Abstract
The DNA of helper-dependent coliphage P4 and the DNA of its helper—P2—show no detectable sequence homology as measured by DNA·DNA hybridization. The lack of cross-hybridization permits direct analysis of P4 as well as of P2 transcription in P4-infected P2 lysogens by RNA·DNA hybridization. P4-transactivated P2 transcription can be detected around 20 min after P4 infection of the P2 lysogen and the rate (per infected cell) of that transcription becomes equal to that of the P4 transcription at the end of the latent period of P4. Furthermore, P4 transcription appears to be stimulated by the presence of the helper. Conceivably, P2 codes for a stimulator of P4 transcription. Rifamycin has been used to investigate the role of the host RNA polymerase during P4 transactivation of P2 transcription. The results exclude the participation of a P4-coded RNA polymerase and indicate that the original host RNA polymerase is responsible for the bulk of P4 and P2 transcription during transactivation.
Keywords: satellite phage, sequence homology, chloramphenicol, rifamycin, transactivation
Full text
PDF
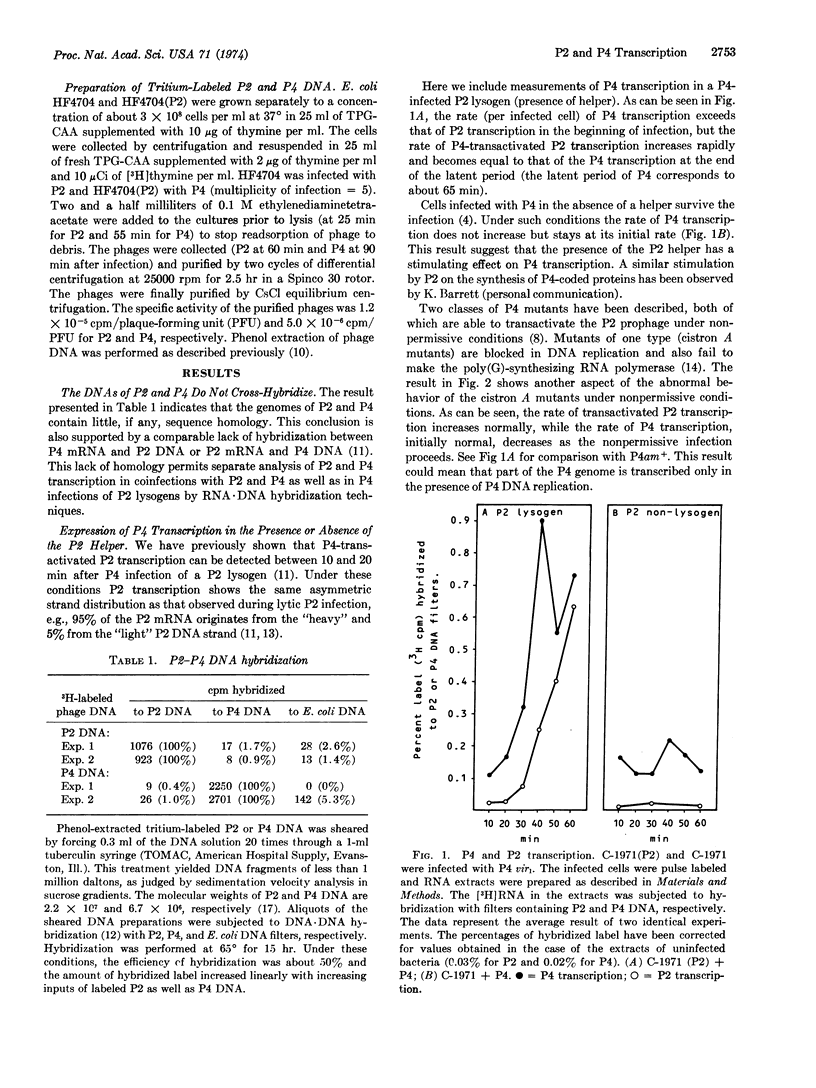
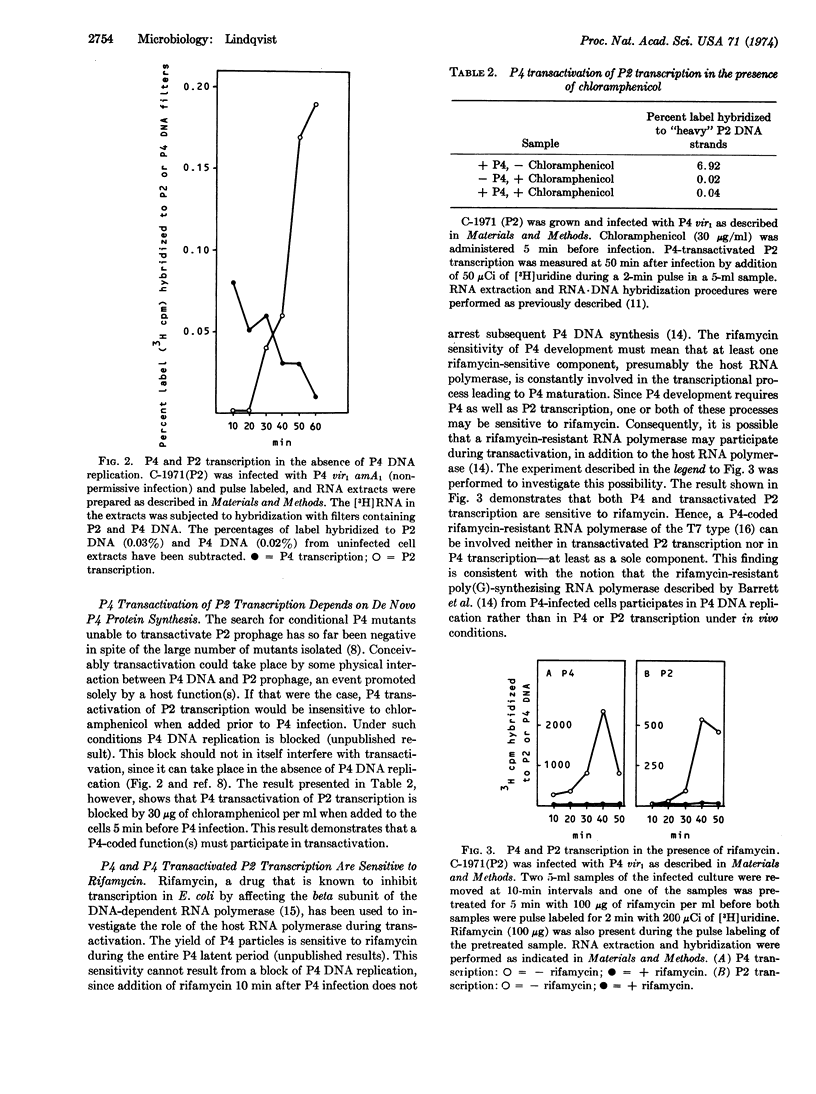

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett K. J., Gibbs W., Calendar R. A transcribing activity induced by satellite phage P4. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2986–2990. doi: 10.1073/pnas.69.10.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett K., Calendar R., Gibbs W., Goldstein R. N., Lindqvist B., Six E. Helper-dependent bacteriophage P4: a model satellite virus and its implications for animal virology. Prog Med Virol. 1973;15:309–330. [PubMed] [Google Scholar]
- Bertani L. E., Bertani G. Preparation and characterization of temperate, non-inducible bacteriophage P2 (host: Escherichia coli). J Gen Virol. 1970 Feb;6(2):201–212. doi: 10.1099/0022-1317-6-2-201. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Geisselsoder J., Mandel M., Calendar R., Chattoraj D. K. In vivo transcription patterns of temperate coliphage P2. J Mol Biol. 1973 Jul 5;77(3):405–415. doi: 10.1016/0022-2836(73)90447-6. [DOI] [PubMed] [Google Scholar]
- Gibbs W., Goldstein R. N., Wiener R., Lindqvist B., Calendar R. Satellite bacteriophage P4: characterization of mutants in two essential genes. Virology. 1973 May;53(1):24–39. doi: 10.1016/0042-6822(73)90462-5. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Schnös M., Simon L. D., Six E. W., Walker D. H., Jr Some morphological properties of P4 bacteriophage and P4 DNA. Virology. 1971 Apr;44(1):67–72. doi: 10.1016/0042-6822(71)90153-x. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Bovre K. Asymmetric transcription of the coliphage P2 genome during infection. Virology. 1972 Sep;49(3):690–699. doi: 10.1016/0042-6822(72)90526-0. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Sinsheimer R. L. Process of infection with bacteriophage phi-X174. XIV. Studies on macromolecular synthesis during infection with a lysis-defective mutant. J Mol Biol. 1967 Aug 28;28(1):87–94. doi: 10.1016/s0022-2836(67)80079-2. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Six E. W. Replication of bacteriophage P4 DNA in a nonlysogenic host. Virology. 1971 Jan;43(1):1–7. doi: 10.1016/0042-6822(71)90218-2. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H. Vegetative DNA of temperate coliphage P2. Mol Gen Genet. 1971;110(2):178–196. doi: 10.1007/BF00332647. [DOI] [PubMed] [Google Scholar]
- Ljungquist E. Interaction of phage P2 DNA with some fast-sedimenting host components during infection. Virology. 1973 Mar;52(1):120–129. doi: 10.1016/0042-6822(73)90403-0. [DOI] [PubMed] [Google Scholar]
- Six E. W., Klug C. A. Bacteriophage P4: a satellite virus depending on a helper such as prophage P2. Virology. 1973 Feb;51(2):327–344. doi: 10.1016/0042-6822(73)90432-7. [DOI] [PubMed] [Google Scholar]
- Six E. W., Lindqvist B. H. Multiplication of bacteriophage P4 in the absence of replication of the DNA of its helper. Virology. 1971 Jan;43(1):8–15. doi: 10.1016/0042-6822(71)90219-4. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Martin K. V., Calendar R. On the sequence similarity of the cohesive ends of coliphage P4, P2, and 186 deoxyribonucleic acid. Biochemistry. 1973 May 22;12(11):2119–2123. doi: 10.1021/bi00735a016. [DOI] [PubMed] [Google Scholar]



