA, Salem AK, et al. No pathogenic mutations identified in the COL8A1 and COL8A2 genes in familial Fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2006;47:3787–90. doi: 10.1167/iovs.05-1635. [DOI] [PubMed] [Google Scholar]
- 56.Mok JW, Kim HS, Joo CK. Q455V mutation in COL8A2 is associated with Fuchs’ corneal dystrophy in Korean patients. Eye (Lond) 2009;23:895–903. doi: 10.1038/eye.2008.116. [DOI] [PubMed] [Google Scholar]
- 57.Li YJ, Minear MA, Rimmler J, et al. Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS One. 2011;6:e18044. doi: 10.1371/journal.pone.0018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gottsch JD, Sundin OH, Liu SH, et al. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2005;46:1934–9. doi: 10.1167/iovs.04-0937. [DOI] [PubMed] [Google Scholar]
- 59.Sony P, Sharma N, Sen S, Vajpayee RB. Indications of penetrating keratoplasty in northern India. Cornea. 2005;24:989–91. doi: 10.1097/01.ico.0000157406.34662.0f. [DOI] [PubMed] [Google Scholar]
- 60.Dandona L, Ragu K, Janarthanan M, et al. Indications for penetrating keratoplasty in India. Indian J Ophthalmol. 1997;45:163–8. [PubMed] [Google Scholar]
- 61.Al-Towerki AE, Gonnah el S, Al-Rajhi A, Wagoner MD. Changing indications for corneal transplantation at the King Khaled Eye Specialist Hospital (1983–2002) Cornea. 2004;23:584–8. doi: 10.1097/01.ico.0000121708.58571.5b. [DOI] [PubMed] [Google Scholar]
- 62.al Faran MF, Tabbara KF. Corneal dystrophies among patients undergoing keratoplasty in Saudi Arabia. Cornea. 1991;10:13–6. [PubMed] [Google Scholar]
- 63.Murre C, Bain G, van Dijk MA, et al. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–35. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 64.Riazuddin SA, McGlumphy EJ, Yeo WS, et al. Replication of the TCF4 intronic variant in late-onset Fuchs corneal dystrophy and evidence of independence from the FCD2 locus. Invest Ophthalmol Vis Sci. 2011;52:2825–9. doi: 10.1167/iovs.10-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Igo RP, Jr, Kopplin LJ, Joseph P, et al. Differing roles for TCF4 and COL8A2 in central corneal thickness and fuchs endothelial corneal dystrophy. PLoS One. 2012;7:e46742. doi: 10.1371/journal.pone.0046742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuot A, Hewitt AW, Griggs K, et al. Association of TCF4 and CLU polymorphisms with Fuchs’ endothelial dystrophy and implication of CLU and TGFBI proteins in the disease process. Eur J Hum Genet. 2012;20:632–8. doi: 10.1038/ejhg.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wieben ED, Aleff RA, Tosakulwong N, et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One. 2012;7:e49083. doi: 10.1371/journal.pone.0049083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hosseini SM, Herd S, Vincent AL, Heon E. Genetic analysis of chromosome 20-related posterior polymorphous corneal dystrophy: genetic heterogeneity and exclusion of three candidate genes. Mol Vis. 2008;14:71–80. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.