SUMMARY
In sexually reproducing multi-cellular organisms, genetic information is propagated via the germline, the specialized tissue that generates haploid gametes. The C. elegans germline generates gametes in an assembly line-like process- mitotic divisions under the control of the stem cell niche produce nuclei that, upon leaving the niche, enter into meiosis and progress through meiotic prophase [1]. Here we characterize the effects of perturbing cell division in the mitotic region of the C. elegans germline. We show that mitotic errors result in a spindle checkpoint-dependent cell cycle delay but defective nuclei are eventually formed and enter meiosis. These defective nuclei are eliminated by programmed cell death during meiotic prophase. The cell death based removal of defective nuclei does not require the spindle checkpoint, but instead depends on the DNA damage checkpoint. Removal of nuclei resulting from errors in mitosis also requires Spo11, the enzyme that creates double-strand breaks to initiate meiotic recombination. Consistent with this, double strand breaks are increased in number and persist longer in germlines with mitotic defects. These findings reveal that the process of initiating meiotic recombination inherently selects against nuclei with abnormal chromosomal content generated by mitotic errors, thereby ensuring the genomic integrity of gametes.
Keywords: C. elegans, germline, mitosis, meiosis, recombination, cell death, DNA damage
RESULTS
Nuclei resulting from mitotic errors are eliminated by cell death in meiotic prophase
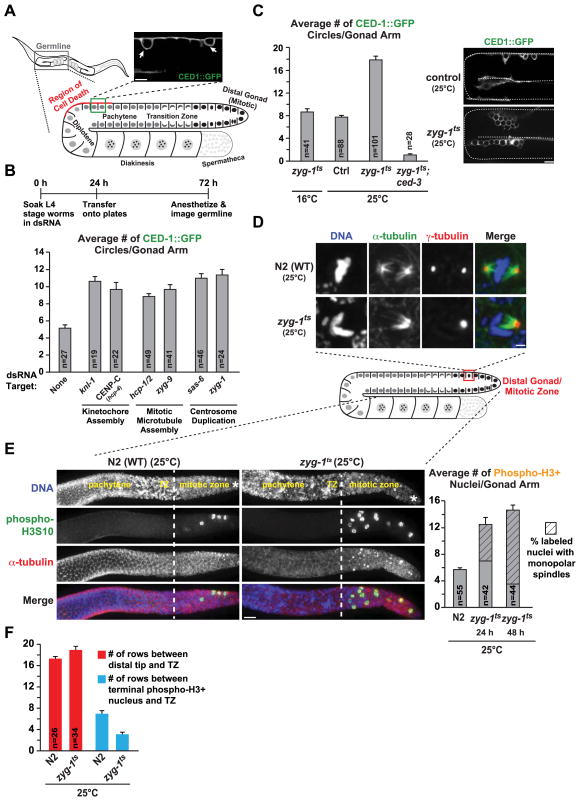
In the C. elegans germline, the cell death pathway targets nuclei near the turn of each gonad arm when nuclei are in the late pachytene stage of meiotic prophase [2] (Fig. 1A). While a significant proportion of meiotic nuclei undergo cell death in unperturbed adult germlines [2], nuclei harboring DNA damage or meiotic errors are preferentially eliminated [3–6]. To determine if the germline cell death pathway targets nuclei that have undergone mitotic errors prior to entry into meiosis, we inhibited key proteins required for chromosome segregation and monitored cell death by visualizing the cell corpse engulfment marker CED-1::GFP [7] (Fig. 1A). Soaking-based RNAi interference (RNAi) was used to deplete the mRNA for six proteins required for three different aspects of mitosis: kinetochore formation (KNL-1 and CENP-CHCP-4;[8]), mitotic microtubule assembly (HCP-1/2 and ZYG-9; [9,10]) and centrosome duplication (ZYG-1 and SAS-6; [11,12])—none of these proteins have previously been reported to function during meiotic prophase. The experimental protocol (Fig. 1B) provides sufficient recovery time after soaking in dsRNA to allow nuclei to migrate from the mitotic zone to the region of cell death [13,14]. Compared to controls, a significant increase in germline cell death was observed following individual inhibition of each component (Fig. 1B).
Figure 1. Perturbation of mitosis in the germline results in an increase in cell death.
(A) Gonad schematic and representative image of nuclei undergoing programmed cell death marked by CED-1::GFP (arrows). Bar, 5 μm.
(B) Quantification of cell death for the indicated conditions; experimental scheme is above the graph. Error bars are SEM; unpaired t tests show significant difference (p<0.001) between control and each individual inhibition.
(C) Quantification of cell death in zyg-1ts worms and example images. Worms were analyzed at the following times: zyg-1ts;ced-3, 24h after upshift; zyg-1ts at 16°C, adults age-matched to 24h at 25°C; control and zyg-1ts at 25°C the compiled average from 24, 36 and 48h post upshift (see Fig. S1A). Error bars are SEM; control and zyg-1ts at 25°C are significantly different (p<0.0001; unpaired t-test). Bar, 10 μm.
(D) Immunofluorescence images of the mitotic zone of extruded gonads labeled for DNA, α-tubulin and γ-tubulin 24h post upshift; partial z stack projections. Bar, 2 μm.
(E) Images and quantification of phospho-H3S10 positive nuclei in gonads 24h post upshift; the prevalence of monopolar spindles in the labeled nuclei is also plotted. Errors bars are SEM. Asterisk denotes the distal end of the gonad, dotted white line denotes the end of the mitotic zone/beginning of the transition zone, based on nuclear morphology. Bar, 10 μm.
(F) Quantification of the data presented in (E). Error bars are SEM; unpaired t test shows no significant difference (p=0.062) in the size of the TZ but a significant difference (p<0.0001) in the distance between the last phospho-H3 positive nucleus and the TZ.
(See also Figures S1 & S2).
To investigate the relationship between mitotic errors and cell death, we first sought to develop a consistent means for perturbing germline mitoses without directly affecting subsequent meiotic events. Inhibition of centrosome duplication is ideally suited for this purpose; centrosomes are critical for mitosis but are inactivated upon meiotic entry and degraded as nuclei progress through the germline [15]. To disrupt centrosome duplication, we capitalized on a temperature-sensitive mutation in the kinase ZYG-1 (zyg-1(b1), referred to here as zyg-1ts; [11]). When shifted to the restrictive temperature for 24 – 48 hours, zyg-1ts worms displayed a robust increase in germline cell death compared to controls (Fig. 1C, S1A). To validate that the observed CED1::GFP circles represented programmed cell death, we analyzed zyg-1ts;ced-3(n717) double mutants; CED-3/caspase is required for all programmed cell death. In the double mutant, the number of CED-1::GFP circles was reduced to nearly zero (Fig. 1C). We additionally performed shorter interval/transient upshifts to confirm that the increase in cell death seen upon ZYG-1 inactivation is a consequence of errors in mitosis (Fig. S1A,B). Thus, conditional inhibition of centrosome duplication is a convenient and reproducible means to induce mitotic errors that result in a robust elevation of programmed cell death in meiotic prophase.
Nuclei resulting from errors in mitosis progress into meiosis after a delay
To assess the mitotic errors generated by the perturbations described above, we immunostained extruded gonads to visualize chromosomes, microtubules and centrosomes. In zyg-1ts worms, defective mitotic figures were observed in the mitotic zone, the most notable of which were monopolar spindles (Fig. 1D); less frequently observed (~20%) defects include abnormalities in spindle structure and unaligned or lagging chromosomes. Defective mitotic figures were also observed following RNAi-mediated inhibition of kinetochore assembly or mitotic microtubule formation (Fig. S1C) and micronuclei, defined as small, round DAPI-stained dots, were evident in zyg-1ts germlines (Fig. S1E).
To determine if the observed defects perturbed cell cycle progression, we measured the mitotic index by labeling mitosis-associated phosphorylation of histone H3 (phospho-H3S10). Compared to controls, both zyg-1ts upshift and the other mitotic perturbations led to a significant increase in the number of phospho-H3S10 nuclei in the germline (Fig. 1E, S1D). In zyg-1ts worms, the number of phospho-H3S10 positive nuclei did not further increase between 24 and 48 hours after ZYG-1 inactivation suggesting that induction of mitotic errors leads to a 2.5 – 3-fold increase in the duration of mitosis rather than a permanent mitotic arrest. Consistent with this, the number of nuclear rows prior to the transition zone (defined by DNA morphology or phosphorylation of SUN-1 [16] as a marker for meiotic entry) in zyg-1ts worms was similar to controls (Fig. 1F, S2A); however phospho-H3 positive nuclei were observed closer to the transition zone (Fig. 1F). Additionally, the size of the transition zone was not altered in zyg-1ts worms (Fig. S2B). We also labeled zyg-1ts mutant gonads for HTP-3, a synaptonemal complex protein whose recruitment requires the pre-meiotic S-phase-associated loading of cohesin [17]. In zyg-1ts worms, HTP-3 localization was identical to that seen in controls; HTP-3 was also observed on morphologically aberrant nuclei (Fig. S2C, arrow). Injection of fluorescent nucleotides to pulse label S-phase nuclei confirmed that ZYG-1 inhibition did not grossly perturb transit of nuclei through the germline (Fig. S2D–F). Collectively, these observations suggest that mitotic errors prolong M phase but nuclei with incorrect chromosomal complements are eventually formed and enter the meiotic program.
The spindle checkpoint is required for the cell cycle delay but is dispensable for mitotic defect-induced cell death
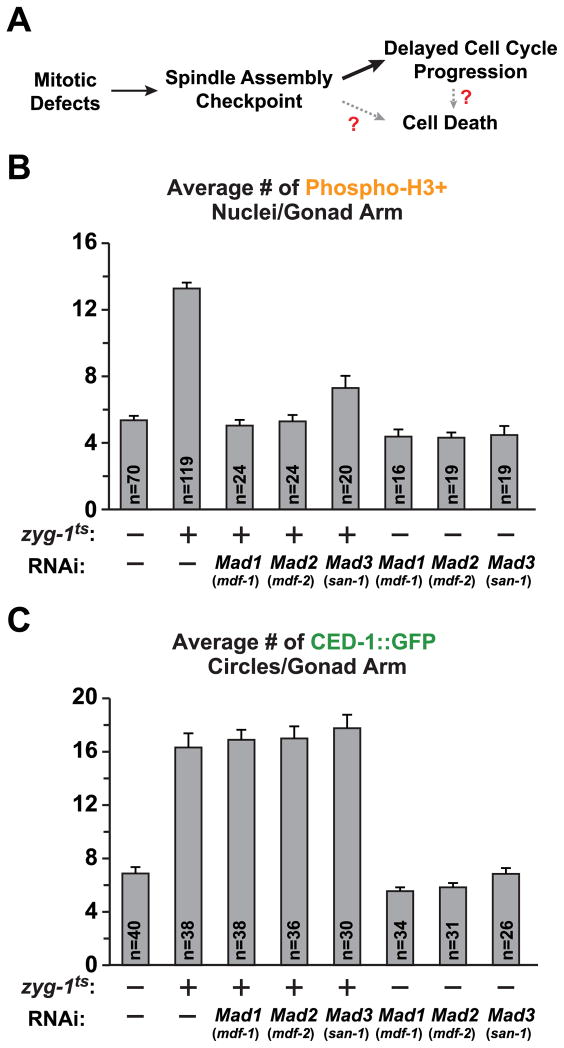
We next tested whether activation of the spindle checkpoint underlies the observed cell cycle delay and marks defective nuclei for elimination (Fig. 2A). For this purpose, we inhibited the spindle checkpoint components Mad1MDF-1, Mad2MDF-2 and Mad3MDF-3. Inhibition of the spindle checkpoint eliminated the zyg-1ts-induced mitotic delay (Fig. 2B) but had no effect on the elevation in cell death (Fig. 2C). In control worms with no induced mitotic defects, inhibition of the spindle checkpoint did not significantly affect either the number of phospho-H3S10 nuclei or the number of nuclei undergoing cell death (Fig. 2B,C). Thus, the spindle checkpoint is responsible for the mitotic delay triggered by division defects but is not required for the subsequent elimination of defective nuclei by programmed cell death.
Figure 2. The spindle checkpoint delays cell cycle progression but is not required for the increase in cell death.
(A) Schematic summarizing the function of the spindle checkpoint in delaying the cell cycle and hypothetical links between checkpoint activation and cell death.
(B) Quantification of phospho-H3S10 positive nuclei for the indicated conditions. Soaking RNAi was performed as in Fig. 1B. Errors bars are SEM.
(C) Quantification of cell death following inhibition of the spindle checkpoint. Error bars are SEM.
The DNA damage checkpoint is required for the increase in cell death observed following defective mitoses in the germline
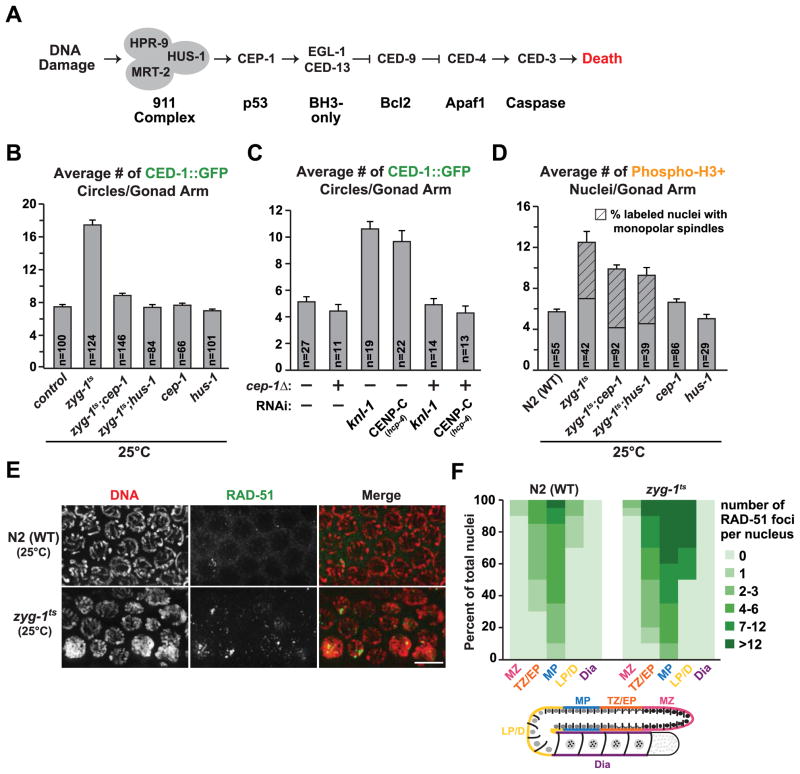
We next focused on the DNA damage checkpoint, which has been well studied in the C. elegans germline (Fig. 3A; [18]). We first tested p53CEP-1, the DNA damage responder directly upstream of the canonical cell death pathway. A null mutant of p53CEP-1, cep-1(gk138), is viable but fails to elevate cell death following induction of DNA damage [19]. A comparison of zyg-1ts;cep-1(gk138) double mutants to zyg-1ts mutants alone at the restrictive temperature revealed that loss of p53CEP-1 suppressed the elevation in cell death (Fig. 3B). Loss of p53CEP-1 also suppressed the elevated cell death observed following inhibition of the kinetochore proteins KNL-1 and CENP-CHCP-4 (Fig. 3C). Similar to loss of p53CEP-1, loss of HUS-1, which functions upstream of p53CEP-1 [20], also suppressed the cell death induced by ZYG-1 inhibition (Fig. 3B). The zyg-1ts;cep-1(gk138) and the zyg-1ts;hus-1(op241) double mutants showed a mild reduction in the number of phospho-H3S10 positive nuclei (Fig. 3D). However this reduction cannot underlie the suppression of cell death, as inhibition of the spindle assembly checkpoint had no effect on cell death despite restoring the number of phospho-H3S10 nuclei to control levels (Fig. 2B,C).
Figure 3. The DNA damage checkpoint mediates the elevation in cell death induced by defective mitoses.
(A) Simplified schematic of the DNA damage checkpoint pathway in the C. elegans germline.
(B) Quantification of cell death following inhibition p53CEP-1 and HUS-1, in both control and zyg-1ts worms, 24 – 48h post upshift. Error bars are SEM.
(C) Quantification of cell death for the indicated conditions (experimental scheme Fig. 1B). Error bars are SEM.
(D) Quantification of phospho-H3S10 positive nuclei and the prevalence of monopolar spindles 24h post upshift (control and zyg-1ts data same as Fig. 1E). Error bars are SEM; unpaired t test shows high significance (p<0.001) between zyg-1ts and the double mutants.
(E) Immunofluorescence images of the late pachytene region of extruded gonads stained with DAPI and RAD-51; partial z stack projection. Bar, 5 μm.
(F) Quantification of the RAD-51 foci for control and zyg-1ts 48h post upshift. Each gonad was divided into 5 zones (MZ=mitotic zone; TZ/EP=transition zone/early pachytene; MP=mid-pachytene; LP/D=late pachytene/diplotene; Dia=diakinesis); at least 10 gonads per genotype were analyzed.
(See also Figure S3).
Mitotic errors likely generate nuclei with incorrect chromosomal complements and, as pairing and the initial licensing step in synaptonemal complex formation are dependent on homology [21], could lead to pairing/synapsis defects. To assess if unpaired/unsynapsed chromosomes were present in pachytene stage nuclei following ZYG-1 inhibition and contributed to the increased cell death, we immunostained for HTP-3 and SYP-1, which co-localize between paired, synapsed chromosomes [22]. This analysis revealed that the majority of the chromosomes were synapsed in zyg-1ts worms, however in a small percentage (~15%) of pachytene nuclei, a single unsynapsed chromosome was observed (Fig. S3A; we additionally performed FISH for Chr. V, Fig. S3B). To determine the extent to which unsynapsed chromosomes contributed to the observed cell death, we inhibited the synapsis checkpoint protein PCH-2, which is required for elevated cell death in the presence of an unsynapsed pairing center that is associated with its cognate zinc finger protein [6]. Consistent with the low frequency of asynapsis, the increased cell death observed in zyg-1ts worms was only mildly reduced by a pch-2(tm1458) null mutant (Fig. S3C). We suspect that self-synapsis, which is observed in the absence of pairing and results in activation of the DNA damage checkpoint [23], may underlie the majority of DNA damage-triggered cell death observed following mitotic errors. The above results lead us to conclude that cell division defects in the mitotic zone are primarily detected by the DNA damage checkpoint, which activates programmed cell death to trigger removal of defective nuclei.
Spo11 is required for the persistent double-strand breaks and the increase in cell death observed following defective mitoses
The DNA damage checkpoint typically senses broken DNA ends. We therefore immunostained for RAD-51—the RecA family protein that concentrates at the sites of double-strand breaks to promote strand invasion—to test whether nuclei with mitotic errors displayed abnormal numbers of DNA double-strand breaks. In an unperturbed germline, RAD-51 foci transiently appear in meiotic nuclei undergoing recombination; in the presence of DNA damage or recombination defects, RAD-51 foci increase in both frequency and persistence [24]. ZYG-1 inhibition caused a significant increase in both the number of nuclei with RAD-51 foci as well as the average number of RAD-51 foci per nucleus (Fig. 3E,F). Accumulation of RAD-51 in zyg-1ts gonads was restricted to meiotic nuclei, however foci extended well into the late pachytene stage of meiotic prophase, where they were not detectable in controls (Fig. 3F).
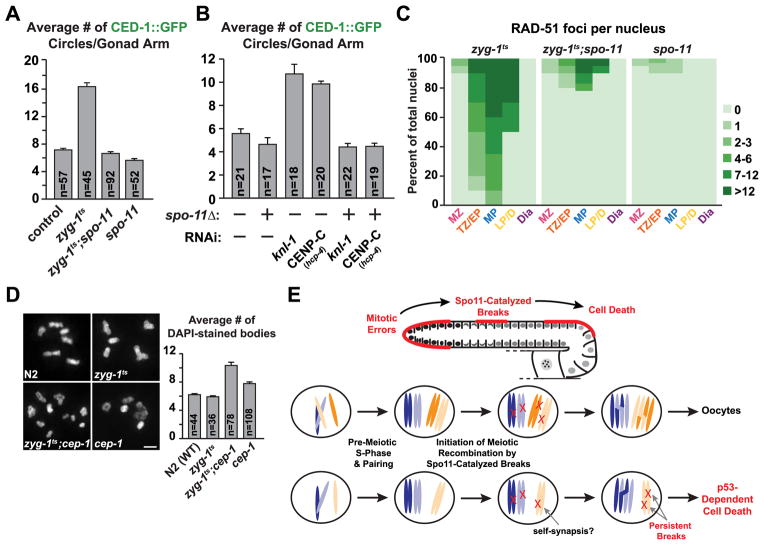
As RAD-51 foci were not observed prior to the transition zone, which marks the start of homologous recombination, we tested if recombination itself was required for the double-strand breaks and increase in cell death seen in meiotic nuclei resulting from defective mitoses. To this end, we inhibited Spo11, the topoisomerase-related enzyme required to catalyze the formation of double-strand breaks [25]. Analysis of zyg-1ts;spo-11(ok79) double mutants revealed a complete suppression of the elevated cell death observed in zyg-1ts worms (Fig. 4A). Removal of Spo11 also suppressed the elevated cell death seen following RNAi-mediated inhibition of the kinetochore components KNL-1 and CENP-CHCP-4 (Fig. 4B). Inhibition of Spo11 had no effect on the mitotic delay, as the number of phospho-H3S10 nuclei in the zyg-1ts;spo-11(ok79) double mutants was similar to that in the zyg-1ts mutant alone (Fig. S4A). Finally, the double-strand breaks visualized as RAD-51 foci were markedly decreased in zyg-1ts;spo-11(ok79) double mutants compared to the zyg-1ts mutant alone (Fig. 4C). Residual breaks were present in a small percentage of nuclei in the zyg-1ts;spo-11(ok79) double mutants, likely representing damage created during pre-meiotic replication of highly aberrant nuclei. However, as cell death was suppressed to near-control levels in the double mutant worms, we conclude that the Spo11-mediated double-strand breaks that initiate recombination are required for the increase in cell death observed following defects in mitosis.
Figure 4. Meiotic double-strand breaks are responsible for the elevated cell death observed following errors in mitosis.
(A) Quantification of cell death following inhibition of spo-11 in both control and zyg-1ts 24h post upshift. Error bars are SEM.
(B) Quantification of cell death following inhibition of spo-11 in both control and indicated mitotic inhibitions (experimental scheme, Fig. 1B). Error bars are SEM.
(C) Quantification of the RAD-51 foci for the indicated genotypes 48h post upshift (zyg-1ts values are the same as in Fig. 3F).
(D) Representative images and quantification of the chromosome content of oocytes for control, zyg-1ts, zyg-1ts;cep-1 and cep-1 72h post upshift. Errors bars are SEM; unpaired t test shows high significance (p<0.0001) between zyg-1ts and the double mutant. Bar, 2 μm.
(E) Schematic summarizing the role of Spo11-catalyzed breaks in the detection of mitotic errors that occurred in the distal gonad. The individual nucleus cartoons below show a normal nucleus with 2 pairs of homologous chromosomes (top) and a defective nucleus in which one of the chromosomes is missing due to missegregation (bottom). For simplicity, potential self-synapsis that would result in accumulation of DNA double-strand breaks is suggested but not depicted in the cartoon.
(See also Figure S4).
Cell death in response to errors in mitosis prevents the production of aneuploid oocytes
To assess whether the increase in cell death following mitotic errors prevents the generation of aneuploid oocytes, we measured the number of chromosomes (scored as discernable DAPI-stained bodies) present in oocytes 72 hours post upshift (Fig. 4D). In both control and zyg-1ts oocytes, the average number of DAPI-stained bodies was 6, which is the expected number of recombined bivalent chromosomes. In contrast, in the zyg-1ts;cep-1(gk138) double mutants where mitotic defect-induced cell death was suppressed, the average number of oocytes DAPI-stained bodies was ~10, indicating a high degree of aneuploidy. In addition, at a semi-permissive temperature (18.5°C), zyg-1ts;cep-1(gk138) double mutants exhibited synthetic embryonic lethality (Fig. S4B). Thus, the increase in cell death observed following mitotic errors serves a protective function in the germline by preventing the production of oocytes with an improper complement of chromosomes unable to support embryonic viability.
CONCLUSION
Although the mechanisms that protect against errors in meiosis have received much attention [26], how the germline protects itself against mitotic errors remains relatively uncharacterized. The work presented here shows that in the germline, errors in mitosis activate the spindle checkpoint and delay cell cycle progression but do not prevent entry into meiosis. Nuclei harboring errors accumulate unrepaired Spo11-dependent meiotic double strand breaks and are eliminated by DNA damage checkpoint-mediated cell death (Fig. 4E). As recent work suggests that in the absence of proper pairing Spo11-dependent breaks continue to form [27], the number of breaks created in nuclei harboring mitotic errors are likely amplified to ensure triggering of the DNA damage response. The excess number of Spo11-mediated double strand breaks relative to the number of crossovers [28], proposed to aid in homology assessment, could also aid in marking nuclei with prior mitotic errors.
How mitotic defects are linked to cell death is a topic of great interest, particularly in the context of chromosomal instability and cancer [29]. The idea that an abnormal mitosis triggers DNA damage is a common theme emerging from this work [30]. The data presented here supports this idea by showing that in the context of the germline, the purposeful induction of breaks essential for recombination also functions via the DNA damage response to weed out mitotic errors, thereby preventing the production of aneuploid gametes.
EXPERIMENTAL PROCEDURES
C. elegans strains
All strains were maintained at 16°C. Mutant alleles used were zyg-1(b1), ced-3(n717), cep-1(gk138), hus-1(op241), spo-11(ok79) and pch-2(tm1458). The CED-1::GFP and H2b::mCherry transgene insertions are from strains MD701 and OD95, respectively. For upshift experiments, mid-to-late L4 larval stage worms were placed at 25°C for the indicated time.
RNA Interference & Live imaging
Production of dsRNA, soaking RNAi and live imaging of the gonad were performed as previously described [31].
Immunofluorescence of fixed gonads
Extrusion and fixation of the gonad was done as previously described [16]. Primary antibodies used: α-tubulin (DM1α – Sigma), γ-tubulin (A. Desai), phospho-HistoneH3S10 (Sigma), HTP-3 (gift of A. Dernburg), RAD-51 (Novus Biologicals), SYP-1 (gift of A. Villeneuve) and phospho-SUN-1S8 (gift of V. Jantsch). Images were recorded on a Deltavision microscope; Z-stacks (0.2-μm sections) were deconvolved using softWorRx (Applied Precision), analyzed in ImageJ and imported into Adobe Photoshop CS4 (Adobe) for further processing.
Supplementary Material
HIGHLIGHTS.
Mitotic errors in the C. elegans germline cause increased programmed cell death
Increased germline cell death protects against the formation of aneuploid oocytes
The spindle checkpoint delays progression but defective nuclei eventually enter meiosis
Unrepaired meiotic double-strand breaks trigger DNA damage checkpoint-dependent death
Acknowledgments
We thank Abby Dernburg, Anne Villeneuve and Verena Jantsch for antibodies, the Caenorhabditis Genetics Center for strains and Becky Green for comments on the manuscript. This work was supported by an NIH grant (GM074215) to A.D. D.S. was supported in part by an Institutional NRSA Award from NIGMS to the UCSD Genetics Training Program (T32 GM008666). A.D. and K.O. receive salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kimble J, Crittenden SL. Germline proliferation and its control. Wormbook. 2005 doi: 10.1895/wormbook.1.13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–22. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 3.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Molecular Cell. 2000;5:435–43. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 4.MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Development. 2002;16:2428–42. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Developmental Cell. 2003;5:463–74. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 6.Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310:1683–6. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 8.Desai A, Rybina S, Müller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes & Development. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheeseman IM, McLeod I, Yates JR, Oegema K, Desai A. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Current Biology. 2005;15:771–777. doi: 10.1016/j.cub.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Matthews LR, Carter P, Thierry-Mieg D, Kemphues K. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. Journal of Cell Biology. 1998;141:1159–68. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell KF, Caron C, Kopish KR, Hurd DD, Kempheus KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–58. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- 12.Dammermann A, Müller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Developmental Cell. 2004;7:815–29. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analysis of the mitotic region in the Caenorhabditis elegans adult germ line. Molecular Biology of the Cell. 2006;17:3051–61. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J. Differential timing of S phase, X chromosome replication and meiotic prophase in the C. elegans germ line. Developmental Biology. 2007;308:206–21. doi: 10.1016/j.ydbio.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Mikeladze-Dvali T, von Tobel L, Stmad P, Knott G, Leonhardt H, Schermelleh L, Gönczy P. Analysis of centriole elimination during C. elegans oogenesis. Development. 2012;139:1670–9. doi: 10.1242/dev.075440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, La Volpe A, Jantsch V. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genetics. 2013;9:e1003335. doi: 10.1371/journal.pgen.1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Developmental Cell. 2008;14:263–74. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Craig AL, Moser SC, Bailly AP, Gartner A. Methods for studying the DNA damage response in the Caenorhabditis elegans germ line. Methods in Cell Biology. 2012;107:321–52. doi: 10.1016/B978-0-12-394620-1.00011-4. [DOI] [PubMed] [Google Scholar]
- 19.Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of p53 tumor suppressor is required for DNA damage-induced apoptosis. Current Biology. 2001;11:1722–7. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L, Gartner A, Vidal M, Hengartner MO. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Current Biology. 2002;12:1908–18. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 21.MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–50. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colaiácovo MP. The many facets of SC function during C. elegans meiosis. Chromosoma. 2006;115:195–211. doi: 10.1007/s00412-006-0061-9. [DOI] [PubMed] [Google Scholar]
- 23.Harper NC, Rillo R, Jover-Gill S, Assaf ZJ, Bhalla N, Dernburg AF. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Developmental Cell. 2011;21:934–47. doi: 10.1016/j.devcel.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- 25.Keeney S. Mechanism and control of meiotic recombination initiation. Current Topics in Developmental Biology. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 26.Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Current Biology. 2006;16:R217–228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes & Development. 2013;27:873–86. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Perez E, Colaiácovo MP. Distribution of meiotic recombination events: talking to your neighbors. Current Opinion in Genetics & Development. 2009;19:105–112. doi: 10.1016/j.gde.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. Journal of Clinical Investigation. 2012;122:1138–43. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. Journal of Cell Biology. 2012;199:871–81. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green RA, Kao HL, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K, Wang S, Starr DA, Hyman AA, Schedl T, Desai A, Piano F, Gunsalus KC, Oegema K. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–82. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.