Abstract
To study the clinicopathological and genomic characteristics of Xp11.2 translocation renal cell carcinoma (Xp11.2 RCC) in adults, we analyzed 9 Xp11.2 RCCs, confirmed by transcription factor E3 (TFE3) immunohistochemistry, in patients aged ≥20 years. TFE3 expression was also determined in 12 cases of alveolar soft part sarcoma (ASPS) served as a positive control. Comparative genomic hybridization (CGH) was used to investigate genomic imbalances in all Xp11.2 RCC cases. Most of our Xp11.2 RCC patients (5/9) presented with TNM stages 3-4, and 6 patients died 10 months to 7 years after their operation. Histologically, Xp11.2 RCC was composed of a mixed papillary nested/alveolar growth pattern (8/9). Immunostaining showed that all Xp11.2 RCC and ASPS cases had strong TFE3 expression and high positive ratios for p53 and vimentin. However, there were significant differences in the expression of AMACR (p<0.001), AE1/AE3 (p=0.002), and CD10 (p=0.024) between the 2 diseases. CGH profiles showed chromosomal imbalances in all 9 Xp11.2 RCC cases; gains were observed in chromosomes Xp11 (6/9), 7q20-25, 12q25-31 (5/9), 7p16-24 (4/9), 8p12-13, 8q20-21, 16q20-22, 17q25-26, 20q22-23 (4/9), and losses occurred frequently on chromosomes 3p12-16, 9q31-32, 14q22-24 (4/9). Our Conclusions show Xp11.2 RCC that occur in adults may be aggressive cancers, the expressions of AMACR, CD10, AE1/AE3 are helpful in the differential diagnosis between Xp11.2 RCC and ASPS, and CGH assay is a useful complementary method for confirming the diagnosis of Xp11.2 RCC.
Keywords: Xp11.2 translocation, renal cell carcinoma, alveolar soft part sarcoma, comparative genomic hybridization, chromosome imbalance
Introduction
The concept of Xp11.2 translocation renal cell carcinoma (Xp11.2 RCC) was accepted as a distinctive entity in the 2004 World Health Organization renal tumor classification. It accounts for approximately 20-70% of pediatric and adolescent renal neoplasms [1-7] and has recently been reported in adult patients [8,9]. In this article, we investigate 9 Xp11.2 RCC patients aged ≥20 years. All cases were confirmed by transcription factor E3 (TFE3) immunohistochemistry (IHC), a specific and sensitive marker of neoplasms with TFE3 gene fusions, which can be applied to archival material [10]. TFE3 expression was also determined in 12 cases of alveolar soft part sarcoma (ASPS) and the ASPL-TFE3 fusion gene served as a positive control [11]. This study adds to the previously reported clinicopathological characteristics and immunophenotypes, and using comparative genomic hybridization (CGH), we investigate genomic imbalances in Xp11.2 RCC.
Materials and methods
Specimens
Nine Xp11.2 RCC paraffin-embedded tissues were retrieved from the archives in the Department of Pathology, Shihezi University School of Medicine. Clinicopathologic data for these cases were collected from their medical records (Table 1). Sections (3-μm thick) were stained with hematoxylin and eosin and colloidal iron. Inclusion criteria were moderate-to-strong immunoreactivity for TFE3 and a highly sensitive (97.5%) and specific (99.6%) marker of Xp11 RCC [10]. The expression of TFE3 proteins in 12 cases of ASPS was confirmed by IHC, and specimens with the ASPL-TFE3 fusion gene were considered positive controls. CGH was used to investigate genomic imbalances in all Xp11.2 RCC cases.
Table 1.
Clinical characteristics of 9 adult Xp11 translocation renal cell carcinoma cases
| Case | Age/Sex/Laterality | Stage (Tumor Diameter, Comment) | Follow-up |
|---|---|---|---|
| 1 | 31/F/R | pT3M0N0 stage 3 (11.5 cm primary, renal vein invasion) | Died 6 years after operation |
| 2 | 25/F/L | pT3M0N0 stage 2 (9.8 cm primary) | Died 9 years after operation |
| 3 | 55/M/L | pT2M0N0 stage 2 (6 cm primary) | Died 7 years after operation |
| 4 | 30/F/R | pT3M0N0 stage 3 (20 cm primary, invaded into perinephric tissue, renal sinus) | Survival 10 years after operation |
| 5 | 32/F/R | pT1M0N1 stage 3 (6.5 cm primary, 2/2 lymph nodes positive, 1/2 retroperitoneal nodal metastasis) | Died 3 years after operation |
| 6 | 43/M/L | pT2M1N1 stage 4 (8 cm primary, 4/4 lymph nodes positive, lung metastasis) | Developed liver, bone metastasis at 6 months; |
| Died 10 months after operation | |||
| 7 | 75/M/L | pT1M0N0 stage 1 (5.5 cm, primary) | Died 3 years after operation |
| 8 | 72/M/L | pT2M0N0 stage 2 (8.5 cm primary) | Survival 4 years after operation |
| 9 | 56/M/R | pT2M0N9 stage 2 (7.5 cm primary) | Not available |
Immunohistochemistry
IHC staining was performed on formalin-fixed, paraffin-embedded, tissue sections by using heat-induced epitope retrieval or pepsin digestion (Envision detection system, Dako, CA, USA), according to the manufacturer’s instructions. The following standard antibodies and dilutions were used: TFE3 (catalog no., sc-5958; Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:600), Cytokeratin AE1/AE3 (Dako; 1:100), CD10 (GT200410; Dako; 1:100), AMACR (13H4; Dako; 1:100), Vimentin (Vim3B4; Dako; 1:100), and p53 (DO-7; Dako; 1:100). Pretreatment for all antibodies consisted of steaming in a citrate buffer, except for TFE3 wherein EDTA buffer was used.
DNA extraction
Total DNA was extracted from the 9 samples by using a standard phenol/chloroform extraction method. DNA quality was checked on a 1% agarose gel, and the amount of extracted DNA was measured spectrophotometrically at 260 nm (impurity and ratio of DNA to non-DNA were also crosschecked at 280 nm). Extractions were stored at -80°C until they were labeled by nick translation.
Comparative genomic hybridization
CGH was performed according to the manufacturer’s protocol (Vysis, Inc., Downers Grove, IL, USA). Briefly, labeling reactions were performed with 1 μg DNA and a nick translation labeling kit (Vysis, Inc.) in a volume of 50 μl containing the following: 0.1 mmol/L of a dNTP pool containing 0.3 mmol/L each of dATP, dGTP and dCTP; 0.1 mmol/L dTTP; 0.2 mmol/L fluorescein isothiocyanate (FITC)-dUTP (for the experimental sample) or cyanine 3 (Cy3)-dUTP (for the 46, XY karyotype); and nick translation buffer and nick translation enzyme. The probe size was determined by separation on a 1% agarose gel. Metaphase slides were denatured at (73°C±1°C for 5 min in 70% methanamide/2×SSC and dehydrated in an ethanol series (70%, 85%, and 100%). The hybridization mixture consisted of approximately 200 ng Spectrum Green labeled test DNA and 200 ng Spectrum Red total genomic reference DNA co-precipitated with 10 μg of human Cot-1 DNA (Invitrogen, California, USA) and dissolved in hybridization buffer before hybridization to metaphase chromosomes. The probe mixtures were denatured at 73°C for 5 min and then competitively hybridized to the denatured normal metaphase chromosomes in a humid chamber at 37°C for 3 days. After washing, chromosomes were counterstained with 4’,6-diamidino-2-phenylindole-2 HCl (DAPI II; Vysis Inc.) and embedded in an anti-fading agent to reduce photo bleaching.
Microscopy and digital image analysis
A fluorescence microscope equipped with appropriate filters (DAPI, FITC, and Cy3) was used to visualize the signals. For each hybridization panel, raw images from at least 5 metaphases were captured through a computer driven CCD camera and analyzed with the ISIS image software (Carl Zeiss Inc., Goettingen, Germany). Chromosomes were identified by their DAPI banding patterns. Threshold levels of 1.25 and 0.8 were used to score gains and losses, respectively. High-level amplification was indicated by a ratio greater than 1.5. All centromeres, as well as chromosome p35-36, and the heterochromatic regions of chromosomes Y, 16, 19, and 22 were excluded from further analysis because these regions can yield unreliable hybridization owing to incompletely suppressed repetitive DNA sequences. Positive and negative controls provided comparisons for evaluating hybridization and interpretation of the data. Normal female DNA (labeled green) was used as a negative control and normal male DNA was used for reference (labeled red). The intensity profiles for this experiment were within the threshold values, as determined by image analysis. DNA from the MPE600 cell line (with known genetic aberrations that are easy to detect by comparative genomic hybridization) was used as a positive control (labeled green), and normal male DNA was used as a reference.
Statistical analysis
A bilateral exact probability test was applied to analyze differences between 2 groups. All data were analyzed using SPSS17.0. A p value <0.05 was considered statistically significant.
Results
Clinical features
The clinical characteristics of 9 cases are summarized in Table 1. The male:female ratio was 5:4. The mean age at diagnosis was 43 years (range, 25-75 years). The tumors were staged using the 2009 American Joint Committee on Cancer (AJCC) staging criteria. The carcinomas frequently presented at an advanced stage. The median tumor diameter was 9.26 cm (range, 5.5-20 cm). Nodal metastases were identified in 2 of 9 cases when perirenal lymph nodes were evaluated histologically. Several of the carcinomas had distinctive clinical presentations. In case no. 7, the tumor was heavily calcified on the initial computed tomography (CT) scan. Given the tumor’s calcified appearance, it was first thought to be a renal tuberculoma. In case no. 1, also heavily calcified, the carcinoma oppressed the adrenal gland, leading to obesity and hypertension. In addition, patients presented with crura (case no. 7), flank pain (case no. 4), and hematuria (cases no. 2, 3, 4) as well, which are more classic symptoms of RCC.
Histopathology
All tumors demonstrated morphology typical of that described for Xp11 RCC. The tumors showed a nested and alveolar architecture, and papillary features (Figure 1A) were focally identified. The architecture was both nested and papillary in 6 cases, predominantly nested in 2 cases, and predominantly papillary in 1 case. The neoplastic cells were polygonal and had voluminous cytoplasm, a distinct cell border, and vesicular chromatin. Prominent nucleoli with predominantly clear cytoplasm (Figure 1B) were seen in 4 cases, predominantly eosinophilic and clear cytoplasm was seen in 4 cases, and well-developed areas of eosinophilic cytoplasm were seen in 1 case. Psammomatous calcifications were present in 7 cases (Figure 1C) and were numerous and widespread in 2 cases. Neoplastic cell metastasis to the lymph nodes occurred in 2 cases (Figure 1D).
Figure 1.
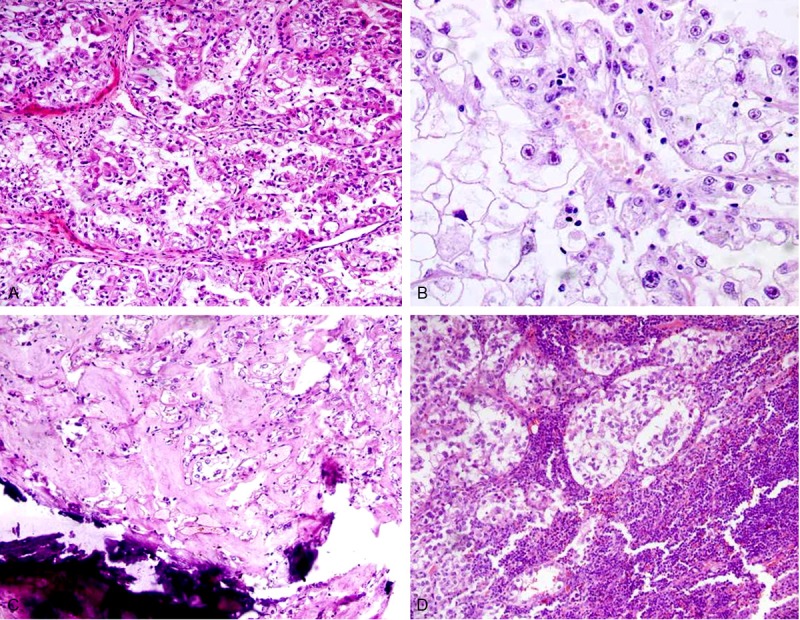
Microscopic findings of Xp11.2 RCC. A: Neoplastic cells intermingled with clear and eosinophilic cytoplasm proliferate in a papillary/nested growth pattern (×200). B: Voluminous tumorous cells with clear cytoplasm and prominent nucleoli proliferate in a nested pattern (×200). C: Psammomatous calcifications are seen in the stroma (×100). D: Neoplastic cell metastasis to the retroperitoneal lymph nodes (×100).
Immunohistochemical analysis
The IHC findings of 9 cases of Xp11.2 RCC and 12 cases of ASPS are summarized in Table 2. All tumors demonstrated nuclear labeling for TFE3 protein by IHC as an inclusion criterion for this study (Figure 2A, 2B). All Xp11.2 RCC cases were positive for the papillary RCC (PRCC) marker antigen AMACR (Figure 2C); in contrast, all 12 ASPS were AMACR negative (p<0.001). Six of 9 Xp11.2 RCC cases were either focally immunoreactive or positive for cytokeratin AE1/AE3, while all 12 ASPS were negative (p=0.002). Seven of 9 Xp11.2 RCC cases were positive for the renal tubular marker CD10 (Figure 2D), and only 33.3% (4/12) cases of ASPS partly expressed CD10 (p=0.024). Both Xp11.2 RCC and ASPS were highly positive for p53 and vimentin.
Table 2.
Immunohistological features of Xp11.2 renal cell carcinoma (RCC) and alveolar soft part sarcoma (ASPS)
| Antigen | Xp11.2 RCC (%) | ASPS (%) | p value |
|---|---|---|---|
| TFE3 | 9 (100) | 12 (100) | |
| AMACR | 9 (100) | 0 (0.0) | <0.001 |
| CD10 | 8 (88.9) | 4 (33.3) | 0.024 |
| CK | 6 (66.7) | 0 (0.0) | 0.002 |
| Vimentin | 7 (77.8) | 7 (58.3) | 0.642 |
| p53 | 6 (66.7) | 10 (88.3) |
Figure 2.
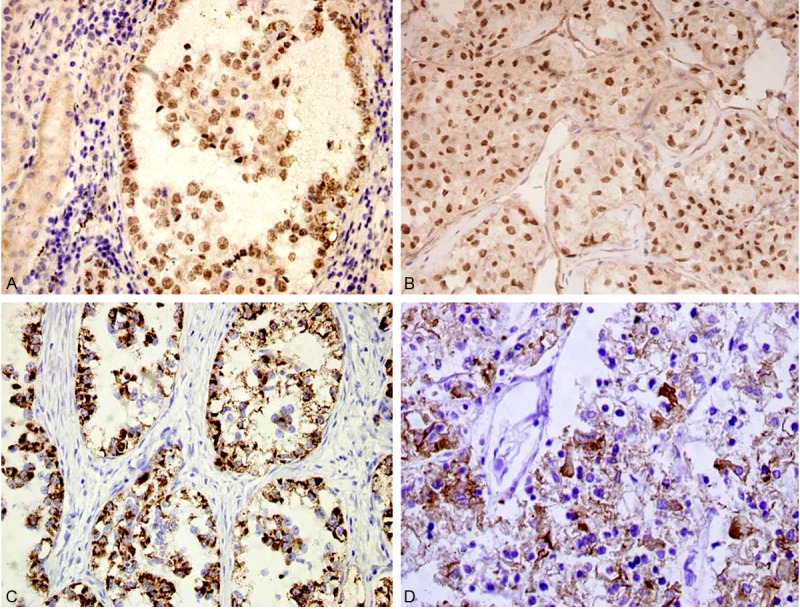
Immunohistochemical findings. A: Xp11.2 RCC shows diffuse intense nuclear labeling for TFE3. The adjacent benign renal parenchyma is negative for TFE3 (×200). B: ASPS shows diffuse intense nuclear labeling for TFE3 (×200). C: Xp11.2 RCC shows diffuse cytoplasm immunoreactivity with AMACR (×200). D: Xp11.2 RCC shows cell membrane immunoreactivity with CD10 (×200)
Comparative genomic hybridization findings
The CGH profiles showed chromosomal imbalance in all 9 cases (Table 3; Figure 3), with 68 gains and 40 losses. The mean numbers of aberrations per tumor sample were 8.1 gains and 5 losses.
Table 3.
Chromosome aberrations in Xp11.2 renal cell carcinoma (RCC)
| Chromosome number | Gain | Number (n=9) | Loss | Number (n=9) |
|---|---|---|---|---|
| 1 | 1q21 | 1 | ||
| 2 | 2q24 | 2 | ||
| 3 | 3p12-14 | 4 | ||
| 5 | 5q21-23 | 3 | ||
| 7 | 7p21-22 | 4 | ||
| 7q21-31 | 5 | |||
| 8 | 8p12 | 4 | ||
| 8q21 | 4 | |||
| 9 | 9q31-32 | 4 | ||
| 12 | 12q24-ter | 5 | ||
| 13 | 13q14-21 | 2 | ||
| 14 | 14q22-24 | 4 | ||
| 16 | 16q21-22 | 4 | 16p12-13 | 3 |
| 17 | 17p12-13 | 2 | ||
| 17q25-ter | 4 | |||
| 19 | 19p13 | 2 | ||
| 20 | 20q13-ter | 4 | ||
| X | Xp11 | 6 | ||
| Xq21 | 2 |
Figure 3.
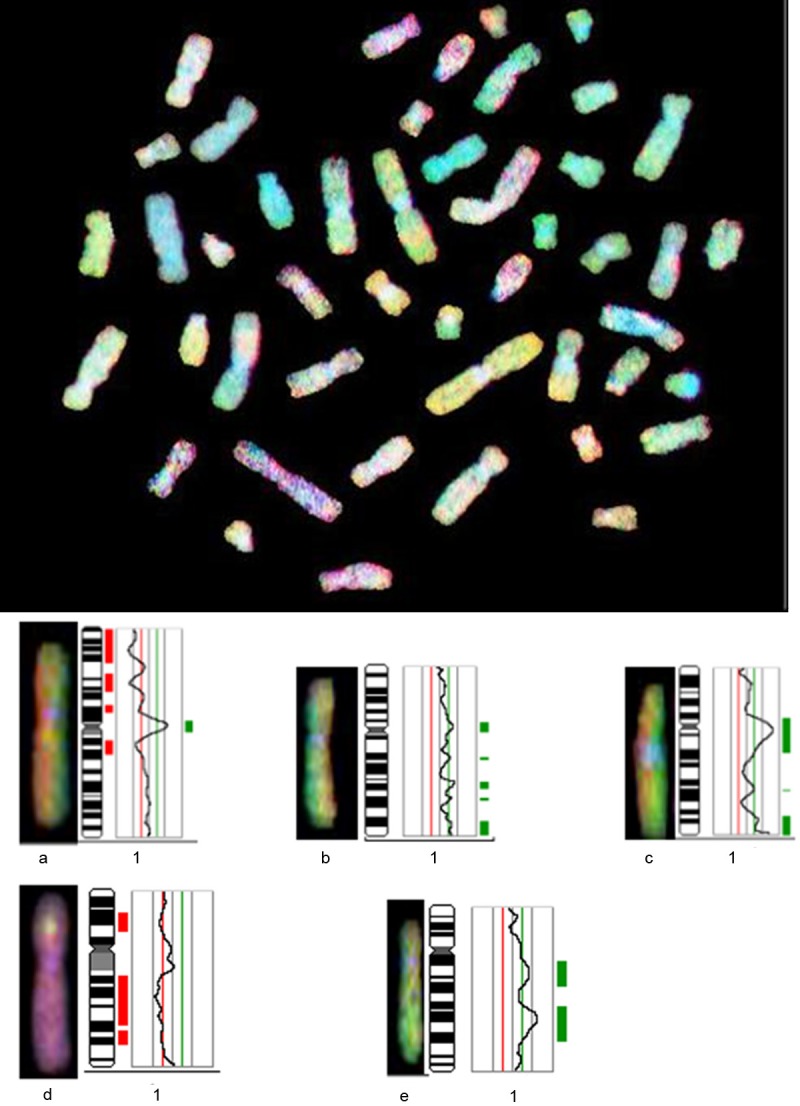
Comparative genomic hybridization profile of chromosome 1. Green to red fluorescent thresholds (represented by the green/red line) are 0.8 and 1.25, respectively. The curve shows the DNA copy number statues. Curves to the left of the red line indicate losses; curves to the right indicate gains; a, b, c, d, and e represent Xp11.2 RCC cases 1, 2, 3, 4, and 7, respectively.
Discussion
RCC associated with Xp11.2 translocations/TFE3 gene fusions is very rare. This tumor frequently occurs in children [5-7,12,13], but rarely in adults [6,8,9,14]. In children and young adults, Xp11.2 RCC is believed to be indolent even whendiagnosed at an advanced stage with regional lymph node metastasis and without distant metastasis. The current study reveals that Xp11.2 RCC is inherently more aggressive in adults than in children [6,8,9,15-17]. In our group, the age of the Xp11.2 RCC patients ranged from 25 to 75 years (mean, 40.6 years); 5 of 9 cases presented with stages 3-4, and 6 patients died 10 months to 7 years following their operation. Our report demonstrates that Xp11.2 RCC in adults behaves in a more aggressive fashion than in pediatric patients. However, there seems to be clinical heterogeneity even in adults [8], and its clinical and/or molecular basis remains to be interpreted.
The distinctive morphology of Xp11.2 RCC, a carcinoma composed of cells with abundant clear or eosinophilic cytoplasm growing with a nested and papillary architecture and forming psammoma bodies, suggests that the diagnosis on routine hematoxylin and eosin sections may overlap significantly with clear cell RCC (CCRCC) and PRCC in adults. The expression of CD10, vimentin, CD117, AMACR, CK7, Cathepsin K, and TFE3 are helpful in the differential diagnosis of Xp11.2 RCC, CCRCC, and PRCC [4,18,19]. Other neoplasms that should be included in the differential diagnosis are chromophobe RCC, collecting duct carcinoma, mucinous tubular and spindle cell carcinoma, sarcomatoid carcinoma, CCPRCC, epithelioid angiomyolipoma, and renal carcinoma t(6;11)(p21;q12-13)1. However, we decided to examine the relationship between Xp11.2 RCC and ASPS. ASPS is a rare soft tissue sarcoma, occasionally presenting in the kidney [11]. Both Xp11.2 RCC and ASPS possess the t(X;17)(p11.2;q25) chromosomal translocation that forms the ASPL-TFE3-fusion gene, which shows moderate-to-strong immunoreactivity with the TFE3 antibody [10,11,20]. Histologically, both tumors can form a nested and alveolar architecture [6,8,11,18,21,22]. Our study found that there are significant differences in the expression of AMACR (p<0.001), AE1/AE3 (p=0.002), and CD10 (p=0.024) in Xp11.2 RCC and ASPS cases. Therefore, these 3 antibodies may be useful in the differential diagnosis of these 2 diseases.
The molecular genetics of Xp11.2 RCC are summarized in Table 4 [8,18,21,23-27]. There are 8 TFE3 gene fusions partners reported to date; the molecular identity of 5 of these are known (62.5%): PRCC, polypyrimidine tract-binding protein-associated splicing factor (PSF), ASPL, non-POU domain-containing octamer-binding (NONO; p54nrb), andclathrin heavy-chain (CLTC) genes, situated on chromosomes 1q21, 1p34, 17q25, Xq12, and 17q23, respectively. The other 3 novel chromosomal translocations situated on chromosomes 3, 10, and 19 have been identified; however, the partner genes remain unknown [8,18,21,23-27]. The ASPL-TFE3 fusion protein binds to the MET promoter and strongly activates it [28]. Similarly, the PSF-TFE3 and NONO-TFE3 fusion proteins also bind to this promoter [24,28,29]. Compared with chromosomal translocations, other chromosome abnormality reports are rare. Altinok et al. found chromosome 7, 8, 12, and 17 trisomy; gain of the X chromosome; and loss of the Y chromosome in 4 cases of Xp11.2 RCC by fluorescence in situ hybridization (FISH) [3]. Deletion of 3p25-26 was reported in 1 case [30,31], and 1 case of a 3-year-old child with Xp11.2 RCC was found coexistent with a von Hippel-Lindau (VHL) gene mutation [30].
Table 4.
Reported cytogenetic abnormalities involving Xp11.2 translocation RCC
| Cytogenetic translocations involving Xp11.2 translocation RCC | Other genetic abnormalities | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Chromosome Translocation | Gene Fusion | Neoplasm | Source, year | Chromosome or gene aberrations | Source, year |
| t(X;1)(p11.2;q21) | PRCC-TFE3 | RCC | Argani et al, 16 2007 | t(X;1)(p11.2;p34) coexistent VHL gene mutation | Parast et al, 2004 |
| t(X;1)(p11.2;p34) | PSF-TFE3 | RCC | Argani et al, 16 2007 | ||
| t(X;17)(p11.2;q25) | ASPL-TFE3 | RCC | Argani et al, 16 2007 | ||
| inv(X)(p11.2;q12) | NONO-TFE3 | RCC | Argani et al, 16 2007 | chromosome 7, 8, 12, 17 trisomy, +add(X), loss of the Y | Altinok et al, 2005 |
| t(X;17)(p11.2;q23) | CLTC-TFE3 | RCC | Argani et al, 8 2003 | ||
| t(X;3)(p11.2;q23) | Unknown | RCC | Argani et al, 16 2007 | ||
| t(X;10)(p11.2;q23) | Unknown | RCC | Dijkuizen et al, 1995 | deletion of 3p25-26 | Bruder et al, 2004 |
| t((X;19)(p11.2;q13.1) | Unknown | RCC | Armah et al, 2009 | ||
As there are so many chromosomal translocation subtypes, it is relatively complex to identify Xp11.2 RCC by conventional cytogenetics and RT-PCR. The break-apart FISH assay on paraffin-embedded tumor tissue may be a helpful ancillary technique in small biopsies or fine-needle aspiration materials for Xp11.2 RCC [32-34], but it cannot find other chromosomal changes. When compared to conventional cytogenetics and FISH, CGH is a convenient and rapid method for screening for chromosomal genomic changes, and application of these technique aids our understanding of the molecular basis of Xp11.2 RCC.
In this preliminary study, we undertook genome-wide screening to detect genetic changes associated with the clinical parameters of primary Xp11.2 RCC. We detected DNA gains and losses in all 9 cases investigated. Furthermore, gains were more common than losses. Gains (in order of frequency) were detected at chromosomes Xp11 (6/9), 7q21-31, 12q24-ter (5/9), 7p21-22 (4/9), 8p12, 8q21, 16q21-22, 17q25, 20q13-ter (4/9), 5q21-23 (3/9), and 17p12-13 (2/9), and losses occurred frequently on chromosome 3p12-14, 9q31-32, 14q22-24 (4/9), 16p12-13 (3/9) and 2q24, 13q14-21, 19p-13 (2/9). Our study showed that 6 of 9 cases have chromosome Xp11 gains in the region of the TFE3 gene. Interestingly, in this series, 1 of these 6 cases lost the 1q21 region, which is related to chromosome translocation t(X;1)(p11.2;q21), and the PRCC gene is located in this region [18]; 2 of these cases lost the 19p13 region related to the chromosome translocation type t(X;19)(p11.2;q13.1) [18]. Four cases gained chromosome 17q25, which is a classical chromosome translocation type t(X;17)(p11.2;q25) and forms the ASPL-TFE3 fusion gene [18]. These results provide a clue to the chromosome translocation and gene fusion. The CGH assay may be a useful complementary method to confirm Xp11.2 RCC diagnosis.
Our study also showed some regions with a high frequency of chromosomal abnormalities. The 7q21-31 loci was a frequently amplified in Xp11.2 RCC patients (5/9), suggesting that it is associated with carcinogenesis. MET is an oncogene, which maps onto chromosome 7q31 and codes for a receptor tyrosine kinase. Argani et al. suggests that MET tyrosine kinase or mTOR kinase may be a potential therapeutic target in the future, and our study supports this hypothesis.
Other high-frequency regions containing chromosomal abnormalities include the gain of 12q24-ter (5/9), 7p21-22 (4/9), and 8p12, 8q21, 16q21-22, 17q25, 20q13-ter (4/9) and losses of chromosome 3p12-14, 9q31-32, 14q22-24 (4/9), and 16p12-13 (3/9). This region may provide further clues to improve our understanding of the molecular basis of Xp11.2 RCC (Table 5). For example, hypoxia-inducible factor 1 (HIF-1) is located in the 14q22-24 region. This gene encodes the alpha subunit of transcription factor HIF-1, which is a heterodimer composed of an alpha and a beta subunit. HIF-1 functions as a master regulator of cellular and systemic homeostatic response to hypoxia by activating transcription of many genes, including those involved in energy metabolism, angiogenesis, apoptosis, and other genes whose protein products increase oxygen delivery or facilitate metabolic adaptation to hypoxia. HIF-1 thus plays an essential role in embryonic vascularization, tumor angiogenesis, and the pathophysiology of ischemic disease. HIF-1 may be a potential therapeutic target for Xp11.2 RCC in the future.
Table 5.
Gene loci in Xp11.2 translocation RCC chromosomal abnormalities
| Chromosomal abnormality region | Gene loci |
|---|---|
| +12q24-ter | ALDH2, PTPN11, NOS1, HNF1A, UBC |
| +7p21-22 | HGF, ABCB1, PON1, CYP3A5, CYP3A4, EPO, SERPINE1 |
| +8p12 | WRN, BRG1, ADRB3, FGFR1, IDO1 |
| +8q21 | NBN |
| +16q21-22 | E-cadherin, CETP, MMP2, NDO1, HP |
| +17q25 | BIRC5, GRB2, ASPL |
| +20q13-ter | CEBPB, PTPN1, AURKA, GNAS |
| -3p12-14 | GPR27 |
| -9q31-32 | ABCA1, TXN |
| -14q 22-24 | BMP4, FOS, PSEN1, HIF-1 |
| -16p12-13 | HBA2, HBA1, TSC2 |
In conclusion, adult Xp11.2 RCC has the potential to be an aggressive cancer that requires morphologic distinction from CCRCC, PRCC, and ASPS. The expressions of TFE3, AMACR, CD10, and CK are helpful in the differential diagnosis of Xp11.2 RCC. CGH analysis revealed novel genomic imbalances in primary Xp11.2 RCC and may not only be a useful complementary method to confirm Xp11.2 RCC diagnosis, but also deepen our understanding of the molecular basis of Xp11.2 RCC. Our study demonstrates that CGH is a reliable tool for detecting alterations in large, critical chromosomal regions in Xp11.2 RCC. Further analysis to map genes to specific regions is underway in our laboratory and is aimed at determining the contributions of these genes to the development of Xp11.2 RCC.
Acknowledgements
This work is Supported by grants from the National Natural Science Foundation of China (NSFC, No. 81060209, 81160322) and from the International S&T Cooperation Program of China (2010DFB34100).
Disclosure of conflict of interest
None.
References
- 1.Kuroda N, Mikami S, Pan CC, Cohen RJ, Hes O, Michal M, Nagashima Y, Tanaka Y, Inoue K, Shuin T, Lee GH. Review of renal carcinoma associated with Xp11.2 translocations/TFE3 gene fusions with focus on pathobiological aspect. Histol Histopathol. 2012;27:133–140. doi: 10.14670/HH-27.133. [DOI] [PubMed] [Google Scholar]
- 2.Hammerschmied C, Denzinger S, Argani P, Otto W, Blaszyk H, Walter B, Hartmann A. Xp11.2/TFE3 translocation tumors are infrequent in unselected sporadic renal cell carcinoma (RCC) in young patients. Diagn Mol Pathol. 2007;203:311. [Google Scholar]
- 3.Altinok G, Kattar MM, Mohamed A, Poulik J, Grignon D, Rabah R. Pediatric renal carcinoma associated with Xp11.2 translocations/TFE3 gene fusions and clinicopathologic associations. Pediatr Dev Pathol. 2005;8:168–180. doi: 10.1007/s10024-004-9106-3. [DOI] [PubMed] [Google Scholar]
- 4.Argani P, Hicks J, De Marzo AM, Albadine R, Illei PB, Ladanyi M, Reuter VE, Netto GJ. Xp11 translocation renal cell carcinoma (RCC): extended immunohistochemical profile emphasizing novel RCC markers. Am J Surg Pathol. 2010;34:1295–1303. doi: 10.1097/PAS.0b013e3181e8ce5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruder E, Moch H. [Pediatric renal cell carcinoma] . Der Pathologe. 2004;25:324–327. doi: 10.1007/s00292-004-0699-0. [DOI] [PubMed] [Google Scholar]
- 6.Armah HB, Parwani AV. Xp11.2 translocation renal cell carcinoma. Arch Pathol Lab Med. 2010;134:124–129. doi: 10.5858/2008-0391-RSR.1. [DOI] [PubMed] [Google Scholar]
- 7.Argani P, Ladanyi M. Recent advances in pediatric renal neoplasia. Adv Anat Pathol. 2003;10:243–260. doi: 10.1097/00125480-200309000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Argani P, Olgac S, Tickoo SK, Goldfischer M, Moch H, Chan DY, Eble JN, Bonsib SM, Jimeno M, Lloreta J, Billis A, Hicks J, De Marzo AM, Reuter VE, Ladanyi M. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–1160. doi: 10.1097/PAS.0b013e318031ffff. [DOI] [PubMed] [Google Scholar]
- 9.Higa B, Flanigan RC, Picken MM. Clinicopathological features of 22 cases of TFE3 renal carcinoma from adult patients. Modern Pathology. 2008;21:161a–161a. [Google Scholar]
- 10.Argani P, Lal P, Hutchinson B, Lui MY, Reuter VE, Ladanyi M. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol. 2003;27:750–761. doi: 10.1097/00000478-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Pang LJ, Chang B, Zou H, Qi Y, Jiang JF, Li HA, Hu WH, Chen YZ, Liu CX, Zhang WJ, Li F. Alveolar soft part sarcoma: a bimarker diagnostic strategy using TFE3 immunoassay and ASPL-TFE3 fusion transcripts in paraffin-embedded tumor tissues. Diagn Mol Pathol. 2008;17:245–252. doi: 10.1097/PDM.0b013e31815d68d7. [DOI] [PubMed] [Google Scholar]
- 12.Ramphal R, Pappo A, Zielenska M, Grant R, Ngan BY. Pediatric renal cell carcinoma: clinical, pathologic, and molecular abnormalities associated with the members of the mit transcription factor family. Am J Clin Pathol. 2006;126:349–364. doi: 10.1309/98YE9E442AR7LX2X. [DOI] [PubMed] [Google Scholar]
- 13.Winarti NW, Argani P, De Marzo AM, Hicks J, Mulyadi K. Pediatric renal cell carcinoma associated with Xp11.2 translocation/TFE3 gene fusion. Int J Surg Pathol. 2008;16:66–72. doi: 10.1177/1066896907304994. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda N, Katto K, Tanaka Y, Yamaguchi T, Inoue K, Ohara M, Mizuno K, Hes O, Michal M, Lee GH. Diagnostic pitfall on the histological spectrum of adult-onset renal carcinoma associated with Xp11.2 translocations/TFE3 gene fusions. Med Mol Morphol. 2010;43:86–90. doi: 10.1007/s00795-008-0423-x. [DOI] [PubMed] [Google Scholar]
- 15.Chapman-Fredricks JR, Cioffi-Lavina M, Reyes C, Goldberg J, Gomez-Fernandez C, Jorda M. Expression of TFE3 Protein in Adult Renal Cell Carcinoma. Modern Pathology. 2010;23:183a–183a. [Google Scholar]
- 16.Trilla E, Mir C, De Torres I, Pamizo J, Bestard JE, Salvador C, Lopez-Pacios MA, Orsola A, Cecchini LL, Busquets CR, Morote J. 139 Clinicopathological Characterization of Adult Renal Cell Carcinoma with Xp11 Translocation (Fusion Gen Tfe3) European Urology Supplements. 2009;8:155. [Google Scholar]
- 17.Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda N, Mikami S, Pan CC, Cohen RJ, Hes O, Michal M, Nagashima Y, Tanaka Y, Inoue K, Shuin T, Lee GH. Review of renal carcinoma associated with Xp11.2 translocations/TFE3 gene fusions with focus on pathobiological aspect. Histol Histopathol. 2012;27:133–140. doi: 10.14670/HH-27.133. [DOI] [PubMed] [Google Scholar]
- 19.Zou H, Pang LJ, Hu WH, Li F, Li HA, Jiang JF, Liang WH, Sun ZZ, Wang C, Lang JY. [Study on clinicopathologic features and immunophenotype of 114 cases of renal cell carcinoma] . Zhonghua Bing Li Xue Za Zhi. 2008;37:726–731. [PubMed] [Google Scholar]
- 20.Moyano S, Aguilera P, Petit A, de Alava E, Mascaro JM, Palou J, Ferrando J, Alos L. Alveolar soft part sarcoma presenting with cutaneous metastases: report of a case with immunohistochemical and molecular characterization. J Am Acad Dermatol. 2009;61:117–120. doi: 10.1016/j.jaad.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 21.Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, Reuter VE, Garvin AJ, Perez-Atayde AR, Fletcher JA, Beckwith JB, Bridge JA, Ladanyi M. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–192. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang WD, Heo SH, Choi YD, Choi HS, Kim SM. Alveolar soft part sarcoma of the uterine cervix in a woman presenting with postmenopausal bleeding: a case report and literature review. Eur J Gynaecol Oncol. 2011;32:359–361. [PubMed] [Google Scholar]
- 23.Argani P, Antonescu CR, Couturier J, Fournet JC, Sciot R, Debiec-Rychter M, Hutchinson B, Reuter VE, Boccon-Gibod L, Timmons C, Hafez N, Ladanyi M. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21) Am J Surg Pathol. 2002;26:1553–1566. doi: 10.1097/00000478-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Argani P, Lui MY, Couturier J, Bouvier R, Fournet JC, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23) Oncogene. 2003;22:5374–5378. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 25.Mathur M, Das S, Samuels HH. PSF-TFE3 oncoprotein in papillary renal cell carcinoma inactivates TFE3 and p53 through cytoplasmic sequestration. Oncogene. 2003;22:5031–5044. doi: 10.1038/sj.onc.1206643. [DOI] [PubMed] [Google Scholar]
- 26.Argani P, Ladanyi M. Translocation carcinomas of the kidney. Clin Lab Med. 2005;25:363–378. doi: 10.1016/j.cll.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Armah HB, Parwani AV, Surti U, Bastacky SI. Xp11.2 translocation renal cell carcinoma occurring during pregnancy with a novel translocation involving chromosome 19: a case report with review of the literature. Diagn Pathol. 2009;4:15. doi: 10.1186/1746-1596-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuda M, Davis IJ, Argani P, Shukla N, McGill GG, Nagai M, Saito T, Lae M, Fisher DE, Ladanyi M. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007;67:919–929. doi: 10.1158/0008-5472.CAN-06-2855. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda N, Tamura M, Tanaka Y, Hes O, Michal M, Inoue K, Ohara M, Mizuno K, Lee GH. Adult-onset renal cell carcinoma associated with Xp11.2 translocations/TFE3 gene fusion with smooth muscle stroma and abnormal vessels. Pathol Int. 2009;59:486–491. doi: 10.1111/j.1440-1827.2009.02398.x. [DOI] [PubMed] [Google Scholar]
- 30.Parast MM, Eudy G, Gow KW, Amin M, Shehata B. A unique case of renal carcinoma with Xp11.2 translocations/ TFE3 gene fusions in a 3-year-old child, with coexistent von Hippel-Lindau gene mutation. Pediatr Dev Pathol. 2004;7:403–406. doi: 10.1007/s10024-004-1018-8. [DOI] [PubMed] [Google Scholar]
- 31.Bruder E, Passera O, Harms D, Leuschner I, Ladanyi M, Argani P, Eble JN, Struckmann K, Schraml P, Moch H. Morphologic and molecular characterization of renal cell carcinoma in children and young adults. Am J Surg Pathol. 2004;28:1117–1132. doi: 10.1097/01.pas.0000131558.32412.40. [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Choi Y, Jeong HY, Lee K, Chae JY, Moon KC. Usefulness of a break-apart FISH assay in the diagnosis of Xp11.2 translocation renal cell carcinoma. Virchows Arch. 2011;459:299–306. doi: 10.1007/s00428-011-1127-5. [DOI] [PubMed] [Google Scholar]
- 33.Mosquera JM, Dal Cin P, Metz KD, Perner S, Davis IJ, Fisher DE, Rubin MA, Hirsch MS. Validation of a TFE3 break-apart FISH assay in Xp11.2 translocation renal cell carcinomas. Diagn Mol Pathol. 2008;21:172a–172a. doi: 10.1097/PDM.0b013e31820e9c67. [DOI] [PubMed] [Google Scholar]
- 34.Zhong M, De Angelo P, Osborne L, Keane-Tarchichi M, Goldfischer M, Edelmann L, Yang Y, Linehan WM, Merino MJ, Aisner S, Hameed M. Dual-color, break-apart FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of Xp11 translocation renal cell carcinoma and alveolar soft part sarcoma. Am J Surg Pathol. 2010;34:757–766. doi: 10.1097/PAS.0b013e3181dd577e. [DOI] [PMC free article] [PubMed] [Google Scholar]


