Abstract
Here we describe a method for the detection of Clostridium difficile from stool using a novel low-complexity and rapid extraction process called Heat Elution (HE). The HE method is two-step and takes just 10 minutes, no specialist instruments are required and there is minimal hands-on time. A test method using HE was developed in conjunction with Loop-mediated Isothermal Amplification (LAMP) combined with the real-time bioluminescent reporter system known as BART targeting the toxin B gene (tcdB). The HE-LAMP-BART method was evaluated in a pilot study on clinical fecal samples (tcdB +, n = 111; tcdB −, n = 107). The HE-LAMP-BART method showed 95.5% sensitivity and 100% specificity against a gold standard reference method using cytotoxigenic culture and also a silica-based robotic extraction followed by tcdB PCR to control for storage. From sample to result, the HE-LAMP-BART method typically took 50 minutes, whereas the PCR method took >2.5 hours.
In a further study (tcdB +, n = 47; tcdB −, n = 28) HE-LAMP-BART was compared to an alternative commercially available LAMP-based method, Illumigene (Meridian Bioscience, OH), and yielded 87.2% sensitivity and 100% specificity for the HE-LAMP-BART method compared to 76.6% and 100%, respectively, for Illumigene against the reference method. A subset of 27 samples (tcdB +, n = 25; tcdB −, n = 2) were further compared between HE-LAMP-BART, Illumigene, GeneXpert (Cepheid, Sunnyvale, CA) and RIDA®QUICK C. difficile Toxin A/B lateral flow rapid test (R-Biopharm, Darmstadt, Germany) resulting in sensitivities of HE-LAMP-BART 92%, Illumigene 72% GeneXpert 96% and RIDAQuick 76% against the reference method. The HE-LAMP-BART method offers the advantages of molecular based approaches without the cost and complexity usually associated with molecular tests. Further, the rapid time-to-result and simple protocol means the method can be applied away from the centralized laboratory settings.
Introduction
Clostridium difficile is a Gram positive bacterium that is the most frequent cause of infectious bacterial diarrhea worldwide [1]. C. difficile infection (CDI), which is associated with the use of broad spectrum antibiotics to treat other underlying conditions, results in the release of the two main virulence factors, toxins A and B [2], that result in mild to severe watery diarrhea with CDI [3]–[4].
An epidemic of CDI with continuously increasing rates was seen in the USA, Canada and most of Europe starting around 2000–2002. This epidemic was mostly associated with the hypervirulent ribotype 027/NAP1 B1 strain. This renewed interest in CDI towards pathophysiology, prevention, detection and treatment [5]–[7]. In England, voluntary surveillance showed an increase in CDI from 1990 to a peak of 52,983 reports in 2007. However, following successful implementation of control measures, there were 13,352 reports in 2012, a reduction of 75% [8]. Mandatory surveillance was introduced in April 2007 and confirms this fall in cases. This may be due to the emergence and subsequent decline of hypervirulent strains of C. difficile (e.g., ribotype 027/NAP1, responsible for 55% of isolates ribotyped in 2007/08, but only 12% of isolates in 2010/11) [9]. The 027/NAP1 strain has an expanded repertoire of antibiotic resistance elements, produces greater quantities of toxins, and therefore gives rise to more severe disease with higher mortality rates and an increased probability of relapse following clinical treatment [10]–[11].
Laboratory diagnosis of CDI from stool has traditionally been based on detecting the presence of C. difficile toxin A and/or toxin B proteins in stool by various methods including cytotoxicity assays and Enzyme Immuno Assay (EIA) [12]–[15]. In general, these methods should meet the minimum desirable characteristics of a diagnostic test of sensitivity, specificity, cost-efficiency, rapid results, ease of use preferably by non-expert users and ability to differentiate between toxigenic and non-toxigenic C. difficile. Cell cytotoxicity assays are sensitive, but are extremely time-consuming and their requirement for cell-culture prohibits their application for near-patient rapid testing. EIA tests for one or both toxins are relatively insensitive, detecting only 30% to 70% of CDI-related disease [16]–[20]. Moreover, toxin gene expression is known to be repressed in nutrient rich conditions [21]. With the development of an EIA for glutamate dehydrogenase (GDH), which is a C. difficile cell wall common antigen, the sensitivity for the detection of C. difficile approaches 100%; however, because GDH is ubiquitous for both toxigenic and non-toxigenic strains, specificity of the GDH assay is poor [11]. Therefore, individual EIA tests for either GDH or toxins A or B are considered to be insufficiently specific for diagnosis. However, the combination of GDH and toxin A/B, such as in the C. DIFF QUIK CHEK COMPLETE EIA assay (Techlab, Blacksburg, VA), gives increased diagnostic sensitivity and specificity provided the results for both analytes are concordant [14]–[15]. However, when GDH and toxin results are discordant, additional reflex or discrepant testing using a highly specific molecular-based assay is required [22].
Molecular-based assays for the detection of toxigenic C. difficile in stool offer increased sensitivity over and are as specific as EIA tests [16], [23]. Genes within the C. difficile pathogenicity locus, including those encoding toxins A and/or B (tcdA or tcdB), have been targeted by specific primer sets, usually in a multiplex PCR assay. Identification of these gene sequences in stool samples correlates highly with positive C. difficile toxigenic stool assays [17]. A positive PCR assay is therefore thought to directly indicate the presence of toxigenic C. difficile strains in stool [24]–[26]. Accordingly, new molecular assays such as the Loop-mediated Isothermal Amplification (LAMP) assay (Illumigene C. difficile; Meridian Bioscience, Cincinnati, OH) and the GeneXpert assay (Xpert C. difficile; Cepheid, Sunnyvale, CA), which detect pathogenicity locus region sequences, are being increasingly used for confirmatory testing of EIA tests. Nonetheless, despite greater diagnostic sensitivity and specificity, current molecular methods are too labor-intensive, complex, and/or costly to be of practical use in many clinical laboratories [13], [15], [25]. There is thus a need for a more facile and less costly molecular method to overcome the drawbacks of the current commercial molecular methods.
The fecal sample matrix represents a challenge to sample preparation for molecular methods due to its high inhibitor content. Inhibitors found in stool that are known to affect both PCR and isothermal methods, such as LAMP, include polyphenols [27]–[28], divalent cations [29], acidic polysaccharides [30]–[31], hemoglobin/hemin [32]–[33], phytic acid [34] and bile salts [35]. Current molecular tests commonly rely upon multi-step, time consuming silica-based extraction processes [36]. Whilst these methods can be automated, this adds significant cost particularly where the system is a “fully integrated” platform like the GeneXpert: often too expensive for many clinical settings. An alternative approach employed is to dilute the sample by 200–700 fold so as to reduce the concentration of inhibitors; however, such ‘dilutive’ methods significantly reduce test sensitivity. Hence there is a need for a simple, easy to use and inexpensive method that does not compromise test sensitivity.
To address these needs a novel sample preparation method was developed based on the principle of Heat Elution (HE). In this method a fecal sample swab is added to a column containing a buffered cocktail of resins selected to bind fecal inhibitors of nucleic acid amplification. The column is sealed, shaken and then placed into a collection tube having first broken off a bottom seal on the column which allows eluate to exit the column into the collection tube. The column and collection tube is then placed on a heating block at 100°C for 10 minutes. During this incubation, the C. difficile cells are lysed and the DNA released into the buffer and fecal inhibitors become bound to the inhibitor removal resins. As the column is heated, pressure inside the column builds which results in the gentle elution of the eluate from the column into the collection tube, leaving the inhibitors bound on the column. The use of the inhibitor removal matrix means that the sample ends up being diluted only 50 fold compared to the 200 to 700 fold dilutions used by dilutive methods. The eluate is then used to directly reconstitute lyophilized LAMP-BART reagent. The robustness of this method also means that both unformed, solid and blood containing stool samples can be tested; this means that it is possible to test for C. difficile carriers who are not presenting with symptoms of diarrhea.
The HE method is described herein in conjunction with Loop-mediated Isothermal Amplification (LAMP) employing the BART (bioluminescent assay in real-time) bioluminescent reporter system. The BART reporter system is designed to work with Isothermal Nucleic Acid Amplification Technologies (iNAATs) and has the advantage over fluorescent and turbidity reporter systems of requiring exceptionally simple, robust and low-cost hardware. The LAMP-BART combination has been well characterized, is robust to inhibitors and forms the basis of commercially available molecular test kits [37]–[39].
This article describes a pilot study of the Heat Elution method in combination with a LAMP-BART test for the toxin B gene of C. difficile (tcdB +). HE-LAMP-BART was compared to cytotoxigenic culture done when the samples were obtained and a PCR method developed by Public Health England (PHE) to control for sample degradation with storage. The PHE PCR was a home-brew multiplex PCR method using the easyMAG extraction method from bioMérieux that was modified by the Clinical Microbiology Laboratory at Addenbrooke’s Hospital (PHE), Cambridge, UK for stool extraction.
The test was also compared against a number of commercial tests, including the Illumigene, GeneXpert and RIDAQuick tests. In addition to the comparative performance on clinical stool specimens, we have also described the analytical sensitivity, performance with interferents as well as the inclusivity and exclusivity of the HE-LAMP-BART C. difficile test.
Materials and Methods
Requirement of ethical approval
It was confirmed by the Research Ethics Chair of Cambridgeshire 2 Research Ethics Committee that ethical approval was not required for the study because it was an evaluation of an established nucleic acid amplification technology that did not involve the use of linked samples, access to patient details, any additional sample other than that required for routine C. difficile screening nor would it screen for additional pathogens other than toxigenic C. difficile.
Bacterial strains and culturing
The following C. difficile strains were obtained from the National Collection of Type Cultures: NCTC 13307, NCTC 11205, NCTC 11209 and NCTC 11204. Non-toxigenic C. difficile strain ATCC 43593 was obtained from the American Type Culture Collection. Other organisms used were from an in-house Lumora culture collection. C. difficile strains were cultured anaerobically on blood agar (Oxoid), Brazier’s agar (Oxoid) or in brain heart infusion (Difco). Other organisms were cultured in tryptone soya agar (Oxoid) or buffered peptone water broth (Oxoid).
Stool specimens
Stool specimens sent to PHE Microbiology services-Cambridge from the East of England region were collected over a 17 month period (July 2010 to November 2011) and stored at -20°C. For this evaluation study we sought to include at least 100 known C. difficile positive stool specimens balanced with around the same number of negative specimens. Such a sample size would provide a 95% confidence interval of +/– 5.9% for an estimated test sensitivity (or specificity) of 90%, which is in line with recommended guidelines [40]. Of 218 stool specimens selected consecutively over the study period with sufficient volume to permit multiple testing, 196 were unformed and 22 were solid. The status of these samples had already been determined by VIDAS® Clostridium difficile A & B (bioMérieux, FR), toxigenic cell culture assay, culture and ribotyping, and comprised 111 C. difficile toxin positives and 107 negatives, and provided the gold standard reference test result for benchmarking. A broad range of different ribotypes were observed in the positive specimen group and are detailed below. Specimens were assigned numbers unique to the study and were blinded and aliquoted for the study.
DNA standards
Genomic DNA from C. difficile 630 was either a kind gift from Dr. Trevor Lawley at the Wellcome Trust Sanger Institute, Cambridge or purchased from ATCC.
Heat Elution extraction & LAMP-BART
Heat Elution extraction was performed as follows (Figure 1). Each stool sample was swabbed with a micro ultrafine tipped flocked swab (Puritan, Guilford, ME). The swab was placed in the HE column and mixed by rotating the swab between the fingers. The shaft of the swab was then broken at the breakpoint and the swab sealed inside the column by putting the cap on the column and the sample then mixed in the column by gently shaking. The twist-off tab at the bottom of the column was then snapped off to open the column, which was placed into a collection tube and placed on a heat block at 100°C and incubated for 10 minutes. During this time inhibitors are bound to the inhibitor removal matrix cocktail, cells lysis occurs and liberated DNA is gently eluted in lysis/amplification buffer into the collection tube. The collection tube and column was then removed from the hot block and the column discarded. When cool, 20 µl of eluate from the collection tube was used to reconstitute lyophilized C. difficile LAMP-BART reagent. A further 20 µl of eluate was used to reconstitute Inhibitor Control LAMP-BART reagent. This reagent contains both primers and associated 104 copies of an artificial DNA sequence which has been well characterized for the effect of inhibitors on amplification. These reactions were then loaded onto a 16 well photodiode-based reader from Lumora (known as a PDQ). Data was collected over 90 minutes for each amplification. On the PDQ, light output from the reactions is displayed and positive results are called by an algorithm that looks for peaks in real time. Negative or undetermined results are called by the algorithm at the end of the run. This algorithm did not change over the course of the study and all peak results were confirmed by visual inspection. Each run carried a positive control of C. difficile genomic 630 DNA (ATCC) in lysis/amplification buffer and a negative control of just lysis/amplification buffer.
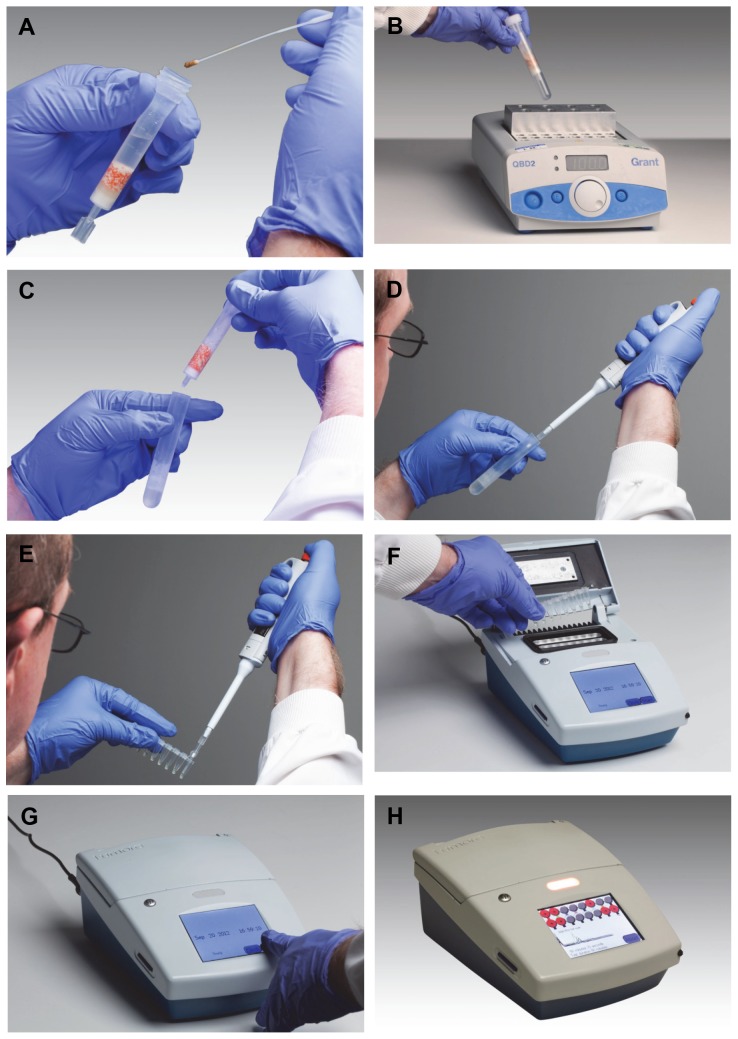
Figure 1. Photographic illustration of the steps of the HE-LAMP-BART.
A. Place fecal swab sample into the HE column, mix, seal and twist off the bottom tab of the column; B. Place the column in a collection tube on a hot block at 100°C and incubate for 10 minutes; C. Remove the collection tube and dispose of the column; D&E. Pipette 20 µl of eluate from the collection tube and use this to reconstitute LAMP-BART freeze dried reagent; F&G. Place reconstituted reagent on PDQ and start run; H. Positive results are called in real time. The person in the photograph has given written informed consent, as outlined in the PLOS consent form, to publication of their photograph.
easyMAG extraction and PHE multiplexed real time PCR
Samples of faecal materials were taken using regular flocked swabs (microRheologics; REF 520CS01). Swabs were broken off into screw-cap Eppendorf tubes containing 1 ml 1x Tris-EDTA (TE) buffer (Becton Dickinson) and vortexed at full speed for 10 s. 500 µl of the resultant suspensions were transferred to lysis tubes (Becton Dickinson) containing glass beads (425–600 µm) and vortexed at full speed for 5 mins. 200 µl of the resultant lysates were added to 2 ml lysis buffer (bioMérieux) and nucleic acids extracted using the Generic 2.0.1 protocol with ‘off-board’ lysis on the easyMAG instrument (bioMérieux). Purified nucleic acids were eluted in 85 µl, of which 5 µl were used as template in 25 µl real-time PCR reactions.
PCR assays contained: 5 µl of nucleic acid sample, 12.5 µl of 2x Platinum Quantitative PCR SuperMix-UDG (Invitrogen), 0.5 µl of MgCl2 (50 mM), 1.0 µl of Internal control oligonucleotide, each of the corresponding forward and reverse primers and corresponding probes at the concentrations given in Table 1, and DNase-free water (Invitrogen), giving a final volume of 25 µl. All primers and probes were purchased from Metabion, Munich, Germany, with the exception of the two minor groove binding (MGB) Taqman® probes which were purchased from Applied Biosystems, Warrington, UK (Sequences are described in Table 1).
Table 1. Primers and probes used for real-time PCR.
| Target | Sequence (5’ → 3’) | Function (concentration) | Product size | Primer/Probe source |
| tcdB | CCAAARTGGAGTGTTACAAACAGGTGTA | Forward primer (800 nM) | 99 bp | This Study |
| GCTTCICCYTCTAGRTTTTCATCAAGTGTA | Reverse primer (400 nM) | |||
| VIC – CCATCTTCTGTACTAA - MGB | Probe (80 nM) | |||
| GDH | GTAATACCAGCTGCATTAGAAAACTC | Forward primer (400 nM) | 89 bp | This Study |
| GGTCCATTAGCAGCCTCACAA | Reverse primer (400 nM) | |||
| 6FAM – TAGATTCAGCAACTTC - MGB | Probe (80 nM) | |||
| Internal | CTCTGCTTTATATTATAAAATTACGGCTG | Forward primer (400 nM) | 104 bp | This Study |
| Control | ACTTTAGTCAAATCATCTTCACTAGTG | Reverse primer (400 nM) | ||
| (IC) | ||||
| ROX – | Probe (100 nM) | |||
| CACATCGATAGATCAAGGTGCCTACAAGC - | ||||
| BHQ2 | ||||
| IC | CTCTGCTTTATATTATAAAATTACGGCTGGGCGTTAAGTGTCACATCGATAGATCAAGGTGCCTACAAGCGAAGTGGCACTAGTGAAGATGATTTGACTAAAGT | This Study |
A specially designed Internal control oligonucleotide DNA (approximately 1000 copies per reaction) was added to real-time PCR reactions as a control for PCR inhibition (Table 1). The control was detected in multiplex real-time PCR reactions concomitantly with the target pathogen nucleic acids. Real-time PCR assays were performed using a Rotor-Gene Q (Qiagen), according to the following profile: 50°C for 120 s, 95°C for 120 s, and then 45 cycles of 95°C for 10 s and 60°C for 60 s. Fluorescence data acquisition was performed at the end of each 60°C incubation period.
The PHE in-house real-time PCR assay was used in this study to provide a direct comparator NAAT for LAMP-BART ensuring any specimen degradation during storage did not unfairly bias the evaluation. While the in-house PCR assay is not used routinely in our diagnostic service (PHE) its performance has been fully assessed and validated. Briefly, 90 consecutive stool specimens sent for C. difficile testing to our laboratory were also processed on the GeneXpert system and our in-house real-time assay. Complete concordance was observed between the two methods, with 17 C. difficile toxigenic positive stools and 73 negative stools, which was also in agreement with our routine diagnostic results. A range of C. difficile isolates with different ribotypes (001, 002, 003, 005, 012, 014, 015, 016, 017, 020, 023, 026, 027, 029, 046, 050, 072, 078, 081, 106, 107, 137, 174) were all detected by the PHE RT PCR assay and it passed a specificity PCR check with a comprehensive bank of chromosomal DNA extracted from bacterial organisms known to populate the intestinal flora. The performance is also assessed every year using an external quality assessment panel obtained from Quality Control for Molecular Diagnostics (www.qcmd.org). A 100% score has been achieved for the last three years (2010, 2011 and 2012) with their QCMD Clostridium difficle DNA EQA Programme (10 samples in each EQA panel).
Illumigene C. difficile LAMP assay
Illumigene C. difficile LAMP assay (Meridian, Cincinnati, OH) was performed on 75 stool samples selected from the PHE multiplex RT-PCR/HE-LAMP-BART method comparison. These particular samples were selected at Lumora as being especially challenging due to either low copy number as indicated by high PHE Ct value, high inhibitor load by delayed inhibitor control peak, coming from solid stool, blood content and less common ribotype. The IllumiPro-10 instrument was used as described by the manufacturer with the exception that formed stools were tested to assess the test’s ability to detect C. difficile in formed stool. Samples were run at the same time on both the Illumigene and HE-LAMP-BART methods. Positive and negative controls were run on the test once a day.
Xpert C. difficile/EPI PCR assay
The Xpert C. difficile/EPI PCR assay (Cepheid, Sunnyvale, CA) was performed on 27 stool samples further selected from the Illumigene/LAMP-BART method comparison for low copy number, high inhibitor load, blood content and less common ribotype. The GeneXpert C. difficile cartridge was used as described by the manufacturer with the exception that formed stools were tested to assess the test’s ability to detect C. difficile in formed stool.
RIDA®QUICK Clostridium difficile Toxin A/B rapid test immunoassay
RIDA®QUICK Clostridium difficile Toxin A/B rapid test (R-Biopharm, Darmstadt, Germany) was performed on the same 27 stool samples used for the GeneXpert comparison as described by the manufacturer.
Interference study for HE-LAMP-BART
Interfering substances likely to be found in stool samples were purchased either from Sigma (Poole, UK) or from retail/pharmacy. Clinically relevant amounts of these substances were added to a negative stool sample spiked with 32 CFU/ reaction equivalent of C. difficile NCTC 13307 extracted by Heat Elution and the eluate run in C. difficile and Inhibitor Control LAMP-BART reagent.
Stool spike detection comparison
A C. difficile-negative unformed stool sample was spiked with 10,000 CFU/ml C. difficile NCTC 13307 and a two-fold dilution series down to 312.5 CFU/ml made using the same negative stool sample. Each level was tested by HE-LAMP-BART, PHE easyMAG plus RT-PCR, GeneXpert, Illumigene and RIDAQuick.
HE-LAMP-BART stool spike Limit of Detection
Twenty replicates of eluate from HE extract of C. difficile negative unformed stool sample spiked with 8 CFU/reaction equivalent C. difficile NCTC 13307 were tested using LAMP-BART.
Comparative analytical sensitivity between LAMP-BART and PHE RT-PCR
A dilution series of C. difficile 630 chromosomal DNA (Sanger Institute, UK) in 10 fold steps down to 100 copies and two fold steps down to 0.5 copies were tested by both LAMP-BART and PHE RT-PCR.
Inclusivity and exclusivity of the C. difficile LAMP-BART test
The inclusivity of the C. difficile LAMP-BART assay was evaluated by testing a panel of 5 C. difficile strains; NCTC 13307, NCTC 11205, NCTC 11209, NCTC 11204 and the non-toxigenic strain ATCC 43593. Bacteria were grown on blood agar plates and single colonies diluted with vortex mixing in the equivalent of 10 ml of Buffered Peptone Water followed by a 90-fold dilution in reaction buffer. The final reaction buffer dilutions were heated at 110°C for 5 minutes. When cool, lysates were tested in singlet with the Lumora C. difficile LAMP-BART assay. In the same way, the exclusivity of the C. difficile LAMP-BART assays was evaluated by testing a panel of 37 bacteria. Bacteria were either grown on TSA or blood agar plates and single colonies, or the equivalent, were transferred to 250 µl of reaction buffer, vortex mixed and heated at 110°C for 5 minutes. When cool, lysates were tested in duplicate with the Lumora C. difficile LAMP-BART assay. The following exclusive organisms were tested with the C. difficile LAMP-BART assay: Aeromonas hydrophilia, Bacillus cereus, Campylobacter jejuni, Citrobacter amalonaticus, Citrobacter freundii, Clostridium perfringens, Clostridium sordellii, Edwardsiella tarda, Enterobacter aerogenes, Enterobacter cloacae, Enterococcus gallinarum, Escherichia coli, Hafnia alvei, Klebsiella oxytoca, Klebsiella pneumoniae, Lactobacillus acidophilus, Listeria grayii, Listeria innocua, Listeria ivanovii, Listeria monocytogenes, Listeria seeligeri, Proteus mirabilis, Proteus vulgaris, Pseudomonas aeruginosa, Pseudomonas fluorescens, Pseudomonas putida, Salmonella enteritidis, Salmonella typhimurium, Serratia liquefaciens, Shigella sonnei, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus agalactiae, Vibrio parahaemolyticus and Yersinia enterocolitica.
Statistical analysis
Sensitivity/specificity and confidence intervals were determined using StatsDirect Statistical Software version 2.7.9 (http://www.statsdirect.com).
Results
Typical HE-LAMP-BART results
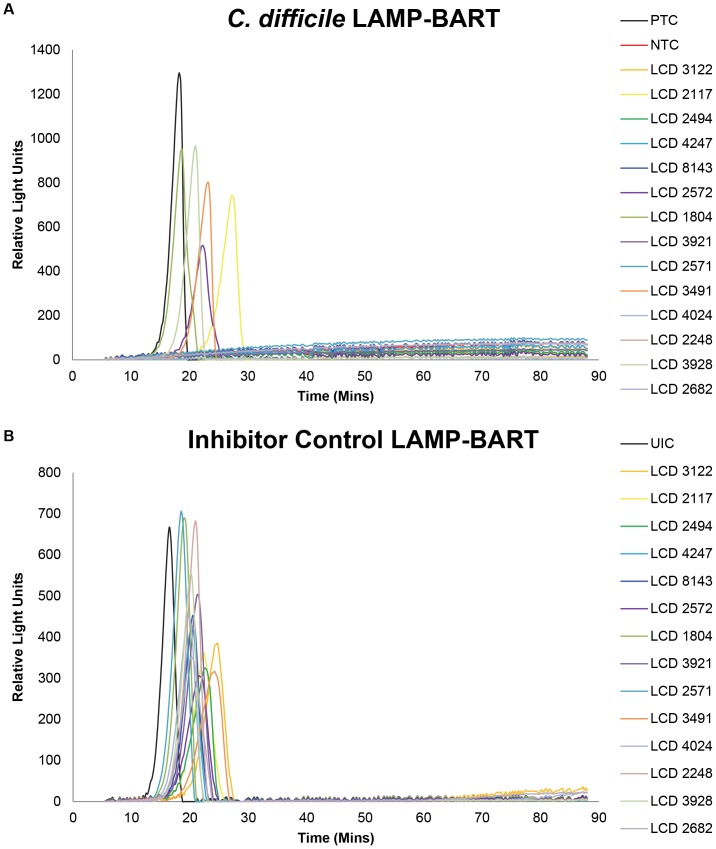
The LAMP-BART results from one of the runs from the comparison with Illumigene are presented in Figure 2 and Table 2. In addition to the C. difficile reactions, the Inhibitor Control results are also displayed. If the Inhibitor Control peak time was later than 35 minutes a retest was necessary if the C. difficile test for that sample was negative. The HE-LAMP-BART is a two-step process that takes typically 30 to 60 minutes from swabbing the stool sample to obtaining a positive peak on the instrument. Within the Illumigene comparison the average LAMP-BART time to result was 37±6.0 minutes.
Figure 2. Example C. difficile HE-LAMP-BART results.
One HE-LAMP-BART run of 14 samples from the Illumigene comparison. Panel A shows the C. difficile LAMP-BART peaks obtained and includes a positive control of 104 copies of C. difficile genomic DNA (PTC) and a no template control (NTC). Panel B shows the Inhibitor Control LAMP-BART results obtained simultaneously from the same set of HE eluates and a buffer uninhibited control (UIC).
Table 2. Example C. difficile HE-LAMP-BART results.
| C. difficile | Inhibitor Control | |||
| Sample | Peak Time (min) | Result | Peak Time (min) | Result |
| PTC | 15.5 | POS | ||
| NTC | NEG | |||
| UIC | 13.75 | POS | ||
| LCD 3122 | NEG | 21.5 | POS | |
| LCD 2117 | 24 | POS | 19.5 | POS |
| LCD 2494 | NEG | 19.5 | POS | |
| LCD 4247 | NEG | 17.5 | POS | |
| LCD 8143 | NEG | 17.75 | POS | |
| LCD 2572 | 19.5 | POS | 18.25 | POS |
| LCD 1804 | 15.75 | POS | 16.25 | POS |
| LCD 3921 | NEG | 18.25 | POS | |
| LCD 2571 | NEG | 15.75 | POS | |
| LCD 3491 | 20 | POS | 20.25 | POS |
| LCD 4024 | NEG | 16.75 | POS | |
| LCD 2248 | NEG | 18 | POS | |
| LCD 3928 | 18 | POS | 17.25 | POS |
| LCD 2682 | NEG | 17.25 | POS | |
Peak time and results called for the example LAMP-BART data set presented in Figure 1. For the C. difficile assay Positive control of 104 copies of C. difficile genomic DNA (PTC) and a no template control (NTC) were used. For the inhibitor control LAMP-BART results a buffer uninhibited control (UIC) was used.
Comparative analytical sensitivity between LAMP-BART and PHE RT-PCR
Both the PHE RT-PCR and LAMP-BART tests reproducibly detected down to 10 copies per amplification reaction of C. difficile 630 genomic DNA in quadruplicate.
Stool spike sensitivity comparison
In this experiment GeneXpert and HE-LAMP-BART gave complete detection down to 5000 CFU/ml, Illumigene detected down to 2500 CFU/ml and PHE RT-PCR detected down to 312.5 CFU/ml. None of the spike levels tested were detected by RIDAQuick Toxin A/B rapid test immunoassay. Although we do not fully understand this result, it could be due to the toxins not being expressed by cultured C. difficile [21] or the dilution of expressed toxin proteins.
Inclusivity and exclusivity of LAMP-BART
C. difficile strains NCTC 13307, NCTC 11205, NCTC 11209, NCTC 11204 were detected by the HE-LAMP-BART assay. Non-toxigenic strain ATCC 43593 was not detected and none of the exclusive organisms tested were detected. In addition, from the fecal samples, 78 of the 111 tcdB harboring strains with ribotypes 001, 002, 003, 005, 011, 012, 013, 014, 015, 020, 023, 026, 027, 046, 050, 078, 081, 126, 150, 258, 351 and 355 were successfully detected by HE-LAMP-BART. The remaining 33 positive isolates were also detected, but reported as sporadic (with respect to their ribotype) as their ribotypes did not match any of the ribotype profiles currently held in our laboratory database.
Interferents
The concentrations of interferents shown in Table 3 did not interfere with the detection of 32 CFU/reaction of C. difficile NCTC 13307 spiked into a C. difficile negative unformed stool sample.
Table 3. Concentrations of substances that do not interfere with the detection of C. difficile by HE-LAMP-BART.
| Interferent | Conc. in stool |
| Anusol | 40% |
| Gygel | 40% |
| 1% Hydrocortisone | 40% |
| KY Jelly | 40% |
| Peptobismol | 40% |
| Vaseline | 40% |
| Preparation H | 40% |
| Stearic acid | 20% |
| Palmitic acid | 20% |
| Haemoglobin | 20% |
| CaCO3 Tabs | 10% |
| Lactulose | 10% |
| Senokot | 10% |
| Benzalkonium Cl | 10% |
| Barium sulphate | 5% |
| Milk of Magnesia | 5% |
| Micoconazole | 20 mg/ml |
| Phenylephrine | 20 mg/ml |
| Metronidazole | 10 mg/ml |
| Vancomycin | 10 mg/ml |
| Imodium | 10 mg/ml |
| Nystatin | 10000 USP u/ml |
| Mucin | 3.5 mg/ml |
| Feminax Ultra | 1.25 mg/ml |
Performance on Clinical Stool Samples
Method comparison between HE-LAMP-BART with gold standard reference & PHE multiplex RT-PCR. A total of 218 stool samples were tested by HE-LAMP-BART C. difficile and compared to the cytotoxic culture reference method, with sample status after -20°C storage confirmed by PHE multiplex RT-PCR following easyMAG extraction (Table 4). 106 out of 111 C. difficile positive samples were positive by HE-LAMP-BART resulting in 5 false negatives and 95.5% sensitivity. HE-LAMP-BART was concordant with the reference and PHE methods for negative samples. Thus, HE-LAMP-BART gave very good agreement with the cytotoxigenic culture reference.
Table 4. Comparison between the reference method and PHE-Cambridge multiplex RT-PCR and HE-LAMP-BART.
| Total | PHE RT-PCR Positive | PHE RT-PCR Negative | HE-LAMP-BART Positive | HE-LAMP-BART Negative | ||
| PHE | Total | 218 | 106 | 112 | 106 | 112 |
| Reference | Positive | 111 | 111 | 0 | 106 | 5 |
| Test/method | Negative | 107 | 0 | 107 | 0 | 107 |
| Sensitivity (95% CI) | 100% (96.73–100%) | 95.5% (89.8–98.52%) | ||||
| Specificity (95% CI) | 100% (96.61–100%) | 100% (96.61–100%) | ||||
Comparison between HE-LAMP-BART and the Illumigene LAMP Methods. On the subset of 75 challenging stool samples (including samples with low copy number, solid stool, samples with high inhibitor load, containing blood and less common ribotype), compared to the cytotoxic culture reference method, PHE RT-PCR gave 100% sensitivity, HE-LAMP-BART gave 87.2% sensitivity compared to 76.6% sensitivity for Illumigene (Table 5) due to 6 and 11 false negatives, respectively. The Illumigene method gave one consistent invalid result for a negative sample with high blood content. Only one sample was detected as C. difficile positive by Illumigene, but not by HE-LAMP-BART; whereas 6 samples were detected as C. difficile positive by HE-LAMP-BART, but negative by Illumigene. However, when the former Illumigene positive/HE-LAMP-BART negative sample was re-extracted and run with C. difficile LAMP-BART in sextuplet, 4 out of the 6 reactions were detected as positive, indicating a low copy number sample close to the sensitivity of the HE-LAMP-BART method. These sensitivity values should be considered in the context of the challenging sample subset tested. All tests showed 100% specificity.
Table 5. Comparison of HE-LAMP-BART and the Illumigene LAMP Methods against the reference method.
| Illumigene | HE-LAMP-BART | PHE Reference Test/Method | |
| Positives | 36 | 41 | 47 |
| Negatives | 38 | 34 | 28 |
| Invalid | 1 | 0 | |
| Total | 75 | 75 | |
| False Positives | 0 | 0 | |
| False Negatives | 11 | 6 | |
| Sensitivity (95% CI) | 76.6% (61.97–87.7%) | 87.2% (74.26–95.17%) | |
| Specificity (95% CI) | 100% (87.66–100%) | 100% (87.66–100%) |
Extension of comparison to a subset from the Illumigene-HE-LAMP-BART study to GeneXpert and RIDAQuick. A further subset of 27 challenging stool samples, including low copy number, high inhibitor load, blood containing and less common ribotype samples from the HE-LAMP-BART vs Illumigene study were also tested by the GeneXpert and RIDAQuick methods. Compared to the cytotoxic culture reference method and PHE RT-PCR results, the following sensitivities were obtained: GeneXpert 96%, HE-LAMP-BART 92%, Illumigene 72% and RIDAQuick 76% (Table 6) due to 1, 2, 7 and 6 false negatives, respectively. For the RIDA Quick Toxin A/B method, 6 out 19 positive results gave weak positive test lines. Again, these sensitivity values should be considered in the context of the challenging sample subset tested and all tests showed 100% specificity.
Table 6. Comparison of a subset of samples from the Illumigene-HE-LAMP-BART study with GeneXpert and RIDAQuick against the reference method.
| PHE Reference Test/Method | PHE RT-PCR | GeneXpert | HE-LAMP-BART | Illumigene | RIDA Quick Toxin A/B | |
| Positive | 25 | 25 | 24 | 23 | 18 | 19 |
| Negative | 2 | 2 | 2 | 2 | 1 | 2 |
| Total | 27 | 27 | 27 | 27 | 27 | 27 |
| False Positive | 0 | 0 | 0 | 0 | 0 | |
| False Negative | 0 | 1 | 2 | 7 | 6 | |
| Invalid | 0 | 0 | 0 | 1 | 0 | |
| Sensitivity | % | 100 | 96 | 92 | 72 | 76 |
| %CI | 86.28–100 | 79.65–99.9 | 73.97–99.02 | 50.61–87.93 | 54.87–90.64 | |
| Specificity | % | 100 | 100 | 100 | 100 | 100 |
| %CI | 15.81–100 | 15.81–100 | 15.81–100 | 15.81–100 | 15.81–100 | |
Discussion
DNA preparation for current commercial molecular tests for C. difficile in stool either rely on robotic extraction that is integral to the test (GeneXpert), robotic or manual Boom style extraction before the test or a multistep manual dilutive sample preparation method involving a 200–700 fold dilution to overcome inhibition by diluting the fecal inhibitors (Illumigene).
The GeneXpert test from Cepheid uses a 2 addition sample preparation procedure consisting of swabbing the stool sample, adding the swab to a small bottle containing Sample Reagent, which is then vortexed and the contents then transferred by pipette to the cartridge. The cartridge is then loaded on the GeneXpert Dx instrument where sample extraction and PCR take place over 45 minutes resulting in detection of most positive samples reported in real time at about 30 minutes. It has been reported that GeneXpert requires 4 minutes technical time and a turnaround time of 45 minutes [41]. The complicated GeneXpert Dx instrument GeneXpert Dx that performs extraction, PCR cycling and fluorescence detection, could therefore be too expensive for certain clinical settings.
The Illumigene test from Meridian uses a 7 step, 4 transfer procedure between specimen collection with a Sample Collection Brush to reconstituting both test and control lyophilized beads reagent. These reaction tubes are transferred to the Illumipro-10 and amplified for 40 minutes with the result being displayed at the end of the run. Illumigene takes 5 minutes, has a turnaround time of 1 hour and does not require costly capital equipment [41].
The HE-LAMP-BART procedure, as described above, has the same number of steps as GeneXpert, has less hands-on time than the Illumigene sample preparation and can report the majority of positive samples within 45 minutes of swabbing. The photodiode PDQ instrument itself is small, has a low-cost of goods and is exceptionally robust.
In the performance comparison between the PHE real time C. difficile PCR and HE-LAMP-BART against the cytotoxigenic culture reference method, the PHE method showed perfect detection of toxigenic C. difficile. Although the PHE method costs $45 per specimen and has significant hands on time for extraction and set up of the PCR assay. HE-LAMP-BART showed very good agreement with the cytotoxigenic reference in detecting 95.5% of the positive samples. The comparison between Illumigene and HE-LAMP-BART on a challenging subset showed that the HE-LAMP-BART method had a sensitivity of 87.2% compared to 76.6% for the Illumigene method. The Illumigene turbidimetric detection method suffered when compared to HE-LAMP-BART in the detection of a blood rich sample. Illumigene has already been reported as giving invalid results in blood rich samples [41]–[42]. In this sample we observed a blood derived particulate in the reaction buffer that would be expected to interfere with detection. HE-LAMP-BART was not inhibited by this blood rich sample and the hemoglobin in the interference study, which already exhibited a strong inhibitor tolerance, showed that a stool sample containing 20% hemoglobin would still be detected. Whilst the GeneXpert method is able to tolerate fecal blood, it is suggested in the kit instructions that fecal particulates can interfere with the method. Due to the design of the Heat Elution columns, such particulates do not interfere with the HE-LAMP-BART method.
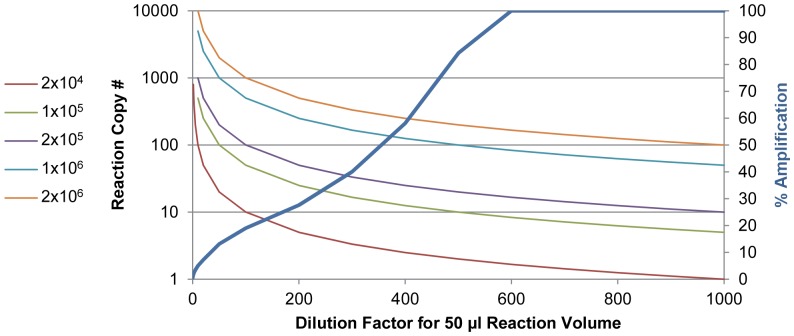
In the smaller study comparing the HE-LAMP-BART results with GeneXpert, RIDAQuick and Illumigene, GeneXpert showed a sensitivity of 96%, followed by HE-LAMP-BART at 92%, RIDAQuick at 76% and Illumigene at 72%. GeneXpert had already been reported to show superior sensitivity to Illumigene [41]. It was unexpected that the RIDAQuick immunoassay showed a higher sensitivity than Illumigene. To understand this we carefully examined all the data for these samples by all methods. Most of these samples displayed late PHE Ct values that became further delayed or failed when given a further 10-fold dilution. We observed a similar pattern by diluting some of these fecal samples by 200, 500 and 700 fold (factors typical of commercial dilutive methods) and running heat lysates on C. difficile LAMP-BART. This suggested that these samples had a low number of C. difficile that when subjected to the Illumigene dilutive method still had inhibitor at a concentration sufficient to slow down or stop amplification such that it could not be detected within the 40 minute amplification time. We have arrived at a model to explain the dilutive method low copy number failure. According to this model, illustrated in Figure 3, in the absence of inhibitor a reaction can show 100% amplification efficiency. At the highest concentration of inhibitor no amplification is seen; but as the fecal sample is given a gradual dilution, a gradual increase in amplification efficiency can occur up to over 700 fold for stool when inhibition becomes negligible. On the other hand, as this dilution proceeds, the DNA copy number also gradually decreases, such that samples of very low C. difficile content will have a very low DNA copy number in the assay. We have determined that the Illumigene assay uses a 300 to 400-fold dilution depending on the volume range of the sample swabbed. In comparison to 500-fold dilution and above, a 300 to 400-fold dilution leaves a sufficient inhibitor concentration to inhibit amplification of a, e.g. 2×104 CFU/gram, sample diluted to assay sensitivity level. In contrast, the inhibitor removal by the Heat Elution process provides an eluate diluted only by 50-fold that can contain 6-8 times more concentrated target due to the lower dilution factor. Thus, the difference in sensitivity of HE-LAMP-BART (87.2%) over Illumigene (76.6%) may have been a result of the lower dilution factor combined with robust inhibitor removal in HE-LAMP-BART. In addition to the use of Heat Elution for LAMP-BART, we have also tested eluate in real time PCR reactions, without any optimization, and had promising results showing detection of all high copy number samples (data not shown). Furthermore, we anticipate that the method would be applicable to other bacterial diarrhea pathogens.
Figure 3. Model describing low copy number non-detection for dilutive methods.
The right hand Y-axis (% amplification) shows the increase in amplification associated with dilution of inhibitors from a fecal sample. A range of starting target concentrations (CFU/gram) is plotted with the left hand Y-axis showing the copy number present in a 50 µl reaction volume (Illumigene). The reduced inhibition and increasing % amplification with increasing dilution is concomitant with a reduction in copy number. At lower CFU/gram samples, the dilution places the copy number per reaction beneath the sensitivity of the assay. HE-LAMP-BART used a 50 fold dilution together with the inhibitor removal such that a higher copy number can be present in the reaction.
The observation above is in agreement with the observation of Viala et al. that some commercial PCR tests reporting false negatives have been attributed to target copy numbers below the sensitivity limits of the tests [43]. When low copy number HE eluates were run in C. difficile LAMP-BART (in sextuplet), positive detection was seen in typically ≤4 out of the 6 replicates. From this result it is clear to see how such low copy number samples with DNA content around the sensitivity limit of the assay could show variable detection when single replicates are run.
Based on testing with spiked fecal samples, Bélanger et al. [44] showed that their cycle thresholds range of 22 to 35 corresponded to 107 to 104 CFU/gram. They further suggested that Ct 35 patients could have been C. difficile carriers exhibiting diarrheal symptoms of another etiology. A number of studies have reported asymptomatic adult carriage of C. difficile in the range 0.5–13% [45]–[47]. This increases to up to 14% in hospitalized patients [48]–[50] and is as high as 52% in long term care facilities [50]–[51]. A study by de Jong et al. [45] concluded that neither PCR nor cytotoxic culture is able to distinguish between CDI and asymptomatic carriage. It has also been suggested that such asymptomatic carriers contribute to the transmission of CDI [50]. However, it seems plausible that samples sub-detectable by HE-LAMP-BART, and indeed Illumigene, may not have been CDI cases; although for ethical reasons this study has not had access to patient information to prove this. A full clinical trial is needed to determine the performance of the HE-LAMP-BART method on stool samples from diagnosed CDI. Compared to one of the latest reports using the Boom method, our method is much simpler [52]. The HE-LAMP-BART method described here is a rapid, facile, real time molecular method with a low cost of goods for the detection of C. difficile that lends itself to use in decentralized environments.
Funding Statement
The authors have no support or funding to report.
References
- 1. Stabler RA, He M, Dawson L, Martin M, Valiente E, et al. (2009) Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. BMC Genome Biol 10: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, et al. (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467: 711–713. [DOI] [PubMed] [Google Scholar]
- 3. Voth DE, Ballard JD (2005) Clostridium difficile toxins: Mechanism of action and role in disease. Clin Microbiol Rev 18: 247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartlett JG (2006) Narrative review: The new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med 145: 758–764. [DOI] [PubMed] [Google Scholar]
- 5. Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nature Rev Microbiol 7: 526–536. [DOI] [PubMed] [Google Scholar]
- 6. Jones AM, Kuijper EJ, Wilcox MH (2013) Clostridium difficile: a European perspective. J Infect 66: 115–28. [DOI] [PubMed] [Google Scholar]
- 7. Stanley JD, Bartlett JG, Dart BW4th, Ashcraft JH (2013) Clostridium difficile infection. Curr Probl Surg 50: 302–37. [DOI] [PubMed] [Google Scholar]
- 8.Health Protection Agency. Voluntary surveillance of Clostridium difficile in England, Wales and Northern Ireland. London; 2013. Available: http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1317138039648 Accessed 22 November 2013.
- 9.Health Protection Agency. Clostridium difficile Ribotyping Network (CDRN) for England and Northern Ireland 2010/11 Annual Report (2012). Available: http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1317133396963 Accessed 22 November 2013.
- 10. Burns DA, Heeg D, Cartman ST, Minton NP (2011) Reconsidering the sporulation characteristics of hypervirulent Clostridium difficile BI/NAP1/027. PLoS ONE 6: e24894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brouwer MSM, Warburton PJ, Roberts AP, Mullany P, Allan E (2011) Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile . PLoS ONE 6: e23014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knetsch CW, Bakker D, de Boer RF, Sanders I, Hofs S, et al. (2011) Comparison of real-time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection. J Clin Microbiol 49: 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ananthakrishnan AN (2011) Clostridium difficile infection: epidemiology, risk factors and management. Nat Rev Gastroenterol Hepatol 8: 17–26. [DOI] [PubMed] [Google Scholar]
- 14. Bartlett JG (2010) Detection of Clostridium difficile infection. Infect Control Hosp Epidemiol 31 Suppl 1S35–S37. [DOI] [PubMed] [Google Scholar]
- 15. Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, et al. (2010) Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J Clin Microbiol 48: 3719–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kvach EJ, Ferguson D, Riska PF, Landry ML (2010) Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J Clin Microbiol 48: 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lalande V, Barrault L, Wadel S, Eckert C, Petit JC, et al. (2011) Evaluation of a loop-mediated isothermal amplification assay for diagnosis of Clostridium difficile infections. J Clin Microbiol 49: 2714–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Novak-Weekley SM, Marlowe EM, Miller JM, Cumpio J, Nomura JH, et al. (2010) Clostridium difficile testing in the clinical laboratory by use of multiple testing algorithms. J Clin Microbiol 48: 889–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quinn CD, Sefers SE, Babiker W, He Y, Alcabasa R, et al. (2010) C. Diff Quik Chek complete enzyme immunoassay provides a reliable first-line method for detection of Clostridium difficile in stool specimens. J Clin Microbiol 48: 603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sloan LM, Duresko BJ, Gustafson DR, Rosenblatt JE (2008) Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J Clin Microbiol 46: 1996–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL (2007) Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol 66: 206–19. [DOI] [PubMed] [Google Scholar]
- 22. Bartlett JG (2010) Clostridium difficile: progress and challenges. Ann N Y Acad Sci 1213: 62–69. [DOI] [PubMed] [Google Scholar]
- 23. Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, et al. (2013) Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis 13: 936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antunes A, Dupuy B (2010) Molecular methods to study transcriptional regulation of Clostridium difficile toxin genes. Methods Mol Biol 646: 93–115. [DOI] [PubMed] [Google Scholar]
- 25. Rupnik M (2010) Clostridium difficile toxinotyping. Methods Mol Biol 646: 67–76. [DOI] [PubMed] [Google Scholar]
- 26. Spigaglia P, Mastrantonio P (2002) Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol 40: 3470–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lawson AJ, Linton D, Stanley J, Owen RJ (1997) Polymerase chain reaction detection and speciation of Campylobacter upsaliensis and C. helveticus in human faeces and comparison with culture techniques. J Appl Microbiol 83: 375–80. [DOI] [PubMed] [Google Scholar]
- 28. Tsai YL, Olson BH (1992) Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol 58: 2292–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Al-Soud WA, Rådström P (1998) Capacity of Nine Thermostable DNA Polymerases To Mediate DNA Amplification in the Presence of PCR-Inhibiting Samples. Appl Environ Microbiol 64: 3748–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Monteiro L, Bonnemaison D, Vekris A, Petry KG, Bonnet J, et al. (1997) Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol 35: 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oikarinen S, Tauriainen S, Viskari H, Simell O, Knip M, et al. (2009) PCR inhibition in stool samples in relation to age of infants. J Clin Virol 44: 211–214. [DOI] [PubMed] [Google Scholar]
- 32. Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K (1994) Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstrains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Foren Sci 39: 362–372. [PubMed] [Google Scholar]
- 33. Al-Soud WA, Rådström P (2001) Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol 39: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thornton CG, Passen S (2004) Inhibition of PCR amplification by phytic acid, and treatment of bovine faecal specimens with phytase to reduce inhibition. J Microbiol Methods 59: 43–52. [DOI] [PubMed] [Google Scholar]
- 35. Lantz PG, Tjerneld F, Hahn-Hägerdal B, Rådström P (1997) Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J Microbiol Meth 28: 159–67. [Google Scholar]
- 36. Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, et al. (1990) Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gandelman OA, Church VL, Moore CA, Kiddle G, Carne CA, et al. (2010) Novel bioluminescent quantitative detection of nucleic acid amplification in real-time. PLoS One 5: e14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kiddle G, Hardinge P, Buttigieg N, Gandelman O, Pereira C, et al. (2012) GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol 12: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M Molecular Detection System for Pathogen and Toxin Testing from 3M Food Safety Products. Available: http://solutions.3m.com/wps/portal/3M/en_US/Microbiology/FoodSafety/product-applications/two/?PC_7_RJH9U5230O0G602RVAFDHB3026000000_nid=KFH17L9JD0beFSQWSZH9VKgl Accessed 22 November 2013.
- 40. Banoo S, Bell D, Bossuyt P, Herring A, Mabey D, et al. (2010) Evaluation of diagnostic tests for infectious diseases: general principles. TDR Diagnostics Evaluation Expert Panel. Nat Rev Microbiol 8: S17–29. [PubMed] [Google Scholar]
- 41. Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM (2012) Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difficile/Epi, and Illumigene C. difficile assays. J Clin Microbiol 50: 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kozak K, Elagin V, Norén T, Unemo M (2011) Targeting the tcdA Gene: Is This Appropriate for Detection of A and/or B Clostridium difficile Toxin-Producing Strains? J Clin Microbiol 49: 2383–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Viala C, Le Monnier A, Maataoui N, Rousseau C, Collignon A, et al. (2012) Comparison of commercial molecular assays for toxigenic Clostridium difficile detection in stools: BD GeneOhm Cdiff, XPert C. difficile and illumigene C. difficile. J Microbiol Methods 90: 83–85. [DOI] [PubMed] [Google Scholar]
- 44. Bélanger SD, Boissinot M, Clairoux N, Picard FJ, Bergeron MG (2003) Rapid detection of Clostridium difficile in feces by real-time PCR. Clin Microbiol 41: 730–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Jong E, de Jong AS, Bartels CJ, van der Rijt-van den Biggelaar C, et al. (2012) Clinical and laboratory evaluation of a real-time PCR for Clostridium difficile toxin A and B genes. Eur J Clin Microbiol Infect Dis 31: 2219–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kato H, Kita H, Karasawa T, Maegawa T, Koino Y, et al. (2001) Colonisation and transmission of Clostridium difficile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J Med Microbiol 50: 720–727. [DOI] [PubMed] [Google Scholar]
- 47. Ryan J, Murphy C, Twomey C, Paul Ross R, Rea MC, et al. (2010) Asymptomatic carriage of Clostridium difficile in an Irish continuing care institution for the elderly: prevalence and characteristics. Ir J Med Sci 179: 245–250. [DOI] [PubMed] [Google Scholar]
- 48. Kyne L, Warny M, Qamar A, Kelly CP (2000) Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med. 342: 390–397. [DOI] [PubMed] [Google Scholar]
- 49. Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN (1998) Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 351: 633–636. [DOI] [PubMed] [Google Scholar]
- 50. Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, et al. (2007) Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis 45: 992–998. [DOI] [PubMed] [Google Scholar]
- 51. Simor AE, Bradley SF, Strausbaugh LJ, Crossley K (2002) Nicolle LE; SHEA Long-Term-Care Committee (2002) Clostridium difficile in long-term-care facilities for the elderly. Infect Control Hosp Epidemiol 23: 696–703. [DOI] [PubMed] [Google Scholar]
- 52. Huang S, Do J, Mahalanabis M, Fan A, Zhao L, et al. (2013) Low cost extraction and isothermal amplification of DNA for infectious diarrhea diagnosis. PLoS One 8: e60059. [DOI] [PMC free article] [PubMed] [Google Scholar]