Abstract
Purpose: Carotenoids are of great interest in many scientific disciplines because of their wide distribution, diverse functions and interesting properties. The present report describes a new natural source for carotenoid production.
Methods: Halorubrum sp., TBZ126, an extremely halophilic archaeon, was isolated from Urmia Lack following culture of water sample on marine agar medium and incubation at 30 °C. Then single colonies were cultivated in broth media. After that the cells were collected and carotenoids were extracted with acetone-methanol (7:3 v/v). The identification of carotenoids was performed by UV-VIS spectroscopy and confirmed by thin layer chromatography (TLC) in the presence of antimony pentachloride (SbCl5). The production profile was analyzed using liquid-chromatography mass spectroscopy (LC-MS) techniques. Phenotypic characteristics of the isolate were carried out and the 16S rRNA gene was amplified using polymerase chain reaction (PCR).
Results: LC-MS analytical results revealed that produced carotenoids are bacterioruberin, lycopene and β-carotene. Bacterioruberin was found to be the predominant produced carotenoid. 16S rRNA analysis showed that TBZ126 has 100% similarity with Halorubrum chaoviator Halo-G*T (AM048786).
Conclusion: Halorubrum sp. TBZ126, isolated from Urmia Lake has high capacity in the production of carotenoids. This extremely halophilic archaeon could be considered as a prokaryotic candidate for carotenoid production source for future studies.
Keywords: Carotenoids, Halorubrum chaoviator, Bacterioruberin, lycopene, β-carotene
Introduction
Microorganisms are a great source of diverse natural products that are candidates for drug development, food and feed additives, and other industrial products.1 Carotenoid pigments are one of these natural products responsible for the yellow, orange, red, and purple colors in a wide variety of plants, animals, and microorganisms.2,3 These compounds are a group of more than 700 naturally occurring pigments. Animals are incapable of producing carotenoids and must obtain them from their diet. Lycopene, α-carotene, ß-carotene, lutein, zeaxanthin, and ß-cryptoxanthin are the most abundant in human plasma.4
Carotenoids have wide applications as colorants, feed additives, antioxidants, anti-tumor and heart disease prevention agents, precursors of vitamin A and enhancers of in vitro antibody production. Hence, they are widely applied in the food, medical, pharmaceutical, and cosmetic industries as dyes and functional ingredients.2,3 ß-carotene, astaxanthin, lutein, canthaxanthin, and lycopene are important examples.5 Functions of the carotenoids are based on their molecular structures including molecular size, solubility, effective number of conjugated double bonds, and presence/absence of functional groups or cyclic ends.6
Chemical synthesis is one of the production methods for carotenoids. However, the consumer preference for natural products, as well as high costs, presence of by-products and damaging effects on the environment have together intensified efforts to identify alternative sources for chemical method. Accordingly different species of bacteria, molds, yeasts and algae have attracted a great interest as alternative biosources.7 The main reason for the interest in using microorganisms is the simplicity in increasing the production by environmental and genetic manipulation.8 In addition, the production of natural colorants through fermentation has a number of advantages, such as cheaper production, higher yields, possibly easier extraction, less batch-to-batch variations and no seasonal variations. The production is flexible and can easily be controlled. Furthermore, the collection of microbial organisms is sustainable and has no negative impact on the environment.9
Halophilic archaea within the phylum Euryarchaeota are extreme halophiles and (mostly) aerobic, generally red-pigmented. The high-salt tolerance of haloarchaea enables their cultivation under non-sterile and thus cost-reducing conditions.10,11 On the other hand, the process to obtain the carotenoids is simple because in lower NaCl concentrations cell lysis is induced12 and consequently extraction could be conducted directly from the cells without any mechanical operation which is required in case of plants. Therefore, the study on production of carotenoids from halophilic haloarchaea could be considered as an alternative commercial source for carotenoids. Carotenoid pigments are particularly prominent in hypersaline ecosystems. Red and orangish color of hypersaline habitats is due to the presence of pigmented microorganisms, including Dunaliella , rich in ß-carotene, Haloarchaea whose main production is bacterioruberin, and halophilic bacteria, such as Salinibacter ruber producing a carotenoid called salinixanthin.12,13
The most members of the family Halobacteriaceae including halophilic archaea have a high content of bacterioruberin, a 50-carbon open chain carotenoid. Other minor carotenoid compounds have been identified at low concentrations in halophilic archaea: lycopersene, cis- and trans-phytoene, cis- and transphytofluene, neo-β-carotene and neo-α-carotene. The low concentrations of these compounds suggest that they may be used as precursors for the synthesis of other carotenoids including lycopene, retinal and the members of the bacterioruberin group. Some species may also produce the ketocarotenoid canthaxanthin in addition to other carotenoids. Raman spectroscopy identified bacterioruberin as the major carotenoid in the halophilic archaea Halobacterium salinarum strains NRC-1 and R1, Haloarcula sodomense, and Halorubrum vallismortis.13
Following our previous report describing isolation and characterization of halophilic bacteria from Urmia Lake in northwest of Iran,14 herein, we report isolation and characterization of a red haloarchaeon producing various carotenoids.
Materials and Methods
Isolation and growth conditions
Specimens were taken from water and soil of Urmia Lake, were delivered to the laboratory in sterilized containers and were cultured immediately on marine agar with various NaCl concentrations. Marine agar medium contained (per liter); MgCl2.7H2O, 8.8 g; Na2SO4, 3.24 g; CaCl2, 1.8 g; KCl, 0.55 g; NaHCO3, 0.16 g; KBr, 0.08 g; SrCl2, 34.0 mg; H3BO3, 22.0 mg; Na2O3Si, 4.0 mg; NaF, 2.4 mg; (NH4)(NO3),1.6 mg; Na2HPO4, 8.0 mg; peptone 5 g; yeast extract, 1 g; agar, 15.0 g and various NaCl concentrations (0-25% w/v). The pH of medium was adjusted at 7.5 before autoclaving. For this purpose, 400 μl of water samples were inoculated on the medium and incubated at 30 °C. Colony growth was first observed after about 7 days. To obtain pure cultures, single red colonies were picked from the plates and were used for Gram-staining and stock preparation, by growing in marine broth in an orbital shaker (Shaking Incubator VS-8480, Korea). Marine broth contained (per liter); MgCl2.7H2O 5.9 g; MgSO4, 3.24 g; CaCl2, 1.8 g; KCl, 0.55 g; NaHCO3, 0.16 g; KBr, 0.08 g; SrCl2, 34.0 mg; H3BO3, 22.0 mg; Na2O3Si, 4.0 mg; NaF, 2.4 mg; (NH4)(NO3), 1.6 mg; Na2HPO4, 8.0 mg; peptone 5 g and yeast extract 1 g and various NaCl concentrations (0-25% w/v). The pH of culture medium was adjusted at 7.5 before autoclaving. Axenic cultures were stored at −70◦C in marine broth supplemented with 30% glycerol.
Extraction of genomic DNA and sequencing of 16S rRNA
DNA was extracted using Corbin “Genomic DNA isolation” protocol with some modification.15 Briefly, cells were lysed by freeze-thaw cycles in liquid nitrogen and then suspended in solution I [Tris 10 mM (pH 7.4), EDTA 1 mM, sodium dodecyl sulphate (SDS) 0.5%, proteinase K 0.1 mg/ml] and lysed by incubation at 37 °C for 1 h. Then, the solution II [0.8 M NaCl and 1% CTAB] was added to the lysates and incubated at 65°C for 20 min and finally genomic DNA was extracted with equal volume of chloroform-isoamylalcohol (24:1 v/v). Nucleic acids were precipitated from the aqueous phase with 0.6 volume of isopropanol. The 16S rRNA gene was amplified using polymerase chain reaction (PCR) using 20F (5´-TCCGGTTGATCCTGCCG-3´)16 and 1530R (5´-AGGAGGTGATCCAACCGCA-3´)17 primers. PCR was performed using a thermal cycler (eppendorf) with a 50-μl reaction containing 1.5 μl MgCl2, 1 μl of each dNTP, 0.5 μl of each primer, 5 μl PCR buffer, 36.5 μl H2O and 1 U of Taq DNA polymerase (Cinnagen, Iran). Initial denaturation was carried out for 3 min at 95 °C. It was followed by 35 cycles of denaturation at 94 °C for 30 sec, annealing at 45 °C for 40 sec and extension at 72 °C for 1.5 min with a further 10 min extension at 72 °C. The amplified DNA fragment was separated using 1% agarose gel electrophoresis, then the DNA fragment was extracted from gel and sequenced by Faza Biotechnology Co. The sequence was compared with reference 16S rRNA gene sequences available in NCBI GenBank database BLAST using blastn and megablast softwares and EzTaxon-e server (http://eztaxon-e.ezbiocloud.net).18
Physiological characterization of the isolate
Gram-staining was performed as described by Gerhardt et al.19 and confirmed according to Dussault.20 To investigate basic physiological characteristics of the isolate, Mac Faddin21 and Barrow et al.22 methods were used for the following tests: oxidase and catalase reactions, phenylalanine deaminase, nitrate reduction, hydrolysis of urea, gelatin, starch, Tween 80 and tyrsosin, H2S and indol production from L-cysteine and tryptophan, respectively. To examine nitrate reduction, 0.2% (w/v) KNO3 was added to the liquid media. Gelatin and starch hydrolysis were tested by flooding cultures on solid media containing 1% (w/v) gelatin and starch, respectively. Hydrolysis of Tween 80 was tested on solid media supplemented with 1% (w/v) Tween 80. Tyrosine hydrolysis was evaluated by appearance of a clear zone on marine agar medium culture containing 5 g/l tyrosine. H2S production was tested in liquid media supplemented with 0.01% (w/v) of L-cysteine. The indicator used in this experiment was a band of paper impregnated with lead-acetate placed in the neck of the tube.
Pigments extraction and analysis
To extract carotenoids, 100 ml of marine broth cultures were centrifuged at 8,000 rpm for 10 min at 4 °C. The supernatant was separated and a mixture of acetone-methanol (7:3 v/v)23 containing butylhydroxytoluene (BHT) (0.1%; as antioxidant) was added to the pellet. The pelleted cells were then frozen and thawed using liquid nitrogen to facilitate extraction and followed by centrifugation at 10,000 × g for 10 min at 4 °C. Successive extractions carried out until both solvent and cells were colorless. The solvent was evaporated under a stream of nitrogen and the pigments were dissolved in 10 ml of acetone (containing 0.1% BHT). Samples were wrapped with aluminum foil to protect them from light. The extracts were stored under nitrogen at -70 °C. Extraction procedures and analysis tests were conducted in dark conditions.
UV-Visible Spectroscopy
Extraction solution UV spectra were recorded at 200-700 nm using a spectrophotometer (Shimadzu UV-1800 Series, Kyoto, Japan). The approximate content of total carotenoids was determined by measuring the optical density of the sample in 495 (λmax of our extraction solution). The total amount of carotenoids was calculated according to Davies.23
Thin-Layer Chromatography
In order to confirm carotenoid pigments contained in the extract, thin-layer chromatography (TLC) was used. For this analysis, the acetone extract was placed on a TLC silica gel GF254 plate (Merck, Darmstadt, Germany) and developed in hexane:acetone (7:3). After development, the individual spots were identified by visibility and spraying with a saturated solution of antimony pentachloride (SbCl5) in chloroform (1:10 v/v).
Liquid chromatography–mass spectrometry (LC-MS)
The extraction solution was centrifuged, and the supernatant was filtered through cellulose acetate filters (25 mm, 0.45 mm; VWR International). Samples were handled on ice and wrapped with aluminum foil to reduce isomerisation and oxidation of carotenoids by light irradiation. Chromatographic separation was performed on an Agilent 1200 series HPLC system including a quaternary pump and a degasser equipped with a G1315B Diode Array Detector. The accompanying Agilent LC Chemstation was employed for instrument control, data acquisition and processing. HPLC analysis was performed using Eurosphere RP-column (100-5 C18 column, 300×4.6 mm Knauer, Germany) by isocratic elution with a flow rate of 0.8 ml/ min. The mobile phase was acetonitrile-dichloromethane-methanol (70:20:10 v/v/v), 20 mM ammonium acetate and 0.1% triethylamine. The temperature was maintained at 20 °C and UV detection was performed at 450 nm. Both the extracts and standards were injected (injection volume: 20 µL) into the reverse phase column and identifications of all trans-isomers were carried out using comparison of retention times and UV spectra of the extracts with standard mixture. The experiment was conducted in triplicate. The mass spectra were recorded in the positive ion mode in the mass range from 300 to 2000 m/z. The mass spectrometer parameters were set as follows: Nebulizer pressure was 40 psi, drying gas flow was 20 liter/min and gas temperature was 250 °C. The capillary voltage was 5000 V. Ions were monitored in the scan mode. The identification of carotenoids were performed by comparing retention time, UV spectra and characteristics of the mass spectra (protonated molecule ([M+H]+) and its MS/MS fragments. All of the carotenoids were monitored at 450 nm with a UV-visible detector.
Results
Morphologic and genotypic characteristics
Cells were Gram-negative and rod-shaped. Strain TBZ126 formed circular and red colonies on marine agar. Strain TBZ126 didn’t grow in the presence of NaCl and could tolerate NaCl up to 30% at 30 °C. It did not grow in acidic medium but tolerated alkaline medium (pH=10). 16S rRNA gene sequence analysis revealed that strain TBZ126 belongs to Halorubrum genus and has 100% similarity with Halorubrum chaoviator Halo-GT.24
Physiological and biochemical characterization
Catalase and oxidase tests were positive and negative, respectively. Nitrate was not reduced to nitrite and indole was produced. Hydrolysis of gelatin and starch were found to be positive whereas Tween 80 was not hydrolysed. Urease activity was found to be positive. Phenylalanine deaminase was negative. H2S formation was positive. Results of physiological and biochemical characteristics of Halorubrum sp. TBZ126 are summarized in Table 1.
Table 1. Physiological and biochemical characteristics of Halorubrum sp. TBZ126. +: positive reaction; -: negative reaction.
| Characteristic | TBZ126 |
| Gram-staining | - |
| Cell shape | Rod |
| Colony | Circle- red |
| Growth in the absence of NaCl | - |
| Catalase | + |
| Oxidase | - |
| Tyrosin Hydrolysis | - |
| Nitrate reduction | - |
| Gelatin Hydrolysis | + |
| Tween 80 Hydrolysis | - |
| Starch Hydrolysis | + |
| Urease | + |
| Indol | + |
| H2S | + |
| Phenylalanine deaminase | - |
UV-VIS spectrum of carotenoids and total carotenoid content
Pigments solution in acetone mixture showed characteristic absorptions of carotenoids (Figure 1).25,26 Britton described that bacterioruberin and its derivatives exhibited the characteristic spectral peaks of red carotenoids at nearly identical absorption maxima at 467, 493, and 527 nm for three fingered peaks and at 370 and 385 nm for two cis peaks.27 As seen in Figure 1, the pigments in the extract solution showed absorption peaks at 469 nm, 494 nm and 526 nm, indicating bacterioruberin is the main component in the extracted sample. The total carotenoid content of Halorubrum sp. TBZ126 was found to be 11280 µg/l.
Figure 1 .
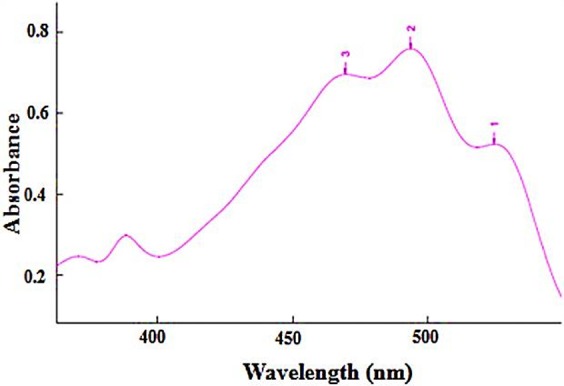
UV-VIS spectrum of acetone extract from Halorubrum sp. TBZ126. The extract corresponds to 100 ml of culture in marine broth medium and 1 ml of the extract was diluted to 10 ml with acetone to give a reading in the spectrophotometer between 0.5 and 0.8 at the wavelength of the middle main absorption maximum of the extract (495 nm). The pigments in the extract solution showed absorption peaks at 469 nm, 494 nm and 526 nm.
Thin-Layer Chromatography
After development of TLC test, four spots were visible. Staining the plate with antimony pentachloride (SbCl5) in chloroform showed only one blue spot23 confirming the presence of carotenoids in the extraction solution.
Liquid chromatography–mass spectrometry (LC-MS)
HPLC analysis of TBZ126 carotenoids revealed 5 distinctive peaks (Figure 2). Carotenoid production is composed of bacterioruberin (peak 1), all-trans-lycopene (peak 2), 13-cis-lycopene (peak 3), all-trans-β-carotenes (peak 4), and all-cis-β-carotene (peak 5).
Figure 2 .
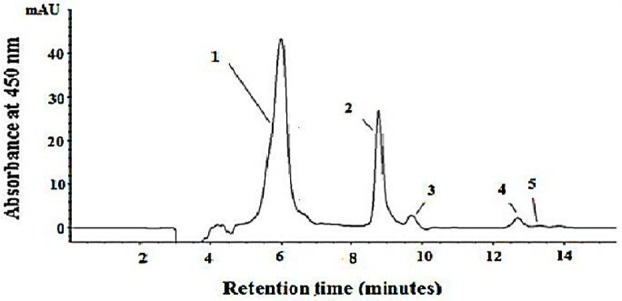
Reverse-phase liquid chromatography of the major carotenoids of Halorubrum sp. TBZ126. Column: 100-5 C18 column (300 by 4.6 mm, Knauer, Germany). Eluent: acetonitrile-dichloromethane-methanol (70:20:10, vol/vol/vol). Flow rate: 0.8 ml/min. Detection: 450 nm. Peak identities: peak 1, bacterioruberin; peak 2, all-trans lycopenes; peaks 3, 13-cis-lycopene; peak 4, all-trans- β-carotenes; peak 5, all-cis- β-carotenes.
The peaks of all trans-isomers of lycopene and β-carotene were identified by comparing the retention times of authentic standards and the UV spectra. Since most of the major cis-isomers of the carotenoids are not available in the market, they were identified by their UV absorption characteristics and also by comparing the obtained chromatograms with the fully investigated isomers of lycopene and β-carotene reported by Müller et al.28 Bacterioruberin was identified by comparing the UV and mass spectra with those reported in literature.29,30 The maximum absorption wavelengths for the carotenoids of interest, detected by diode array detector are 466 nm, 495 nm and 528 nm for bacterioruberin, 448 nm, 474 nm and 505 nm for Lycopene and 425 nm, 455 nm and 482 nm for β-carotene. The UV-VIS spectrum of this main carotenoids present in TBZ126 extract detected by diode array detector are presented in Figure 3. Because the ionization mode was positive, most of the m/z data were [M+H]+ and the mass data of compounds identified are given in Table 2. Derived from the mass fragmentation interpretation, bacterioruberin, lycopene and β-carotene were identified in TBZ126 extract.
Figure 3 .
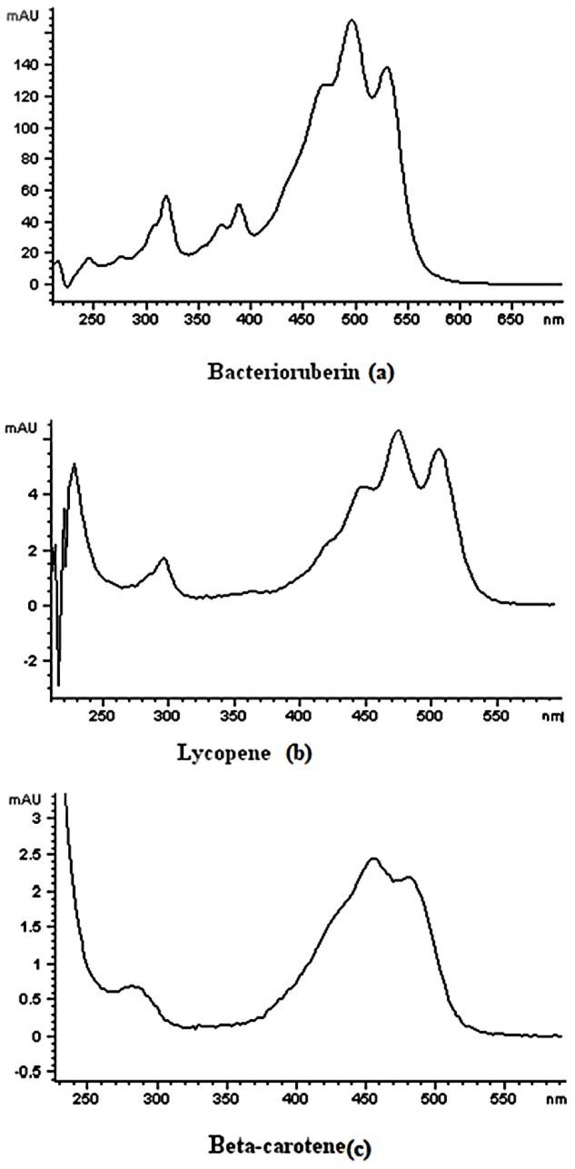
UV–visible absorption spectra of the carotenoids extracted from Halorubrum sp. TBZ126. bacterioruberin (a), lycopene (b) and β-carotene (c) detected by G1315B Diode Array Detector. The maximum absorption wavelengths for the carotenoids of interest are 466 nm, 495 nm and 528 nm for bacterioruberin, 448 nm, 474 nm and 505 nm for Lycopene and 425 nm, 455 nm and 482 nm for β-carotene.
Table 2. Identification of carotenoids from Halorubrum sp. TBZ126 extract elucidated by ESI ion mode showing their molecular mass and molecular formula.
| Identification of carotenoids | Molecular formula | Approximate molecular mass (Dalton) | Fragmentation mass (m/z) | MS/MS fragment ion (m/z) |
| Bacterioruberin | C50H76O4 | 740 | 741 | 723 [M+H-18]+ |
| 705 [M+H-18-18]+ | ||||
| 683 [M+H-58]+ | ||||
| Lycopene | C40H56 | 536 | 537 | 444 [M+H-92]+ |
| ß-carotene | C40H56 | 536 | 537 | 444 [M+H-92]+ |
Discussion
The carotenoid pigments are favorable ingredients owing to high biological activities and potential health benefits with many functions in nutraceuticals, cosmetics and feed industries.31 The use of microorganisms in biotechnology to produce carotenoids is approving by consumer and can help meet the growing demand for these bioactive compounds in the food, feed, and pharmaceutical industries. The major advantages of the biological production of carotenoids are the wide range of their biosynthetic capability and ability to produce only the naturally occurring stereoisomers.32 The marine environment is estimated to be home to more than 80 % of life and yet it remains largely unexplored. Marine microorganisms are an intact source for pigments that can have wide range of applications. In a general evaluation of several thousand colonies isolated from marine sources it was found that 31.3 % were yellow, 15.2 % orange, 9.9 % brown and 5.4 % red or pink.33 However, there are not many reports available on investigation on marine microorganisms as source of natural pigments mainly carotenoids and there is little information in the literature about the carotenoid profile of extremophile microorganisms. In the present study we reported for the first time a new extremely halophilic archaeon isolated from Urmia Lack called Halorubrum sp. TBZ126 capable to produce carotenoid pigments.
Carotenoid-producing microorganisms have been isolated from various extreme environments, such as very low temperatures, high salinity, strong light, acidic and alkaline, and thermophilic conditions. One may hypothesize, based on these evidences, that the oxidative stresses in extreme environments are selective factors associated with pigmented microorganisms, which are able to synthesize antioxidants (i.e., carotenoids) to protect their vital molecules (e.g., proteins and nucleic acids). Carotenoids give the microbial colonies their distinctive color.32
Although there are several reports about carotenoid profile of eubacteria, fewer papers describe carotenoid production from archaea.34 Marshall et al. mentioned that bacterioruberin is a ubiquitous and abundant pinkish-red pigment in moderately to extremely halophilic archaea.35 Jehlicka et al. identified bacterioruberin by Resonance Raman spectroscopy as the major carotenoid in the halophilic archaea Halobacterium salinarum strains NRC-1 and R1, Haloarcula sodomense, and Halorubrum vallismortis. Kelly et al. showed that bacterioruberin was the main carotenoid (85 % of total) of Halobacterium salinarium (an extremely halophilic marine archaeon).29 Ronnekleiv and colleagues reported that Heloferax vokanii contained the (2S,2ʹS)-bacterioruberin (82% of total carotenoid), monoanydrobacterioruberin (7%), (2S,2ʹS)-bisanhydrobacterioruberin (3%), 3,4-dihydromonoanhydrobacterioruberin (2%) and two undecaene C50H74O4 carotenoids (each 2%), the C45-carotenoid (2S)-2-isopentenyl-3,4-dehydrorhodopin (1%) and lycopene (0.3%).30 Mandelli et al. studied carotenoid production by the extremophile microorganisms Halococcus morrhuae, Halobacterium salinarium and Thermus filiformis. The major carotenoid was all-trans-bacterioruberin, accounting for 69% of the carotenoids in Halococcus morrhuae and 68% in Halobacterium salinarium.36
Halorubrum sp. TBZ126, a Gram-negative, aerobic, rod-shaped and extremely halophilic archaeon was isolated from Urmia Lake in Azerbaijan region of Iran. 16S rRNA gene sequence showed that TBZ126 is a new extremely halophilic archaeon related to Halorubrum chaoviator Halo-G*T(AM048786). To choose the appropriate solvent for carotenoid extraction, acetone, methanol, hexane and acetone/methanol (7:3 v/v) were examined, individually with the best recoveries for acetone/methanol (7:3 v/v). The simplicity of carotenoid extraction with acetone/methanol (7:3 v/v) at room temperature gives the strain an advantage over for instance yeast cells which need either be disrupted by mechanical means or treated with DMSO before carotenoid extraction is possible and might prove useful in a future production process. Today the most widely used method for the analysis of carotenoids is reversed phased HPLC equipped with diode array detection (DAD) and MS detection (LC-DAD-MS).37 As shown in Figure 2, the present reverse phase isocratic HPLC method was separated three main carotenoids with good resolution and in a short time (14min). The retention times of Lycopene and β-carotene in the sample were in a good agreement with authentic standards. The UV–Vis spectra of the carotenoids also serve as a helpful source for their identification and the DAD allows the UV–Vis spectrum of each component to be determined on line. Both the wavelengths of maximum absorption (λmax) and the shape of the spectra are feature of each carotenoid.
From the results, it can be concluded that the produced carotenoids of Halorubrum sp. TBZ126 isolated from Urmia Lake, the largest saline lake in the Middle East and the second largest salt water lake on the Earth, were bacterioruberin, lycopene and β-carotene.
Conclusion
As a consequence, this study introduced and opens the way for employment of a novel prokaryotic source of carotenoid production for future use in the pharmaceutical and food industries. Further reports on enhanced carotenoid production in the optimized condition are in program.
Conflict of Interest
The authors report no conflicts of interest.
References
- 1.Stafsnes MH, Dybwad M, Brunsvik A, Bruheim P. Large scale MALDI-TOF MS based taxa identification to identify novel pigment producers in a marine bacterial culture collection. Antonie Van Leeuwenhoek. 2013;103(3):603–15. doi: 10.1007/s10482-012-9844-6. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Sun M, Li Q, Li A, Zhang C. Profiling of carotenoids in six microalgae (Eustigmatophyceae) and assessment of their beta-carotene productions in bubble column photobioreactor. Biotechnol Lett. 2012;34(11):2049–53. doi: 10.1007/s10529-012-0996-2. [DOI] [PubMed] [Google Scholar]
- 3.Cabral MMS, Cence K, Zeni J, Tsai SM, Durrer A, Foltran LL. et al. Carotenoids production from a newly isolated Sporidiobolus pararoseus strain by submerged fermentation. Eur Food Res Technol. 2011;233(1):159–66. [Google Scholar]
- 4.Peng J, Yuan JP, Wu CF, Wang JH. Fucoxanthin, a marine carotenoid present in brown seaweeds and diatoms: metabolism and bioactivities relevant to human health. Mar Drugs. 2011;9(10):1806–28. doi: 10.3390/md9101806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mantzouridou F, Tsimidou MZ, Roukas T. Performance of crude olive pomace oil and soybean oil during carotenoid production by Blakeslea trispora in submerged fermentation. J Agric Food Chem. 2006;54(7):2575–81. doi: 10.1021/jf0526339. [DOI] [PubMed] [Google Scholar]
- 6.Furubayashi M, Umeno D. Directed Evolution of Carotenoid Synthases for the Production of Unnatural Carotenoids. In: Barredo J, editor. Microbial Carotenoids from Bacteria and Microalgae : Methods and Protocols. USA: Springer; 2012. P. 245-53. [DOI] [PubMed]
- 7.Perez-Fons L, Steiger S, Khaneja R, Bramley PM, Cutting SM, Sandmann G. et al. Identification and the developmental formation of carotenoid pigments in the yellow/orange Bacillus spore-formers. Biochim Biophys Acta. 2011;1811(3):177–85. doi: 10.1016/j.bbalip.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Zeni J, Colet R, Cence K, Tiggemann L, Toniazzo G, Cansian R. et al. Screening of microorganisms for production of carotenoids Selección de microorganismos para la producción de carotenoides. CyTA-J Food. 2011;9(2):160–6. [Google Scholar]
- 9.Wang B, Lin L, Lu L, Chen W. Optimization of β-carotene production by a newly isolated Serratia marcescens strain. Electron J Biotechn. 2012;15(6):1–3. [Google Scholar]
- 10.Manikandan M, Pasic L, Kannan V. Optimization of growth media for obtaining high-cell density cultures of halophilic archaea (family Halobacteriaceae) by response surface methodology. Bioresour Technol. 2009;100(12):3107–12. doi: 10.1016/j.biortech.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Andrei AS, Banciu HL, Oren A. Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol Lett. 2012;330(1):1–9. doi: 10.1111/j.1574-6968.2012.02526.x. [DOI] [PubMed] [Google Scholar]
- 12.El-Banna AaE-R, El-Razek AMA, El-Mahdy AR. Isolation, identification and screening of carotenoid-producing strains of Rhodotorula glutinis. Food Nutr (Roma) 2012;3(5):627–33. [Google Scholar]
- 13.Jehlicka J, Edwards HG, Oren A. Bacterioruberin and salinixanthin carotenoids of extremely halophilic Archaea and Bacteria: a Raman spectroscopic study. Spectrochim Acta A Mol Biomol Spectrosc. 2013;106:99–103. doi: 10.1016/j.saa.2012.12.081. [DOI] [PubMed] [Google Scholar]
- 14.Vahed SZ, Forouhandeh H, Hassanzadeh S, Klenk HP, Hejazi MA, Hejazi MS. Isolation and characterization of halophilic bacteria from Urmia Lake in Iran. Mikrobiologiia. 2011;80(6):826–33. [PubMed] [Google Scholar]
- 15.Corbin DR, Greenplate JT, Wong EY, Purcell JP. Cloning of an insecticidal cholesterol oxidase gene and its expression in bacteria and in plant protoplasts. Appl Environ Microbiol. 1994;60(12):4239–44. doi: 10.1128/aem.60.12.4239-4244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enache M, Itoh T, Kamekura M, Teodosiu G, Dumitru L. Haloferax prahovense sp. nov., an extremely halophilic archaeon isolated from a Romanian salt lake. Int J Syst Evol Microbiol. 2007;57(Pt 2):393–7. doi: 10.1099/ijs.0.64674-0. [DOI] [PubMed] [Google Scholar]
- 17.Spangler R, Goddard NL, Thaler DS. Optimizing Taq polymerase concentration for improved signal-to-noise in the broad range detection of low abundance bacteria. PLoS One. 2009;4(9):e7010. doi: 10.1371/journal.pone.0007010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62(Pt 3):716–21. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 19.Gerhardt P, Murray RGE, Wood WA, Krieg NR. Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology; 1994. [Google Scholar]
- 20.Dussault HP. An improved technique for staining red halophilic bacteria. J Bacteriol. 1955;70(4):484–5. doi: 10.1128/jb.70.4.484-485.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mac Faddin JF. Biochemical tests for identification of medical bacteria. Philadelphia: Lippincott Williams & Wilkins; 1976. [Google Scholar]
- 22.Barrow G, Feltham RKA. Cowan and Steel's manual for the identification of medical bacteria. Cambridge: Cambridge university press; 2004. [Google Scholar]
- 23.Davies BH. Chemistry and biochemistry of plant pigments. London: Academic Press; 1976. [Google Scholar]
- 24.Mancinelli RL, Landheim R, Sanchez-Porro C, Dornmayr-Pfaffenhuemer M, Gruber C, Legat A. et al. Halorubrum chaoviator sp. nov., a haloarchaeon isolated from sea salt in Baja California, Mexico, Western Australia and Naxos, Greece. Int J Syst Evol Microbiol. 2009;59(Pt 8):1908–13. doi: 10.1099/ijs.0.000463-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Wang J, Liu Z, Zhao J, Zhou S, editors. Identification of a Gordonia sp. strain producing carotenoids. 2011 International Symposium on IT in Medicine and Education (ITME 2011); 2011; China.
- 26.De La Vega M, Diaz E, Vila M, Leon R. Isolation of a new strain of Picochlorum sp and characterization of its potential biotechnological applications. Biotechnol Prog. 2011;27(6):1535–43. doi: 10.1002/btpr.686. [DOI] [PubMed] [Google Scholar]
- 27.Britton G, Liaaen-Jensen S, Pfander H. Carotenoids. Volume 1B: Spectroscopy. Basel: Birkhäuser Verlag AG; 1995. [Google Scholar]
- 28.Miller A, Pietsch B, Faccin N, Schierle J, Waysek EH. Method for the determination of lycopene in supplements and raw material by reversed-phase liquid chromatography: single-laboratory validation. J AOAC Int. 2008;91(6):1284–97. [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly M, Norgard S, Liaaen-Jensen S. Bacterial carotenoids. 31. C50-carotenoids 5. Carotenoids of Halobacterium salinarium, especially bacterioruberin. Acta Chem Scand. 1970;24(6):2169–82. doi: 10.3891/acta.chem.scand.24-2169. [DOI] [PubMed] [Google Scholar]
- 30.Ronnekleiv M, Liaaen-Jensen S. Bacterial carotenoids 53, C50-carotenoids 23; carotenoids of Haloferax volcanii versus other halophilic bacteria. Biochem Syst Ecol. 1995;23(6):627–34. [Google Scholar]
- 31.Gharibzahedi SMT, Razavi SH, Mousavi SM, Moayedi V. High efficiency canthaxanthin production by a novel mutant isolated from Dietzia natronolimnaea HS-1 using central composite design analysis. Ind Crop Prod. 2012;40:345–54. [Google Scholar]
- 32.Asker D, Awad TS, Beppu T, Ueda K. Isolation, characterization, and diversity of novel radiotolerant carotenoid-producing bacteria. Methods Mol Biol. 2012;892:21–60. doi: 10.1007/978-1-61779-879-5_3. [DOI] [PubMed] [Google Scholar]
- 33.Zobell CE, Feltham CB. Preliminary studies on the distribution and characteristics of marine bacteria. Berkeley: University of California Press; 1934. [Google Scholar]
- 34.Takano H, Asker D, Beppu T, Ueda K. Genetic control for light-induced carotenoid production in non-phototrophic bacteria. J Ind Microbiol Biotechnol. 2006;33(2):88. doi: 10.1007/s10295-005-0005-z. [DOI] [PubMed] [Google Scholar]
- 35.Marshall CP, Leuko S, Coyle CM, Walter MR, Burns BP, Neilan BA. Carotenoid analysis of halophilic archaea by resonance Raman spectroscopy. Astrobiology. 2007;7(4):631–43. doi: 10.1089/ast.2006.0097. [DOI] [PubMed] [Google Scholar]
- 36.Mandelli F, Miranda VS, Rodrigues E, Mercadante AZ. Identification of carotenoids with high antioxidant capacity produced by extremophile microorganisms. World J Microbiol Biotechnol. 2012;28(4):1781–90. doi: 10.1007/s11274-011-0993-y. [DOI] [PubMed] [Google Scholar]
- 37.Breeman RBV. Peer Reviewed: Innovations in Carotenoid Analysis Using LC/MS. Anal Chem. 1996;68(9):299A–304A. [Google Scholar]


