Abstract
Our previous studies identified important molecules involved in lung carcinogenesis through a comprehensive search for the downstream targets of oncogenic KRAS, and these findings suggested that an investigation into the downstream targets of oncogenic KRAS might represent a useful strategy for elucidating the common molecular bases of lung cancer. Among the downstream targets of oncogenic KRAS, a focus was placed on HDAC9, a member of the histone deacetylase family, in the present study because epigenetic modification of DNA or the histone proteins is known to play an important role in carcinogenesis. The immunohistochemical expression of HDAC9 was examined in surgically resected primary lung cancers (130 adenocarcinoma, 49 squamous cell carcinomas, one large cell carcinoma, and 6 small cell carcinomas) and potential associations between its expression level and pathologic factors were analyzed. The results showed that HDAC9 expression levels were lower in lung cancer cells than in non-tumor epithelial cells, and were also significantly lower in adenocarcinomas among the histological types. Moreover, HDAC9 expression levels were significantly lower in adenocarcinomas with lymphatic canal involvement. The restoration of HDAC9 in lung cancer cells losing its expression severely attenuated their growth activity in vitro. These results suggest that HDAC9 may be a suppressor and its downregulation might promote the progression process, especially in lung adenocarcinomas.
Keywords: HDAC9, KRAS, lung cancer
Introduction
Lung cancer is one of the most common causes of cancer-related death in the developed world [1,2]. Even if surgical removal of the primary tumor is successful, the incidence of recurrence is high [1,2]. Although some lung tumors are sensitive to conventional chemotherapeutic agents or certain molecular targeting agents, many are not [3,4]. Thus, further understanding of the molecular basis of lung carcinogenesis is important for developing a novel therapeutic strategy.
Our previous studies identified important molecules involved in carcinogenesis of the lung through a comprehensive search for the downstream targets of oncogenic KRAS [3,5]. Such downstream targets were involved not only in the development of lung cancers with KRAS mutations, but also in those without KRAS mutations [3,5]. These findings indicated that an investigation into the downstream targets of oncogenic KRAS may represent a useful strategy for revealing the common important molecular bases of lung cancer. The present study focused on HDAC9 because independent studies including our previous reports demonstrated that HDAC9 was unregulated in different types of cells undergoing oncogenic RAS-induced senescence [6]. HDAC9 is a class IIa histone deacetylase family member and is thought to regulate gene expression through epigenetic modulation of the chromatin structure by catalyzing the deacetylation of histone proteins [7]. HDAC9 is known to target non-histone proteins, such as forkhead box protein 3, ataxia telangiectasia group D-complementing protein (ATDC), and glioblastoma 1 protein, which are members of pathways implicated in carcinogenesis [7-10]. Aberrant HDAC9 expression has been observed in several types of cancers including medulloblastoma [11], acute lymphoblastic leukemia [12], and cervical carcinoma [13]. In our preliminary study, HDAC9 mRNA expression was found to be upregulated in oncogenic KRAS-transduced cell immortalized airway cell lines (NHBT-T) (Figure 1), which supported the results obtained from our gene chip microarray analysis (Table 1) [14]. Among the lung cancer cell lines examined, some almost lost its expression (Figure 1), which implied its potentially suppressive role in carcinogenesis. These findings prompted us to speculate that aberrant HDAC9 expression may also be involved in lung carcinogenesis.
Figure 1.
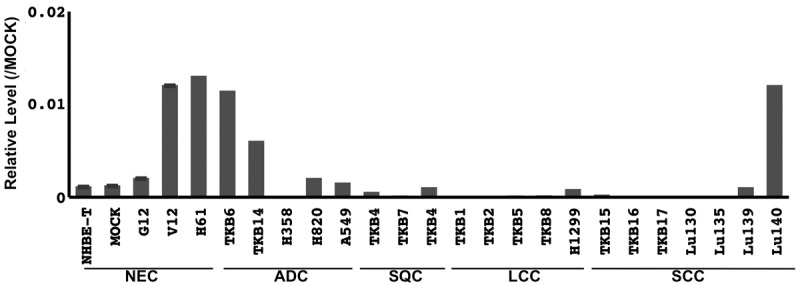
Expression of HDAC9 mRNA in cell lines. The mRNA level of HDAC9 normalized to that of GAPDH was measured by quantitative RT-PCR and plotted in a graph. NEC, non-cancerous airway epithelial cells; ADC, adenocarcinoma cell lines; SQC, squamous cell carcinoma cell lines; LCC, large cell carcinoma cell lines; NSCLC, non-small cell lung carcinoma cell lines.
Table 1.
Downstream targets (DNA binding proteins) upregulated by oncogenic KRAS
| MOCK | KRAS/G12 | KRAS/V12 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Common | Genbank | Map | Signal Value | Flags | Signal Value | Flags | Signal Value | Flags | Ratio V12/MOCK | Ratio V12/G12 |
| TBX3 | NM_016569 | 12q24.1 | 2.591 | P | 2.802 | P | 7.845 | P | 3.027 | 2.800 |
| COPEB | AB017493 | 10p15 | 3.298 | P | 4.015 | P | 12.069 | P | 3.660 | 3.006 |
| ARNT2 | NM_014862 | 15q24 | 0.584 | A | 0.625 | A | 2.034 | P | 3.482 | 3.253 |
| FOS | BC004490 | 14q24.3 | 0.044 | A | 0.088 | A | 1.065 | P | 24.013 | 12.116 |
| HDAC9 | NM_014707 | 7p21.1 | 0.834 | P | 0.883 | P | 3.490 | P | 4.186 | 3.953 |
| EGR1 | NM_001964 | 5q31.1 | 1.796 | P | 1.377 | P | 12.829 | P | 7.144 | 9.314 |
| IRF6 | AU144284 | 1q32.3-q41 | 3.670 | P | 4.825 | P | 13.904 | P | 3.789 | 2.882 |
| ATF3 | NM_001674 | 1q32.3 | 1.084 | P | 1.659 | P | 4.047 | P | 3.733 | 2.440 |
MOCK, empty vector-transduced; KRAS/G12-transduced; KRAS/V12-transduced cells; Common, common gene name; Map, Chromosomal locus; GenBank, gene bank accession number; Flags indicate whether gene expression was present (P) or absent (A). Genes whose signal values were more than 3-fold higher by the KRAS/V12 transduction than by the empty vector transduction or the KRAS/G12 transduction were extracted from our data of a gene chip microarray previously described [14]. The DNA binding molecules sorted are shown in this table.
We here examined the immunohistochemical expression of HDAC9 in surgically resected primary lung cancers and analyzed associations between its expression levels and pathologic factors to investigate the potential role of HDAC9 in lung carcinogenesis.
Materials and methods
Primary lung cancer
All 186 tumors examined (130 adenocarcinoma, 49 squamous cell carcinomas, one large cell carcinoma, and 6 small cell carcinomas) were removed by radical surgical resection at Kanagawa Cardiovascular and Respiratory Center (Yokohama, Japan). Informed consent for research use of the resected materials was obtained from all subjects. Histological types and disease stages were determined according to the international TNM classification system (seventh edition of the UICC) [15].
Culture cells
Human lung cancer cell lines (A549, H358, H2087, H1819, H441, and H1299) and a human embryonic kidney cell line (HEK293T) were purchased from the American Type Culture Collection (ATCC, Manassas, VA). The human lung cancer cell lines, Lu135 and Lu139 were purchased from Riken Cell Bank (Tsukuba, Japan). PC9 and HARA were purchased from Immunobiological Laboratories Co. (Gunma, Japan). TKB5, TKB6, TKB7, TKB8, TKB8, TKB14, and TKB20 were from Dr. H Kamma via Dr. T Yazawa (Kyorin University School of Medicine, Tokyo, Japan).
Quantitative RT-PCR
Total RNA from snap frozen tissue sections and cell lines were extracted using the RNA easy kit (Qiagen). First-strand cDNA was synthesized from total RNA using the SuperScript First-Strand Synthesis System according to the protocols of the manufacturer (Invitrogen, Carlsbad, CA). The cDNA generated was used as a template in real-time PCR with SYBR Premix EXTaq (Takara) and run on a Thermal Cycler DICE real-time PCR system (Takara). The primers used for the detection of HDAC9 and GAPDH were purchased from Takara Bio Inc. The level of HDAC9 mRNA was normalized to the level of GAPDH mRNA.
Immunohistochemistry
The largest tumor sections were cut from formalin-fixed, paraffin-embedded tissue blocks. Sections were deparaffinized, rehydrated, and incubated with 3% hydrogen peroxide, followed by 5% goat serum to block endogenous peroxidase activities and non-immunospecific protein binding. Sections were boiled in citrate buffer (0.01 M, pH 6.0) for 15 minutes to retrieve masked epitopes and were then incubated with a primary antibody against HDAC9 that recognized some variants of HDAC9, such as variant 5, and variant 3 which lacks the histone acetyltransferase (HAT) domain or Ki-67 (DAKO, Ely, UK). Immunoreactivity was visualized using the Envision detection system (DAKO), and nuclei were counterstained with hematoxylin. HADC9 immunohistochemical expression levels were evaluated by a scoring system as described in the Results section. The labeling index of Ki-67 was calculated as the proportion of positive nuclei cells by counting 500-1000 cancer cells. The Ki-67 labeling indices of < 10% and ≥ 10% were classified as low and high levels, according to the results of our previous study [4].
Statistical analysis
Differences in the mean values of the immunohistochemical score for HDAC9 among groups classified based on various clinicopathologic factors were analyzed by one-way ANOVAs. Recurrence curves were plotted using the Kaplan-Meier method, and the absolute risk of recurrence at five years was estimated from these curves. Differences in the disease-free survival (DFS) span and rate were analyzed using the log-rank test. P values less than 0.05 were considered significant. Statistical analyses were performed using SPSS software (SPSS for Windows Version 10.0; SPSS; Chicago, IL, USA).
Cell lines and culture
The immortalized human airway epithelial cell line (16HBE14o, Simian virus 40 (SV40)-transformed human bronchial epithelial cells) described by Cozens AL et al. [16] was kindly provided by Grunert DC (California Pacific Medical Center Research institute). A sub-clone of 16HBE14o cells, described as NHBE-T in this study, was used for subsequent experiments. NHBE-T cells transfected KRAS genes were described elsewhere [14].
Plasmid construction
The complementary DNA of HDAC9 variant 5 (NM_178425.2), and variant 3 (NM_014707.1) which lacks the histone acetyltransferase domain, were PCR-amplified and inserted into the pQCXIP (BD Clontech, Palo Alto, CA) pro-retrovirus vector.
Retroviral-mediated gene transfer
The expression vectors and pCL10A1 retrovirus-packaging vector (IMGENEX, San Diego, CA) were co-transfected into HEK293T cells with Lipofectoamine 2000 reagent (Invitrogen, Carlsbad, CA). Conditioned medium was recovered as a viral solution. The genes were introduced by incubating cells with the viral solution containing 10 μg/ml of polybrene (Sigma, St. Louis, MO). Cells stably expressing the desired genes were selected with 5.0 μg/ml of Puromycin (Invitrogen) for 3 days. Pooled clones were used for biological analyses as follows.
Western blotting
Whole cell lysates were subjected to SDS-polyacrylamide gel electrophoresis, and transferred onto PVDF membranes (Amersham). These membranes were incubated with nonfat dry milk in Tris-buffered saline containing Tween-20 (TBS-T) to block non-immunospecific protein binding, and then with a primary antibody against HDAC9 (ab18790, Abcam, Cambridge, MA) or β-actin (Sigma). After washing with TBS-T, the membranes were incubated with animal-matched HRP-conjugated secondary antibodies (Amersham). Immunoreactivity was visualized with an enhanced chemiluminescence system (Amersham).
Colony formation assays
Cells (2.5×104) were seeded onto a 10 cm culture dish (Iwaki, Tokyo, Japan), and grown for 10 days. Cells were fixed with methanol and Giemsa-stained, and colonies visible in scanned photographs were counted.
Growth curve assays
Cells (2.5×105) were seeded onto a 10 cm culture dish, and grown to a semi-confluent state for the appropriate number of days. Cells were counted, and 2.5×105 cells were seeded again onto a 10 cm dish. Several passages were repeated in the same manner. The sum of population doublings at each point was calculated by the formula ΣPDLn = log2 (countn/2.5×105) + ΣPDLn-1.
Results
Immunohistochemical expression of HDAC9 in primary lung cancer
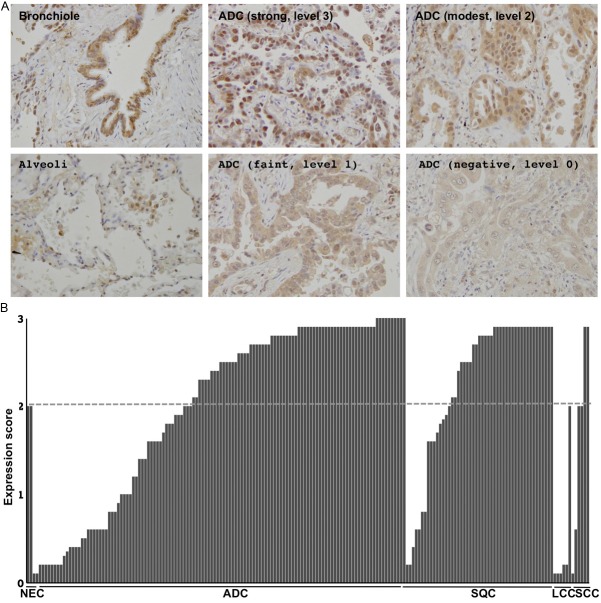
The HDAC9 protein was commonly expressed in the nuclei of non-cancerous lung cells including alveolar epithelial cells, bronchial epithelial cells, and various mesenchymal cells (Figure 2A). Lung cancer cells expressed the HDAC9 protein at various levels. Some tumors expressed the HDAC9 protein at markedly higher levels than those of their non-cancerous counterparts, others expressed it at equivalent or lower level, and the remaining tumors almost lost its expression (Figure 2A). Expression levels were classified into negative (level 0), faint (level 1), modest (level 2), and strong (level 3). The faint level was defined as being weaker than that in the bronchioles and alveoli, but not negative. The modest level was defined as being equivalent to that in the bronchioles and alveoli. The strong level was defined as being markedly higher than that in the bronchioles and alveoli. The score of immunohistochemical expression was determined as an average level in the maximal tumor section (if 30%, 10%, 50%, and 10% of neoplastic cells in the maximal section of the tumor were negative, faint, modest, and strong levels, respectively, the average level was calculated as “1.4 = 0.3×0 + 0.1×1 + 0.5×2 + 0.1×1”). The average levels of all the tumors examined are shown in Figure 2B. Overall, the expression level of the HDAC9 protein was lower in neoplastic than in bronchial and alveolar epithelial cells (Figure 2B).
Figure 2.
Representative photographs from the bronchioles, alveoli, and tumors, in which HDAC9 expression was negative, faint, modest, and strong, are shown (A). HDAC9 expression levels of all the tumors examined are shown on a graph (B). Dashed line indicates the level of non-neoplastic epithelia cells (NTE). ADC, adenocarcinoma; SQC, squamous cell carcinoma; LCC, large cell carcinoma.
Association between the HDAC9 expression and pathologic factors
The HDAC9 expression level differed depending on the histological type. It was significantly lower in adenocarcinomas (Table 2). Moreover, HDAC9 expression levels were significantly lower in adenocarcinomas with lymphatic canal involvement. However, the expression of HDAC9 was not associated with any other factors analyzed, including histological grade, and oncogenic mutations (Table 2). Moreover, it was not significantly correlated with the post-operative disease-free survival rate or span. However, patients with a lower level of HDAC9 protein expression had slightly poorer outcomes (Figure 3).
Table 2.
Correlation between HDAC9 expression levels and pathologic factors
| Expression score | |
|---|---|
| *Histology (186) | |
| ADC (130) | 1.942 ± 1.015 |
| Grade | |
| WEL (79) | 2.069 ± 0.998 |
| MOD (36) | 1.814 ± 1.028 |
| POR (17) | 1.865 ± 1.028 |
| Subtype | |
| BAC (48) | 2.028 ± 1.023 |
| ACI (42) | 1.946 ± 1.031 |
| PAP (10) | 2.110 ± 0.824 |
| SOL (12) | 1.758 ± 1.039 |
| MUC (18) | 1.600 ± 1.069 |
| SQC (49) | 2.279 ± 0.845 |
| Grade | |
| WEL (8) | 2.375 ± 0.858 |
| MOD (26) | 2.337 ± 0.800 |
| POR (15) | 2.127 ± 0.948 |
| LCC (1) | 2.000 ± 0.000 |
| SCC (6) | 1.867 ± 0.845 |
| Vascular involvement/ADC (130) | |
| Present (20) | 1.792 ± 1.060 |
| Absent (110) | 1.990 ± 1.002 |
| *Lymphatic canal involvement/ADC (130) | |
| Present (22) | 1.540 ± 1.078 |
| Absent (108) | 2.069 ± 0.968 |
| MIB1 labeling index/ADC (130) | |
| Low level (< 10%) (57) | 2.053 ± 0.975 |
| High level (≥ 10%) (73) | 1.827 ± 1.030 |
| Oncogenic mutation/ADC (130) | |
| KRAS (10) | 1.800 ± 0.981 |
| EGFR (42) | 1.963 ± 1.093 |
| NONE (78) | 1.944 ± 0.980 |
Significant in a one way ANOVA analysis;
Histology (ADC versus SQC, P = 0.0426); Lymphatic canal involvement/ADC (present versus absent, P = 0.0214); ADC, adenocarcinoma; SQC, squamous cell carcinoma; LCC, large cell carcinoma; WEL, well differentiated; MOD, moderately differentiated; POR, poorly differentiated carcinomas; BAC, bronchioloalveolar carcinoma; ACI, acinar adenocarcinoma; PAP, papillary adenocarcinoma; SOL, solid adenocarcinoma; MUC, mucinous adenocarcinoma; NONE, cases without KRAS or EGFR mutations.
Figure 3.
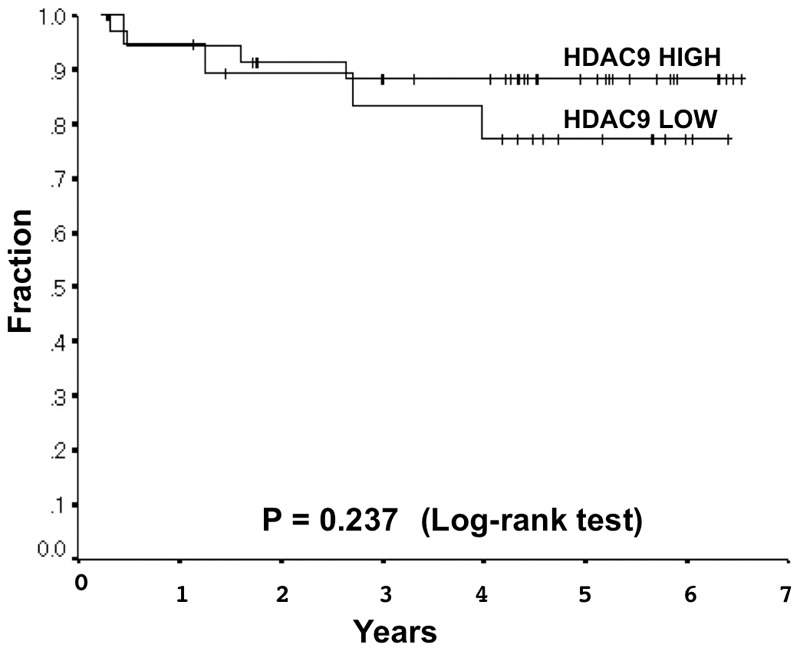
The expression score of < 2.85 and ≥ 2.85 were classified as low and high based on a receiver operating characteristic curve (area under the curve 0.632, 95% confidential interval 0.453-0.810). Associations between HDAC9 levels and disease-free survival in stage I lung adenocarcinomas were analyzed by the Log-rank test (P = 0.3552). Sixty-three patients were used for disease-free survival analysis. Kaplan-Meier survival curves are shown (five-year disease-free survival rates were 77.3% and 88.3% HADC9 low- and high-expressers, respectively.
Effect of the forced expression of HDAC9 on growth activity
The forced expression of both HDAC9 variant 5, and variant 3 which lacks the HAT domain, reduced clonogenicity and prolonged the doubling time in an immortalized airway epithelial cell line (NHBE-T) and some NSCLC cell lines, in which the expression of HDAC9 was lost (A549 and H2087). Representative results from exa- minations on NHEB-T cells are shown (Figure 4). The exogenous expression of HDAC9 was verified by Western blotting (Figure 4).
Figure 4.
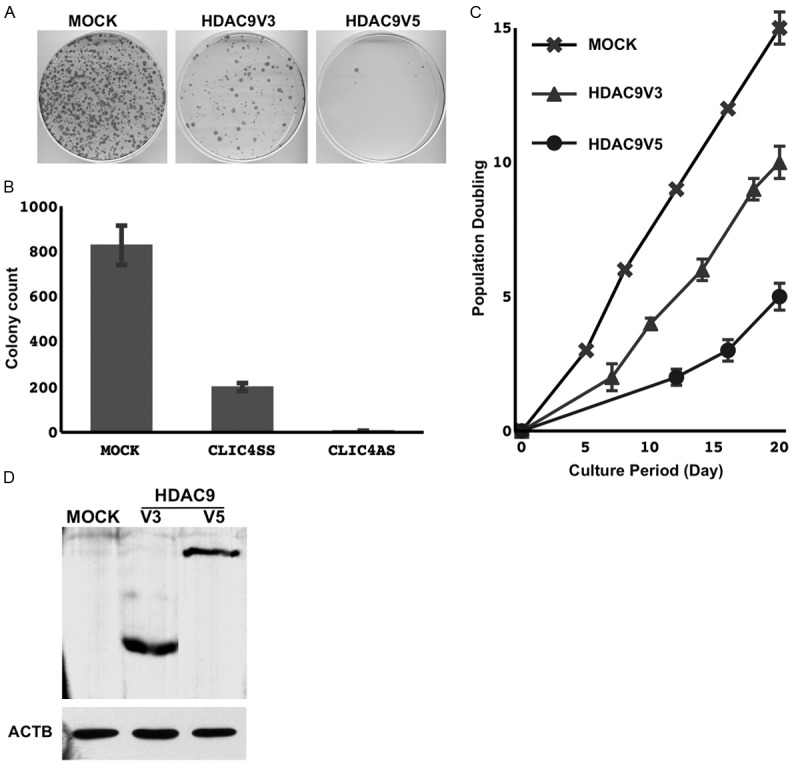
The empty vector (MOCK), HDAC9 transcriptional variant 5, and HDAC9 transcriptional variant 3 were transduced. Following a brief selection for 3 days, the surviving cells were harvested and counted, and 2.0×104 cells were re-seeded on a 10 cm dish. After 14 days, cells were methanol-fixed and Giemsa-stained (A). The means and standard deviations (error bars) of colony counts from triplicate experiments are presented (B). Cells selected were grown and passed several times. Cumulated population doublings are presented as a line chart (C). Cells harvested immediately after the selection process were examined for the expression of HDAC9 by Western blotting (D).
Interestingly, although the suppressive effect on cell growth was markedly stronger in cells transduced with variant 5, and variant 3 lacking the HAT domain, also suppressed cell growth. HADC9 may possess potential properties other than deacetylation activity, which modulate cellular biological activity.
Discussion
The present study focused on HDAC9 and investigated its potential role in lung carcinogenesis. The HDAC9 expression levels were reduced in some lung cancer cell lines (Figure 1). The restoration of HDAC9 in lung cancer cell lines with reduced HDAC9 expression severely attenuated their growth activity (Figure 4), which suggested that HDAC9 might be a tumor suppressor. Consistent with these results, the immunohistochemical expression of HDAC9 was also weaker in primary lung cancer cells than in non-tumorous airway epithelial cells, and was occasionally undetectable in some tumors (Figure 2). HDAC9 levels were significantly lower in adenocarcinomas, especially in adenocarcinomas with lymphatic canal involvement (Table 2). These findings suggest that HDAC9 may function as a tumor suppressor in the progression process of carcinogenesis of lung adenocarcinomas. Although HDAC9 is a downstream target of oncogenic KRAS, HDAC9 levels were not associated with the types of oncogenic mutations (Table 2). The two major oncogenes of KRAS and EGFR are known to transmit the oncogenic signals via a common pathway [17-19]. HDAC9 may be involved in this common pathway. Thus, the aberrant expression of HDAC9 may promote the progression of not only KRAS-mutated lung cancers, but also EGFR-mutated lung cancers.
The human HDAC9 gene is located on chromosome 7p21, a region that has been implicated in various malignancies, including colon cancer, childhood acute lymphoblastic leukemia, peripheral nerve sheath tumor, and gynecological tumors [7-10,12]. The aberrant expression of HDAC9 has been reported and is suggested to play a potential role in human carcinogenesis. Higher HDAC9 expression levels have been associated with poorer outcomes in some tumors, such as lymphocytic leukemia [8] and medulloblastoma [11], implicating its oncogenic role. In contrast, in glioma, its expression was reportedly reduced in tumors with higher malignant activity (glioblastoma) [20]. Thus, the effect of the aberrant expression of HDAC9 on carcinogenesis may differ among various types of malignancies [7]. Markedly deviated levels (very high or very low) of HDAC9 may negatively impact physiological regulation of transcription, which may produce selection pressure and result in the promotion of carcinogenesis. On the other hand, a recent study demonstrated that HDAC9 positively regulated the transcriptional activity of p53, a tumor suppressor, through the deacetylation of ATDC, a p53 binding protein [7,21]. Thus, the loss of HDAC9 is considered to impair the tumor-suppressive function of p53 and promote tumor progression under certain conditions. This is consistent with our suggestion that HDAC9 may play a suppressive role in lung carcinogenesis.
In summary, the results of the present study suggest the potential involvement of HDAC9 in lung carcinogenesis. The downregulation of HDAC9 may promote the progression process, especially in adenocarcinomas. The molecular biological function of HDAC9 has not yet been investigated in detail and remains largely unclear. Further investigations are expected to elucidate the potential role of HDAC9 in carcinogenesis.
Acknowledgements
We especially thank Misa OTARA and Emi Honda (Kanagawa Prefectural Cardiovascular and Respiratory Center Hospital, Yokohama, Japan) for their assistance.
Disclosure of conflict of interest
None.
References
- 1.Hoffman PC, Cohen EE, Masters GA, Haraf DJ, Mauer AM, Rudin CM, Krauss SA, Huo D, Vokes EE. Carboplatin plus vinorelbine with concomitant radiation therapy in advanced non-small cell lung cancer: a phase I study. Lung Cancer. 2002;38:65–71. doi: 10.1016/s0169-5002(02)00144-7. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 3.Okudela K, Woo T, Kitamura H. KRAS gene mutations in lung cancer: particulars established and issues unresolved. Pathol Int. 2010;60:651–660. doi: 10.1111/j.1440-1827.2010.02580.x. [DOI] [PubMed] [Google Scholar]
- 4.Woo T, Okudela K, Yazawa T, Wada N, Ogawa N, Ishiwa N, Tajiri M, Rino Y, Kitamura H, Masuda M. Prognostic value of KRAS mutations and Ki-67 expression in stage I lung adenocarcinomas. Lung Cancer. 2009;65:355–362. doi: 10.1016/j.lungcan.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 5.Okudela K, Yazawa T, Ishii J, Woo T, Mitsui H, Bunai T, Sakaeda M, Shimoyamada H, Sato H, Tajiri M, Ogawa N, Masuda M, Sugimura H, Kitamura H. Down-regulation of FXYD3 expression in human lung cancers: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175:2646–2656. doi: 10.2353/ajpath.2009.080571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason DX, Jackson TJ, Lin AW. Molecular signature of oncogenic ras-induced senescence. Oncogene. 2004;23:9238–9246. doi: 10.1038/sj.onc.1208172. [DOI] [PubMed] [Google Scholar]
- 7.Clocchiatti A, Florean C, Brancolini C. Class IIa HDACs: from important roles in differentiation to possible implications in tumourigenesis. J Cell Mol Med. 2011;15:1833–1846. doi: 10.1111/j.1582-4934.2011.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 9.Morrison BE, D’Mello SR. Polydactyly in mice lacking HDAC9/HDRP. Exp Biol Med (Maywood) 2008;233:980–988. doi: 10.3181/0802-RM-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. Int J Cancer. 2010;127:759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- 11.Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M, Taylor MD, von Deimling A, Pfister S, Witt O. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 12.Moreno DA, Scrideli CA, Cortez MA, de Paula Queiroz R, Valera ET, da Silva Silveira V, Yunes JA, Brandalise SR, Tone LG. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;150:665–673. doi: 10.1111/j.1365-2141.2010.08301.x. [DOI] [PubMed] [Google Scholar]
- 13.Choi YW, Bae SM, Kim YW, Lee HN, Kim YW, Park TC, Ro DY, Shin JC, Shin SJ, Seo JS, Ahn WS. Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int J Gynecol Cancer. 2007;17:687–696. doi: 10.1111/j.1525-1438.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- 14.Okudela K, Yazawa T, Woo T, Sakaeda M, Ishii J, Mitsui H, Shimoyamada H, Sato H, Tajiri M, Ogawa N, Masuda M, Takahashi T, Sugimura H, Kitamura H. Down-regulation of DUSP6 expression in lung cancer: its mechanism and potential role in carcinogenesis. Am J Pathol. 2009;175:867–881. doi: 10.2353/ajpath.2009.080489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. Pathology and Genetics – Tumors of the Lung, Pleura, Thymus and Heart. Lyon: IARC Press; 2004. [Google Scholar]
- 16.Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 17.Reinersman JM, Johnson ML, Riely GJ, Chitale DA, Nicastri AD, Soff GA, Schwartz AG, Sima CS, Ayalew G, Lau C, Zakowski MF, Rusch VW, Ladanyi M, Kris MG. Frequency of EGFR and KRAS Mutations in Lung Adenocarcinomas in African Americans. J Thorac Oncol. 2011 Jan;6:28–31. doi: 10.1097/JTO.0b013e3181fb4fe2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 19.Marks JL, Broderick S, Zhou Q, Chitale D, Li AR, Zakowski MF, Kris MG, Rusch VW, Azzoli CG, Seshan VE, Ladanyi M, Pao W. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 2008;3:111–116. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- 20.Lucio-Eterovic AK, Cortez MA, Valera ET, Motta FJ, Queiroz RG, Machado HR, Carlotti CG Jr, Neder L, Scrideli CA, Tone LG. Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer. 2008;8:243. doi: 10.1186/1471-2407-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Z, Peng L, Radhakrishnan R, Seto E. Histone deacetylase 9 (HDAC9) regulates the functions of the ATDC (TRIM29) protein. J Biol Chem. 2010;285:39329–39338. doi: 10.1074/jbc.M110.179333. [DOI] [PMC free article] [PubMed] [Google Scholar]