Abstract
Dithiobutylamine immobilized on a resin is a useful reagent for the reduction of disulfide bonds. Its ability to reduce a disulfide bond in a protein is enhanced greatly if used along with a soluble strained cyclic disulfide or mixed diselenide, small molecules that relays electrons from the resin to the protein. This relay catalysis system provides distinct advantages over the use of excess soluble reducing agent alone.
Keywords: heterogeneous catalysis, protein, redox chemistry, selenium, selenocystamine, sulfur
For the proper function of many proteins, sulfhydryl groups need to be maintained in a reduced state or disulfide bonds need to be maintained in an oxidized state.[1] In cellulo, this maintenance entails thiol–disulfide interchange reactions, often initiated by a membrane-associated protein and mediated by a soluble protein or peptide (e.g., glutathione).[2] In vitro, small-molecule thiols and disulfides, like those in Scheme 1, can accomplish this task.[3]
Scheme 1.
Disulfides (1–6) and diselenides (7–9) used in this work. Compounds 2, 5, 6, and 9 are racemic mixtures.
Recently, we reported on a novel disulfide-reducing agent, (2S)-2-amino-1,4-dimercaptobutane (dithiobutylamine or DTBA; reduced 1), derived from L-aspartic acid.[4] Like dithiothreitol (DTT; reduced 2), DTBA is a dithiol capable of adopting an unstrained ring upon oxidation.[5] A distinct and untapped attribute of DTBA is the ability of its amino group to act as a handle for facile conjugation. Small-molecule reducing agents typically need to be maintained at millimolar concentrations, and their removal diminishes process efficiency and economy. We reasoned that attaching DTBA to a solid support would enable its removal after disulfide reduction by either filtration or centrifugation.[6]
To test our hypothesis, we choose TentaGel resin as the solid support. This resin consists of hydrophilic poly(ethylene glycol) units grafted onto low-cross-linked polystyrene.[7] We found DTBA immobilized on TentaGel to be a potent disulfide-reducing agent with E°′ = (−0.316 ± 0.002) V (Figures S1 and S2 in the Supporting Information), a value similar to that of soluble DTBA.[4] Immobilized DTBA (10 equiv) was able to reduce cystamine (3) and oxidized β-mercaptoethanol (4) completely (Figures S3 and S4). Immobilized DTBA (10 equiv) was even able to reduce highly stable disulfides such as oxidized DTBA (1) and oxidized DTT (2) with yields of 76% and 68% respectively (Figures S5 and S6). After each procedure, the resin was easily isolated, regenerated, and reused without any observable loss in activity. The latter are not attributes of immobilized reducing agents derived from phosphines, which form recalcitrant phosphine oxides.
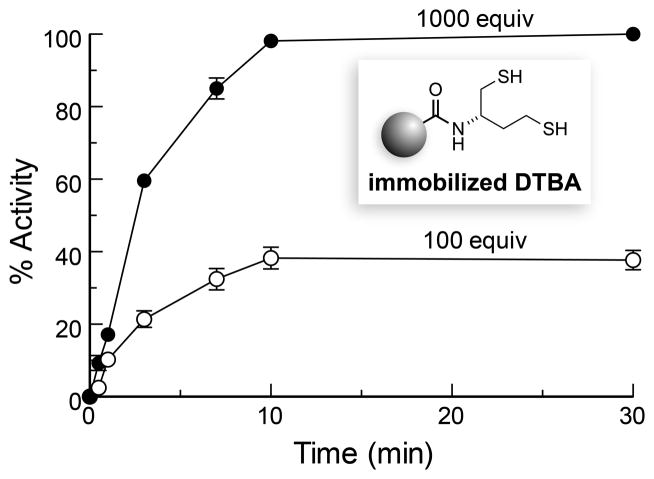
Next, we assessed the ability of immobilized DTBA to reduce a disulfide bond in a folded protein, which can be a challenging task.[8] As the target protein, we choose papain, a cysteine protease.[9] Upon treatment with S-methyl methanethiosulfonate, the active-site cysteine of papain (Cys25) forms a mixed disulfide that has no detectable enzymatic activity.[10] When we incubated the oxidized enzyme with 100 equiv of immobilized DTBA, we found that less than half of papain-Cys25–S–S–CH3 had been reduced after 30 min (Figure 1). Moreover, the rate of reduction for this heterogeneous reaction was slow, ~0.1% of that provided by typical solution-phase reagents,[4,11] and activation ceased after 10 min. When papain was treated with 1,000 equiv of immobilized DTBA, full generation of activity was observed within 10 min (Figure 1). These data indicate that the inefficiency is likely due to a diminished ability of the protein disulfide—in comparison to small-molecule disulfides—to access the sulfhydryl groups of immobilized DTBA.[8a,8c]
Figure 1.
Time-course for the reactivation of papain-Cys25–S–S–CH3 by immobilized DTBA (100 or 1000 equiv) in 0.10 M imidazole–HCl buffer, pH 7.0, containing EDTA (2 mM).
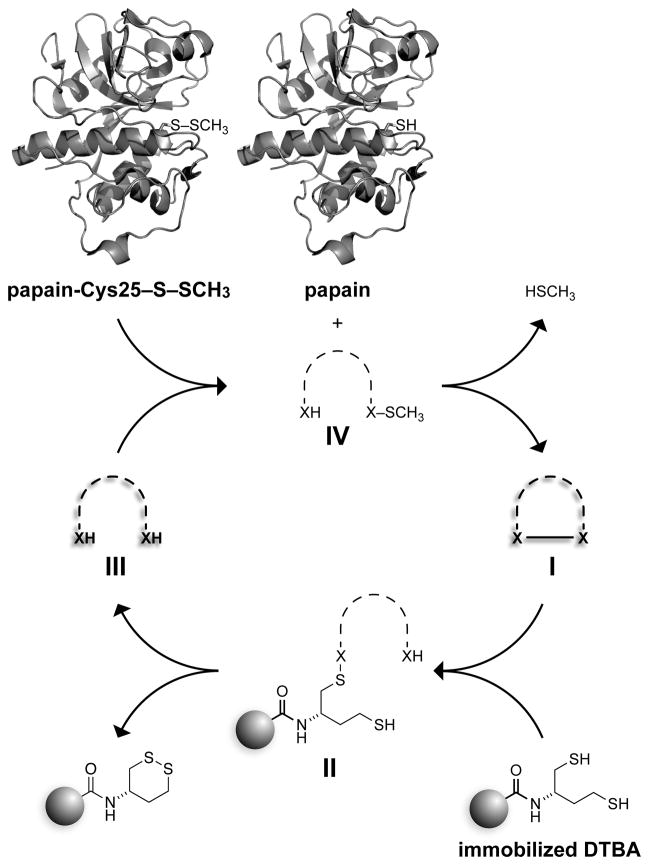
Taking inspiration from cellular thiol–disulfide interchange reactions,[2,12] we reasoned that the utility of immobilized DTBA could be enhanced by a soluble molecule that could “relay” electrons from the resin to the protein (Scheme 2).[13] To test this hypothesis, we incubated papain-Cys25–S–S–CH3 with 100 equiv of immobilized DTBA and 30 mol% of disulfides 1–4 (relative to oxidized protein). Unfortunately, we observed only a slight rate enhancement (Figure 2A).
Scheme 2.
Cycle for relay catalysis of disulfide-bond reduction by soluble thiols (III, X = S) or selenols (III, X = Se). Papain was depicted with the program PyMOL (Schrodinger, Portland, OR) using PDB entry 1ppn.[14]
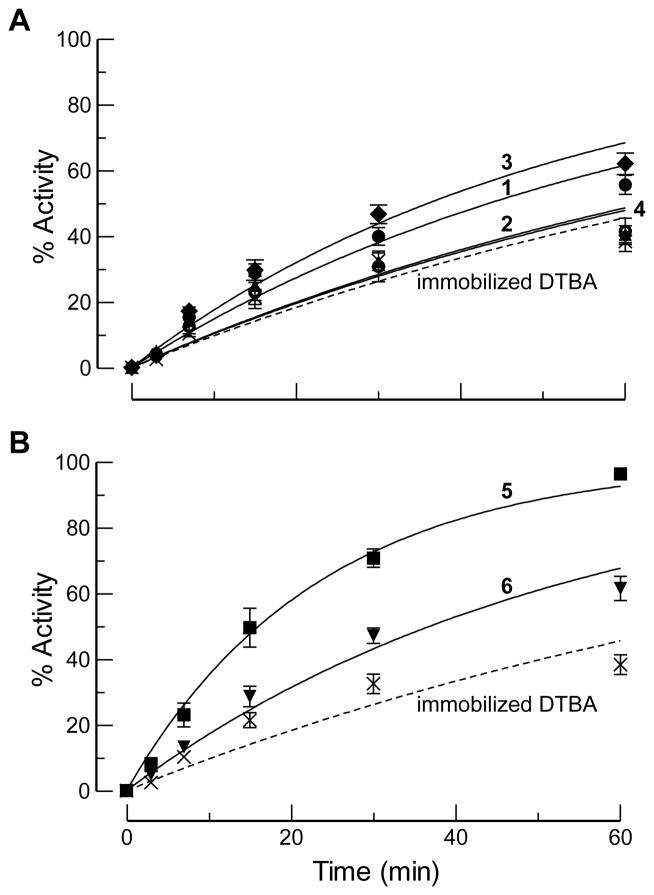
Figure 2.
Time-course for the reactivation of papain-Cys25–S–S–CH3 by immobilized DTBA (100 equiv) and a solution-phase disulfide catalyst (30 mol%). Reactions were performed in 0.10 M imidazole–HCl buffer, pH 7.0, containing EDTA (2 mM). (A) Unstrained disulfide catalysts. Cystamine (3): ; DTBAox (1): ; DTTox (2): ; βMEox (4): . (B) Strained disulfide catalysts. BMCox (5): ; lipoic acid (6): . Data for immobilized DTBA alone are shown in both panels.
Suspecting that the rate of the heterogeneous reaction between immobilized DTBA and unstrained disulfides 1–4 was slow (I→II in Scheme 2),[15] we turned to disulfides 5 and 6, believing that their incipient strain would accelerate the turnover of soluble catalyst. BMCox (5) is a 10-membered cyclic disulfide. Rings of this size suffer transannular strain.[5b,16a,16b] Similarly, cyclic five-membered disulfides (i.e., 1,2-dithiolanes), such as 6, place significant distortion on the preferred CSSC dihedral angle.[5b,17] Hence, the rate constant for the reaction between 1,3-propanedithiol and 1,2-dithiolane is ~650-fold greater than that for the homologated exchange reaction between 1,4-butanedithiol and 1,2-dithiane.[18]
Consistent with our expectations, we found that disulfide 5 provided a significant enhancement in the rate of papain-Cys25–S–S–CH3 reduction. Disulfide 6 was somewhat less effective, as its mixed disulfide (II in Scheme 2) has a greater tendency to partition back to the disulfide (I).[5b] Moreover, in the absence of immobilized DTBA, we found that the reduced form of DTBA regenerates activity faster than does the reduced form of BMC (Figure S8), affirming that the reduction of the soluble disulfide catalyst (I→III in Scheme 2) limits the rate of relay catalysis.
To improve catalytic efficiency further, we considered the use of selenium, which has physicochemical properties similar to those of sulfur. Yet, selenols manifest several desirable attributes as reducing agents in aqueous solution.[19] For example, selenols have pKa values that are typically 3 units lower than those of analogous thiols, significantly enhancing their nucleophilicity near neutral pH and their ability to act as a leaving group.[20] Diselenides also have E°′ values that are typically 0.15 V lower than those of analogous disulfides, making selenols more potent reducing agents. In addition, reactions with selenium as the electrophile can be 104-fold faster than those with sulfur as the electrophile, and might not require strain for efficient turnover. Indeed, there are numerous reports of small-molecule diselenides being used as catalysts for biochemical oxidation reactions.[21] Enzymes, such as thioredoxin reductase,[19h] are known to employ a selenol as a reducing agent. Yet, reported in vitro reduction reactions rarely employ small-molecule selenols, and never diselenols. A practical problem is the high reactivity of selenols with molecular oxygen. We recognized that this problem would be averted in our system, which would generate catalytic selenols in situ (Scheme 2). Because of the efficacy of disulfide 5 (Figure 2B), we were motivated to investigate its seleno congener. Accordingly, we synthesized selenoBMCox (9) as well as selenoDTBAox (7), and we obtained selenocystamine (8), which is available commercially and has demonstrated marked success in mediating thiol–disulfide interchange reactions.[20,21a,21b]
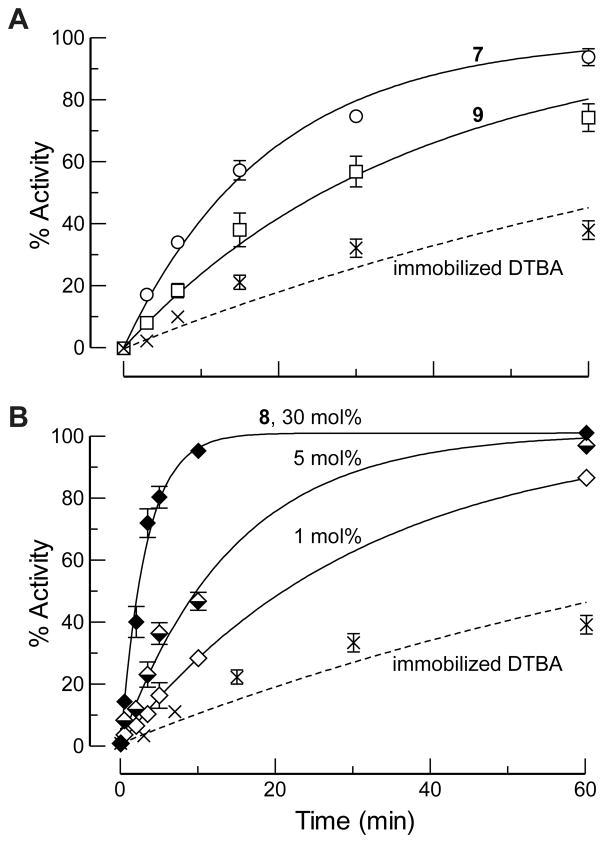
We found that diselenide 7 is superior to its congener 1, and that diselenide 9 performs comparably to its congener 5 (Figure 3A). These two cyclic diselenides were, however, worse catalysts than was acyclic diselenide 8 (Figure 3B). This finding is attributable to the selenylsulfide (II in Scheme 2) generated by the reaction of 7 and 9 (but not 8) with immobilized DTBA tending to partition back to the diselenide (I) rather than to the diselenol (III) needed for catalysis.[18,22] Notably, diselenide 8 led to significant rate enhancements even at low loadings of catalyst.
Figure 3.
Time-course for the reactivation of papain-Cys25–S–S–CH3 by immobilized DTBA (100 equiv) and a solution-phase diselenide catalyst. Reactions were performed in 0.10 M imidazole–HCl buffer, pH 7.0, containing EDTA (2 mM). (A) Cyclic diselenide catalysts (30 mol%). SelenoDTBAox (7): ; selenoBMCox (9): . (B) Selenocystamine (8) as a catalyst. 30 mol%: ; 5 mol%: ; 1 mol%: . Data for immobilized DTBA alone are shown in both panels.
In summary, we have established that the amino group of DTBA allows for its facile conjugation to a resin. This supported reagent was effective at reducing disulfide bonds in small molecules. Unlike soluble reducing agents, immobilized DTBA was easy to recover and reuse. We also demonstrated that the rate of reducing a disulfide bond in a protein can be enhanced markedly when the reduced resin is used in conjunction with a “relay”. In this biomimetic strategy, the resin acts as a repository of electrons that are relayed to a macromolecule via a small-molecule catalyst. The optimal catalysts are strained cyclic disulfides and acyclic diselenides, both of which react with excess immobilized DTBA to form a covalent intermediate that partitions towards reduced catalyst and oxidized resin. Finally, we note that a vast excess of soluble reducing agent is typically used to preserve proteins in a reduced state.[23] Instead, maintenance could require a minute (e.g., sub-micromolar) amount of a soluble catalyst along with immobilized DTBA. We anticipate that the low level of soluble reducing agent would be advantageous in common bioconjugation reactions entailing the S-alkylation of cysteine residues,[24] as well as in many other experimental procedures.
Experimental Section
See the Supporting Information for experimental details.
Supplementary Material
Footnotes
We are grateful to Prof. H. J. Reich for contributive discussions. B.V. was supported by postdoctoral fellowship 289613 (CIHR). This work was supported by grant R01 GM044783 (NIH). This work made use of the National Magnetic Resonance Facility at Madison, which is supported by grants P41 RR002301 and P41 GM066326 (NIH), and the Biophysics Instrumentation Facility, which was established with grants BIR-9512577 (NSF) and S10 RR13790 (NIH).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
John C. Lukesh, III, Department of Chemistry, 1101 University Avenue, University of Wisconsin–Madison, Madison, WI 53706, USA.
Dr. Brett VanVeller, Department of Chemistry, 1101 University Avenue, University of Wisconsin–Madison, Madison, WI 53706, USA
Prof. Ronald T. Raines, Email: rtraines@wisc.edu, Department of Chemistry, 1101 University Avenue, University of Wisconsin–Madison, Madison, WI 53706, USA, Fax: (+1) 1-608-890-2583, Homepage: http://www.biochem.wisc.edu/faculty/raines/lab. Department of Biochemistry, 433 Babcock Drive, University of Wisconsin–Madison, Madison, WI 53706, USA
References
- 1.Buchner J, Moroder L, editors. Oxidative Folding of Peptides and Proteins. The Royal Society of Chemistry; Cambridge, UK: 2009. [Google Scholar]
- 2.a) Kadokura H, Katzen F, Beckwith J. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]; b) Tu BP, Weissman JS. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Depuydt M, Messens J, Collet JF. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]; d) Braakman I, Bulleid NJ. Annu Rev Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- 3.a) Kersteen EA, Raines RT. Antioxid Redox Signal. 2003;5:413–424. doi: 10.1089/152308603768295159. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lees WJ. Curr Opin Chem Biol. 2008;12:740–745. doi: 10.1016/j.cbpa.2008.08.032. [DOI] [PubMed] [Google Scholar]; c) Yamaguchi S, Yamamoto E, Mannen T, Nagamune T. Biotechnol J. 2013;8:17–31. doi: 10.1002/biot.201200025. [DOI] [PubMed] [Google Scholar]
- 4.Lukesh JC, III, Palte MJ, Raines RT. J Am Chem Soc. 2012;134:4057–4059. doi: 10.1021/ja211931f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Houk J, Whitesides GM. J Am Chem Soc. 1987;109:6825–6836. [Google Scholar]; b) Burns JA, Whitesides GM. J Am Chem Soc. 1990;112:6296–6303. [Google Scholar]; c) Lees WJ, Whitesides GH. J Org Chem. 1993;58:642–647. [Google Scholar]
- 6.Leznoff CC. Acc Chem Res. 1977;11:327–333. [Google Scholar]
- 7.a) Rapp W, Zhang L, Bayer E. In: Innovations and Perspectives in Solid-Phase Synthesis. Epton R, editor. Mayflower; Birmingham: 1990. pp. 205–210. [Google Scholar]; b) Quarrell R, Claridge TDW, Weaver GW, Lowe G. Molec Divers. 1995;1:223–232. doi: 10.1007/BF01715526. [DOI] [PubMed] [Google Scholar]
- 8.a) Scouten WH, Firestone GL. Biochim Biophys Acta. 1976;453:277–284. doi: 10.1016/0005-2795(76)90274-9. [DOI] [PubMed] [Google Scholar]; b) Amos RA, Fawcett SM. J Org Chem. 1984;49:2637–2639. [Google Scholar]; c) Woycechowsky KJ, Hook BA, Raines RT. Biotechnol Progr. 2003;19:1307–1314. doi: 10.1021/bp0257123. [DOI] [PubMed] [Google Scholar]; d) Bienvenu C, Greiner J, Vierling P, Di Giorgio C. Tetrahedron Lett. 2010;51:3309–3311. [Google Scholar]; e) Miralles G, Verdie P, Puget K, Maurras A, Martinez J, Subra G. ACS Comb Sci. 2013;15:169–173. doi: 10.1021/co300104k. [DOI] [PubMed] [Google Scholar]
- 9.a) Schechter I, Berger A. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]; b) Pickersgill RW, Harris GW, Garman E. Acta Crystallogr, Sect B. 1992;48:59–67. doi: 10.1107/s0108768191006663. [DOI] [PubMed] [Google Scholar]
- 10.Kenyon GL. Biochemistry. 1975;14:766–771. doi: 10.1021/bi00675a019. [DOI] [PubMed] [Google Scholar]
- 11.a) Singh R, Whitesides GM. J Org Chem. 1991;56:2332–2337. [Google Scholar]; b) Lees WJ, Singh R, Whitesides GM. J Org Chem. 1991;56:7328–7331. [Google Scholar]
- 12.a) Hwang C, Sinskey AJ, Lodish HF. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]; b) Ohtsu I, Wiriyathanawudhiwong N, Morigasaki S, Nakatani T, Kadokura H, Takagi H. J Biol Chem. 2010;285:17479–17487. doi: 10.1074/jbc.M109.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Van Laer K, Hamilton CJ, Messens J. Antioxid Redox Signal. 2013;18:1642–1653. doi: 10.1089/ars.2012.4964. [DOI] [PubMed] [Google Scholar]
- 13.For an electron “relay” through proteins, see: Wang M, Gao J, Müller P, Giese B. Angew Chem Int Ed. 2009;48:4232–4234. doi: 10.1002/anie.200900827.
- 14.Harris GW, Pickersgill RW, Howlin B, Moss DS. Acta Crystallogr B. 1992;48(Pt 1):67–75. doi: 10.1107/s0108768191006663. [DOI] [PubMed] [Google Scholar]
- 15.a) Bachrach SM, Walker CJ, Lee F, Royce S. J Org Chem. 2007;72:5174–5182. doi: 10.1021/jo070578s. [DOI] [PubMed] [Google Scholar]; b) Baldus IB, Grater F. Biophys J. 2012;102:622–629. doi: 10.1016/j.bpj.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Brown HC, Borkowski M. J Am Chem Soc. 1952;74:1894–1902. [Google Scholar]; b) Woycechowsky KJ, Wittrup KD, Raines RT. Chem Biol. 1999;6:871–879. doi: 10.1016/s1074-5521(00)80006-x. [DOI] [PubMed] [Google Scholar]
- 17.Fava A, Iliceto A, Camera E. J Am Chem Soc. 1957;79:833–838. [Google Scholar]
- 18.Singh R, Whitesides GM. J Am Chem Soc. 1990;112:6304–6309. [Google Scholar]
- 19.a) Gunther WHH, Salzman MN. Ann NY Acad Sci. 1972;192:25–43. doi: 10.1111/j.1749-6632.1972.tb52573.x. [DOI] [PubMed] [Google Scholar]; b) Mugesh G, du Mont WW, Sies H. Chem Rev. 2001;101:2125–2179. doi: 10.1021/cr000426w. [DOI] [PubMed] [Google Scholar]; c) Jacob C, Giles GI, Giles NM, Sies H. Angew Chem Int Ed. 2003;42:4742–4758. doi: 10.1002/anie.200300573. [DOI] [PubMed] [Google Scholar]; d) Shchedrina VA, Novoselov SV, Malinouski MY, Gladyshev VN. Proc Natl Acad Sci USA. 2007;104:13919–13924. doi: 10.1073/pnas.0703448104. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Arner ESJ. Exp Cell Res. 2010;316:1296–1303. doi: 10.1016/j.yexcr.2010.02.032. [DOI] [PubMed] [Google Scholar]; f) Ruggles EL, Snider GW, Hondal RJ. In: Selenium: Its Molecular Biology and Role in Human Health. 3. Hatfield DL, Berry MJ, Gladyshev VN, editors. Springer; New York, NY: 2011. pp. 73–84. [Google Scholar]; g) Hondal RJ, Ruggles EL. Amino Acids. 2011;41:73–89. doi: 10.1007/s00726-010-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Metanis N, Beld J, Hilvert D. Patai’s Chemistry of Functional Groups. John Wiley & Sons; 2011. [DOI] [Google Scholar]; i) Nauser T, Steinmann D, Koppenol WH. Amino Acids. 2012;42:39–44. doi: 10.1007/s00726-010-0602-7. [DOI] [PubMed] [Google Scholar]; j) Hondal RJ, Marino SM, Gladyshev VN. Antioxid Redox Signal. 2013;18:1675–1689. doi: 10.1089/ars.2012.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Singh R, Whitesides GM. J Org Chem. 1991;56:6931–6933. [Google Scholar]; b) Singh R, Kats L. Anal Biochem. 1995;232:86–91. doi: 10.1006/abio.1995.9956. [DOI] [PubMed] [Google Scholar]; c) Steinmann D, Nauser T, Koppenol WH. J Org Chem. 2010;75:6696–6699. doi: 10.1021/jo1011569. [DOI] [PubMed] [Google Scholar]
- 21.a) Chen Y, Maret W. Eur J Biochem. 2001;268:3346–3353. doi: 10.1046/j.1432-1327.2001.02250.x. [DOI] [PubMed] [Google Scholar]; b) Singh R, Maloney EK. Anal Biochem. 2002;304:147–156. doi: 10.1006/abio.2002.5624. [DOI] [PubMed] [Google Scholar]; c) Beld J, Woycechowsky KJ, Hilvert D. Biochemistry. 2008;47:6985–6987. doi: 10.1021/bi8008906. [DOI] [PubMed] [Google Scholar]; d) Walewska A, Zhang MM, Skalicky JJ, Yoshikami D, Olivera BM, Bulaj G. Angew Chem Int Ed. 2009;48:2221–2224. doi: 10.1002/anie.200806085. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Beld J, Woycechowsky KJ, Hilvert D. Biochemistry. 2009;48:4662–4662. [Google Scholar]; f) Beld J, Woycechowsky KJ, Hilvert D. J Biotechnol. 2010;150:481–489. doi: 10.1016/j.jbiotec.2010.09.956. [DOI] [PubMed] [Google Scholar]; g) Steiner AM, Woycechowsky KJ, Olivera BM, Bulaj G. Angew Chem Int Ed. 2012;51:5580–5584. doi: 10.1002/anie.201200062. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Metanis N, Hilvert D. Angew Chem Int Ed. 2012;51:5585–5588. doi: 10.1002/anie.201109129. [DOI] [PubMed] [Google Scholar]
- 22.Iwaoka M, Takahashi T, Tomoda S. Heteroat Chem. 2001;12:293–299. [Google Scholar]
- 23.a) Shafer DE, Inman JK, Lees A. Anal Biochem. 2000;282:161–164. doi: 10.1006/abio.2000.4609. [DOI] [PubMed] [Google Scholar]; b) Walker JM, editor. The Protein Protocols Handbook. Humana Press; Totowa, New Jersey: 2002. [Google Scholar]
- 24.Hermanson GT. Bioconjugate Techniques. 3. Academic Press; New York, NY: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.