Abstract
The microtubule-based mitotic spindle segregates chromosomes during cell division. During chromosome segregation, the centromeric regions of chromosomes build kinetochores that establish end-coupled attachments to spindle microtubules. Here, we used the C. elegans embryo as a model system to examine the crosstalk between two kinetochore protein complexes implicated in temporally distinct stages of attachment formation. The kinetochore dynein module, which mediates initial lateral microtubule capture, inhibited microtubule binding by the Ndc80 complex, which ultimately forms the end-coupled attachments that segregate chromosomes. The kinetochore dynein module directly regulated Ndc80, independently of phosphorylation by Aurora B kinase, and this regulation was required for accurate segregation. Thus, the conversion from initial dynein-mediated, lateral attachments to correctly oriented, Ndc80-mediated end-coupled attachments is actively controlled.
The 4-subunit Ndc80 complex, whose Ndc80 subunit harbors direct microtubule-binding activity, is the central component of the microtubule end-coupled attachments that segregate chromosomes on mitotic spindles (1, 2). In metazoans, initial lateral capture of microtubules by dynein motors localized to kinetochores kinetically accelerates the formation of end-coupled attachments and ensures their correct orientation (3-7). How kinetochores transition from an initial laterally bound state to the final end-coupled state is unclear.
The kinetochore dynein module is composed of the 3-subunit RZZ (Rod/Zw10/Zwilch) complex, which recruits dynein to kinetochores via Spindly (Fig. 1A;(7-9)). Formation of end-coupled microtubule attachments was assessed during the first division of the C. elegans embryo by visualizing chromosome dynamics (Fig. 1B) and by quantifying the kinetics of spindle pole separation (Fig. 1C; (10, 11)). Removal of Spindly (SPDL-1 in C. elegans) was nearly equivalent to removal of NDC-80 (Fig 1B,C). As expected (7, 12), the failure to establish end-coupled attachments resulting from SPDL-1 depletion was suppressed by co-depletion of RZZ (Fig. 1B); the double inhibition exhibited only the mild delay in end-coupled attachment formation expected for loss of kinetochore dynein. Thus, RZZ inhibits the formation of NDC-80-mediated microtubule attachments and relief of this inhibition requires SPDL-1.
Fig. 1. RZZ inhibits microtubule attachment formation at kinetochores independently of Aurora B phosphorylation of NDC-80.
(A) Cartoon depiction of the kinetochore dynein module.
(B) Chromosome segregation phenotypes for the indicated conditions. GFP∷H2b (arrow) and GFP∷γ-tubulin (arrowhead) signals are indicated. Bar, 5 μm.
(C) Plot of spindle pole separation kinetics, which serves as a readout for kinetochore-microtubule attachment formation. Error bars are SEM with a 95% confidence interval.
(D) Chromosome segregation phenotypes for the indicated conditions (see also Fig. S1). Bar, 5 μm.
Aurora B kinase inhibits microtubule binding of Ndc80 by phosphorylating its basic tail (13-15). To determine whether RZZ inhibits NDC-80 by promoting Aurora B-mediated phosphorylation of NDC-80, we created an RNAi-resistant transgenic system (Fig. S1A,B) to replace endogenous NDC-80 with transgene-encoded NDC-80WT or a phosphorylation-resistant NDC-804A mutant (Fig. S1C,D;(13)). NDC-80WT and NDC-804A mutant both rescued the severe chromosome segregation defect and embryonic lethality of NDC-80 depletion (Fig 1D; Fig. S1E,F). Furthermore, NDC-80WT and NDC-804A were equally sensitive to inhibition by RZZ—following SPDL-1 depletion, both exhibited severe chromosome segregation (Fig. 1D) and pole separation (Fig. S1G) defects indicative of a failure to form end-coupled attachments. Thus, Aurora B-mediated phosphorylation of the NDC-80 tail is not required for RZZ to inhibit NDC-80-mediated end-coupled attachments.
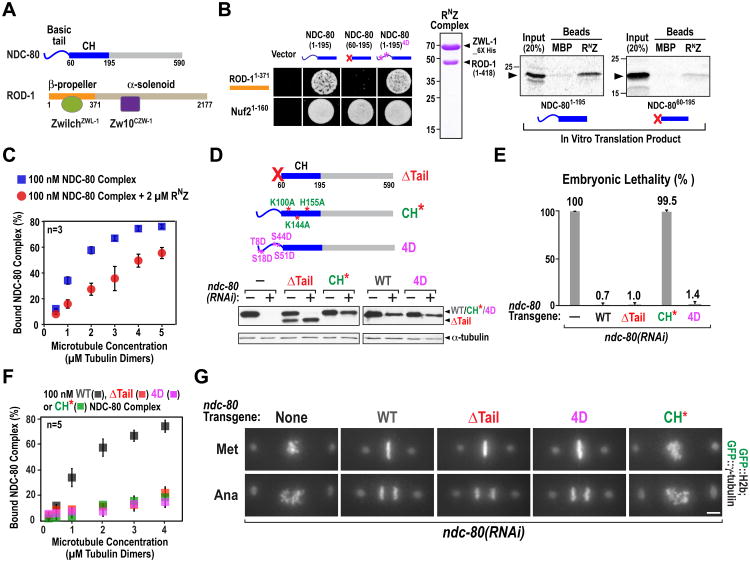
We next tested if the RZZ complex directly interacts with NDC-80 and inhibits its microtubule binding activity. ROD-1 has an N-terminal β-propeller domain that binds to ZwilchZWL-1 followed by an extended α-solenoid that binds Zw10CZW-1 (Fig. 2A; (16)). The N-terminal β-propeller domain of ROD-1 and the N-terminal microtubule-binding region of NDC-80 interacted in a yeast two hybrid assay (Fig. 2B; Fig. S2A). Deletion of the basic tail of NDC-80 abolished its interaction with ROD-1 without affecting binding to its Nuf2HIM-10 partner; in contrast, mimicking an Aurora B phosphorylated NDC-80 tail by mutation of 4 target sites to aspartic acid (4D) did not affect the ROD-1 interaction (Fig. 2B). Binding assays with a partially reconstituted RZZ complex comprised of the N-terminus of ROD-1 and ZwilchZWL-1 (termed RNZ) confirmed a direct tail-dependent interaction between RNZ and NDC-80 (Fig. 2B). To test if the ROD-1–NDC-80 interaction regulates NDC-80 microtubule binding, we used reconstituted C. elegans NDC-80 complex (Fig. S2B) to perform microtubule co-sedimentation assays in the presence or absence of purified RNZ complex. RNZ suppressed NDC-80 complex binding to microtubules (Fig. 2C; Fig. S2C). RNZ did not associate with microtubules on its own, excluding competition for lattice binding sites as a mechanism underlying this suppression (Fig. S2D).
Fig. 2. RZZ interacts with and inhibits NDC-80.
(A) Schematics of NDC-80 and the RZZ complex.
(B) (left) NDC-80 tail-dependent yeast 2-hybrid interaction with ROD-1; Nuf2HIM-10 served as a positive control (Fig. S2). (right) Recombinant RNZ complex and autoradiogram of binding assay with RNZ complex-coated beads and in vitro translated S35-labeled NDC-80 N-terminus. Maltose Binding Protein (MBP)-coated beads are negative controls.
(C) Microtubule cosedimentation with 100 nM NDC-80 complex, indicated concentration of taxol-stabilized microtubules, 2 μM RNZ complex, and 5 μM BSA. Binding was quantified using anti-NDC-80 immunoblots of Supernatant (S) and Pellet (P) fractions. (see also Fig. S2).
(D) (top) Schematic of mutations engineered in NDC-80 (Fig. S3). (bottom) Anti-NDC-80 immunoblot showing replacement of endogenous NDC-80 by transgene-encoded NDC-80 WT & variants (see also Fig.S1).
(E) Embryo viability analysis for the indicated conditions. Error bars are SD of embryo lethality per worm. >10 worms and >1000 embryos were scored per condition.
(F) Microtubule cosedimentation, as in (C), with indicated NDC-80 complex variants (Fig.S2).
(G) Chromosome segregation phenotypes for the indicated conditions. Bar, 5 μm.
ROD-1 binding to the NDC-80 basic tail may mask an electrostatic interaction required for the NDC-80 complex to bind to microtubules (17-19). To test this, we analyzed 3 mutant forms of NDC-80 in vitro and in vivo: a tail deletion (NDC-80ΔTail), an Aurora B phosphorylation-mimicking tail mutant (NDC-804D), and a calponin homology (CH) domain mutant (NDC-80CH*) in which 3 basic residues were changed to alanine (Fig. 2D; Fig. S3A (20)). NDC-80CH*, NDC-80ΔTail and NDC-804D mutations all inhibited reconstituted NDC-80 complex microtubule binding to the same extent in vitro (Fig. 2F; Fig. S2B; (15, 21-23)). However, only the NDC-80CH* mutant resulted in embryonic lethality (Fig. 2E). Consistent with the lack of embryonic lethality, NDC-80ΔTail and NDC-804D were able to segregate chromosomes whereas NDC-80CH* was defective in chromosome segregation (Fig. 2G). In pole-tracking analysis, NDC-80ΔTail overlapped NDC-80WT (Fig. S3B); in addition, removal of the microtubule-binding Ska complex (24) did not enhance the phenotype of NDC-80ΔTail (Fig. S4). Thus, although the basic NDC-80 tail was required for microtubule binding in vitro, it was not required to form end-coupled kinetochore-microtubule attachments in the C. elegans embryo. Therefore, RZZ cannot inhibit NDC-80 in vivo by masking an electrostatic interaction between the tail and the microtubule lattice.
To test whether the ROD-1-NDC-80 tail interaction was required for RZZ inhibition we analyzed the effect of depleting SPDL-1 (to trigger persistent RZZ-mediated inhibition) in embryos in which endogenous NDC-80 was replaced by NDC-80WT, NDC-80ΔTail or NDC-804D. Consistent with the tail-dependence of the ROD-1-NDC-80 interaction, the failure to form end-coupled attachments following SPDL-1 depletion was completely suppressed in embryos expressing NDC-80ΔTail (Fig. 3A,B). In contrast, the NDC-804D mutant that still interacted with ROD-1 (Fig. 2B) but failed to bind to microtubules in vitro (Fig. 2F), was as sensitive to SPDL-1 depletion as NDC-80WT (Fig. 3A; Fig. S3C). Thus, the ability of NDC-80 to be regulated by RZZ correlates with its tail-dependent interaction with ROD-1 and not with in vitro microtubule binding.
Fig. 3. Tail-deleted NDC-80 is resistant to regulation by RZZ.
(A) Chromosome segregation phenotypes for the indicated conditions. Bar, 5 μm.
(B) Plot of spindle pole separation kinetics, as in Fig. 1C.
(C) Experimental strategy for analyzing the effect of different ndc-80 transgenes in an ndc-80(tm5271) mutant background (Fig. S5). Graph on the right shows the percentage of homozygous mutant larvae that develop into fertile adults producing viable embryos. In the absence of a transgene, all homozygous mutants arrest as larvae (Fig. S5).
(D) Fate of ndc-80(tm5271) larvae for the indicated conditions. n is the number of homozygous ndc-80(tm5271) worms/larvae analyzed. See also Fig. S5.
In NDC-80WT embryos, SPDL-1 depletion prevented formation of end-coupled attachments, whereas RZZ depletion only delayed attachment formation owing to the absence of dynein-mediated acceleration of microtubule capture (Fig. 3A,B; (7)). If NDC- 80ΔTail is resistant to RZZ inhibition, the dramatic difference in phenotypic severity between RZZ and SPDL-1 depletion observed with NDC-80WT should be lost in the presence of NDC-80ΔTail. Both visualization of chromosome segregation and quantitative pole tracking confirmed this prediction (Fig. 3A,B). Because the tail was not required for the formation of end-coupled attachments (Fig. 2G,S3B), binding of RZZ to the NDC-80 tail probably prevents the adjacent functionally critical CH domain from interacting with microtubules.
We next employed a genetic approach to analyze phenotypes observed following replacement of endogenous NDC-80 with transgene-encoded variants. We analyzed the fate of embryos homozygous for ndc-80(tm5271) (Fig. S5A) derived from a heterozygous mother that also harbored homozygous ndc-80 transgene insertions (Fig. 3C). While the ndc-80WT, ndc-804D and the ndc-804A transgenes supported development of ndc-80(tm2571) larvae into fertile adults, the ndc-80ΔTail transgene did not (Fig. 3C). The phenotypes observed in the presence of NDC-80ΔTail included late larval arrest, bursting at the vulva, and absence of a germline leading to sterility (Fig. 3D; Fig. S5B). Similar phenotypes are observed in C. elegans mutants that compromise the spindle checkpoint, a well-studied pathway ensuring accurate chromosome segregation (25). Because NDC-80ΔTail exhibited normal checkpoint signaling (Fig. S6A) and synthetic embryonic lethality with checkpoint inhibition (Fig. S6B), the observed phenotypes were not due to a defect in checkpoint signaling. Thus, NDC-80ΔTail exhibited a spectrum of defects associated with reduced accuracy of chromosome segregation in C. elegans and failed to rescue the lethality caused by the ndc-80 mutation. Because similar severity defects were not observed with NDC-804D, which inhibits NDC-80 complex microtubule binding activity in vitro, the origin of these defects is most likely due to loss of NDC-80 regulation by RZZ. A mutant in ROD-1 that selectively disrupts regulation of NDC-80 will be necessary to identify and characterize to confirm this conclusion.
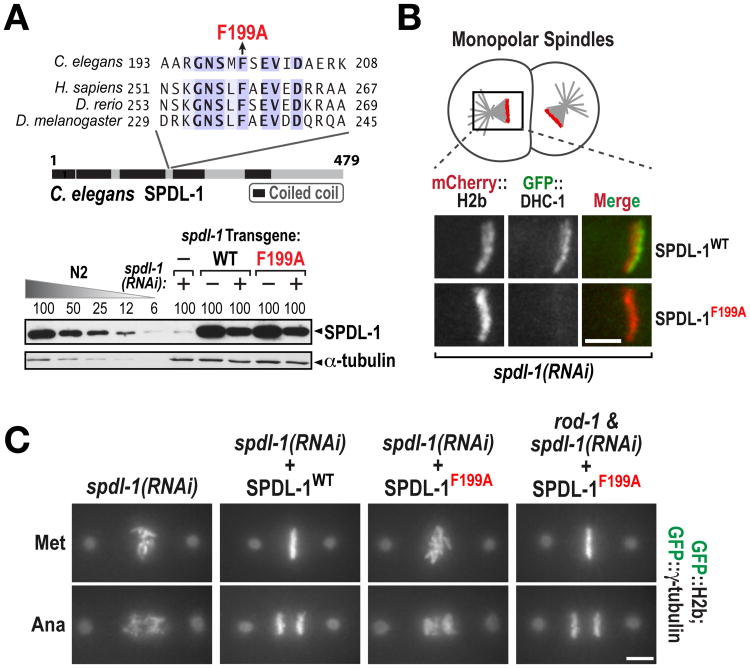
To understand how RZZ inhibition of NDC-80 was alleviated by SPDL-1, we focused on the conserved motif in Spindly family proteins required for dynein recruitment (Fig. 4A; (26)). We replaced endogenous SPDL-1 with SPDL-1WT or a single amino acid mutant in the motif required for dynein binding (SPDL-1F199A; Fig. 4A;Fig. S7A-D). Analogous to human cells, SPDL-1F199A localized to kinetochores (Fig. S7C) but abrogated dynein recruitment (Fig. 4B). SPDL-1F199A exhibited a chromosome segregation defect of similar severity to removal of SPDL-1 (Fig. 4C; Fig. S8). In addition, the severe chromosome segregation defect of SPDL-1F199A was suppressed by co-depletion of RZZ (Fig. 4C; Fig. S7E). Thus, dynein recruitment by SPDL-1 turns off RZZ-mediated inhibition of end-coupled attachment formation (Fig. S9).
Fig. 4. Dynein recruitment by SPDL-1 is required to turn off RZZ-mediated inhibition of NDC-80.
(A) (top) The mutation (F199A) engineered in the Spindly motif. (bottom) Anti-SPDL-1 immunoblot, as in Fig. S1D. α-tubulin is a loading control.
(B) Analysis of dynein heavy chain (DHC-1) recruitment to unattached kinetochores of monopolar spindles generated by depleting the centriole duplication kinase ZYG-1 (Fig. S7). Bar, 5 μm.
(C) Chromosome segregation phenotypes for the indicated conditions. See also Fig. S7 and Fig. S8. Bar, 5 μm.
Regulation of Ndc80 by RZZ represents an Aurora B-independent mechanism for the control of kinetochore-microtubule attachments. We postulate that this regulation prevents NDC-80-mediated attachments from occurring during an initial laterally attached state to minimize the potential for merotely, where a single kinetochore becomes erroneously linked to both spindle poles and results in missegregation (27). We suggest that dynein activity reduces the probability of merotely by coordinating chromosome orientation with activation of NDC-80, either via inducing a change in RZZ conformation or dissociating RZZ from the kinetochore (Fig. S9). Because both the kinetochore dynein module and the Ndc80 complex are conserved throughout metazoans, the mechanism we elucidate here is also likely to be conserved.
Supplementary Material
Acknowledgments
We thank the Japanese National BioResource Project for the ndc-80 deletion strain, K. Corbett for helpful discussions, and B. Green for comments on the manuscript. The data described here are tabulated in the main paper and the supplementary materials. This work was supported by an NIH grant (GM074215) to A.D; A.D. and K.O. receive salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 2.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scaërou F, et al. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J Cell Sci. 1999;112(Pt 21):3757–3768. doi: 10.1242/jcs.112.21.3757. [DOI] [PubMed] [Google Scholar]
- 6.Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gassmann R, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karess R. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 2005;15:386–392. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai A, et al. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barisic M, et al. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol Biol Cell. 2010;21:1968–1981. doi: 10.1091/mbc.E09-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 14.Deluca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 15.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Civril F, et al. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 2010;18:616–626. doi: 10.1016/j.str.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umbreit NT, et al. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci U S A. 2012;109:16113–16118. doi: 10.1073/pnas.1209615109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 22.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2012 doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welburn JPI, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt JC, et al. The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein KK, Davis ES, Hays T, Golden A. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics. 2007;175:107–123. doi: 10.1534/genetics.106.059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gassmann R, et al. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.