Abstract
Dynamic transitions in the epigenome have been associated with regulated patterns of nuclear organization. The accumulating evidence that chromatin remodeling is implicated in circadian function prompted us to explore whether the clock may control nuclear architecture. We applied the 3C-derived 4C technology (Chromosome Conformation Capture on Chip) in mouse embryonic fibroblasts (MEFs) to demonstrate the presence of circadian long-range interactions, using the clock-controlled Dbp gene as bait. The circadian genomic interactions with Dbp are highly specific and are absent in MEFs whose clock is disrupted by ablation of the Bmal1 gene. We establish that the Dbp circadian interactome contains a wide variety of genes and clock-related DNA elements. These findings reveal a previously unappreciated circadian and clock-dependent shaping of the nuclear landscape.
INTRODUCTION
Circadian rhythms govern a large variety of physiological and metabolic functions1–3. The molecular mechanisms that underlie circadian rhythmicity are organized as an intricate and hierarchical network of transcriptional-translational loops4,5. This coordinated circadian machinery confers rhythmicity to a remarkable portion of the transcriptome. Approximately 10% of genomic transcripts present circadian fluctuations of their levels in many tissues or synchronized cells in culture6. A number of transcription factors have been implicated in the regulation of circadian gene expression. In mammals, the core circadian transcription factors include the transcriptional activators CLOCK and BMAL1, which heterodimerize and drive transcription of Per1–3, Cry1–3, Rev-Erbα and β (these are also known as Nr1d1 and Nr1d2) and Ror a, b, or c genes. PER and CRY proteins complexes inhibit CLOCK-BMAL1-driven transcriptional activity, thus giving rise to an autoregulatory transcriptional feedback loop. REV-ERB and ROR are nuclear receptors with opposed transcriptional activities, and dictate the expression of circadian genes such as Bmal1 (also known as Arntl). Indeed, the core clock exerts control over the circadian expression of other transcription factors such as Dbp, Tef, Nfil3 (or E4BP4), Id2, Hlf or Pgc1α (or Ppargc1a), which in turn impose circadian rhythmicity to the expression of downstream genes.
Accumulating evidence shows that chromatin remodeling events are involved in circadian regulation7,8. At a molecular level, CLOCK-BMAL1 and PER-CRY complexes have been shown to interact with several epigenetic modifiers such as JARID1a, EZH2, MLL1, WDR5, CBP-p300, and some histone deacetylases including the NAD+ dependent enzyme SIRT1 9–14. Both genetic and pharmacological approaches have shown that histone modifiers are implicated in circadian control, indicating that their coordinated actions are necessary to fine tune a dynamic circadian epigenome. Genome wide studies mostly consisting on ChIP-Seq analyses on livers harvested from mice in a circadian fashion demonstrate that the histone modifications which are introduced by the circadian epigenetic modifiers are indeed rhythmic at many of the circadian gene promoters. These include rhythmic changes in histone acetylation at H3 K9 and K14 and methylation at H3 K4 and K27 (Refs. 13,15,16), paralleling rhythmic recruitment of Pol II 13,17.
While these studies provided convincing evidence on the role of chromatin remodeling in circadian function, they do not explore whether the nuclear topological organization is influenced by the clock. As the functional compartmentalization of the nuclear interior is being unraveled18–20, the spatial positioning of genes and regulatory elements is becoming increasingly recognized as an important epigenetic regulatory layer 21–24. Hence, the three-dimensional (3D) folding of chromosomes inside of the nucleus has been extensively investigated by Fluorescence in situ Hybridization (FISH) and Chromosome Conformation Capture (3C) techniques25. The outcomes of these studies reveal a non-random distribution of interphase chromosomes in chromosome territories and in topologically associating domains with common epigenetic marks26–28. Positioning configurations of chromosomes and genes diverge between cell types, and they can vary in response to physiological processes such as transcriptional reprogramming, development or disease 29. In this respect, the circadian clock provides an ideal framework to study the interplay between genome organization and a physiologically dynamic program. Moreover, a potential regulatory role for the circadian clock in coordinating the nuclear high-order structure has been lacking. In this study we set out to investigate the contribution of the circadian clock to fine tuning temporal changes in genome organization. To do so, we sought to explore the circadian genomic interactome of the clock-controlled gene Dbp in mouse embryonic fibroblasts (MEFs). We found that the genomic interactions at the Dbp locus change paralleling the circadian cycle progression and the expression of the gene, delineating a Dbp circadian interactome. Remarkably, the Dbp circadian interactome was dependent on intact clock machinery, as it was not observed in Bmal1-deficient MEFs. Interestingly, the Dbp interactome enclosed other circadian genes, and was enriched in functionally-related genes.
RESULTS
Circadian long-range genomic interactions
We have applied the 3C-derived technique, 4C (Chromosome Conformation Capture on Chip)30,31, to detect preferential interactions of the Dbp gene with other loci in the genome during the circadian cycle. The Dbp gene was selected because its robust circadian expression dictated by rhythmic CLOCK:BMAL1 binding to E-boxes DNA elements located on its promoter and coding sequence 32,33. More precisely, we designed the bait for the 4C experiment in a region within intron 2 containing two E-boxes (Supplementary Figure 1A)34. Wild type mouse embryonic fibroblasts (MEFs) were synchronized along the circadian cycle with dexamethasone (DEX). Cyclic expression of Dbp displayed a peak 22 hours after synchronization (Circadian Time 22; CT22), and follows a very robust pattern of circadian expression 32,34 (Figure 1A). As expected, the oscillation is abolished when the circadian core machinery is perturbed, as shown in Bmal1−/− MEFs (Figure 1A, Supplementary Figure 1B). We sought to follow the Dbp circadian interactome in a high temporal resolution during the circadian cycle. Cells were harvested every four hours from CT22, which is the peak of Dbp expression, to CT34, corresponding to the trough. A twelve hours later time point, CT46, corresponding to the peak of the second expression cycle, was also included (Figure 1A, blue arrows). 4C analyses on Dbp genomic locus the selected time points were performed as described previously 31,35. About 75% of the positive probes on average mapped to chromosome 7, which allocates the Dbp locus. This observation is consistent with the spatial organization of the genome into chromosome territories, a known feature of almost all eukaryotic cells 27,36,37. Based on this notion, we adapted the 4C protocol to enhance the detection of interchromosomal (trans) contacts (see ref.35 and Methods for further explanation).
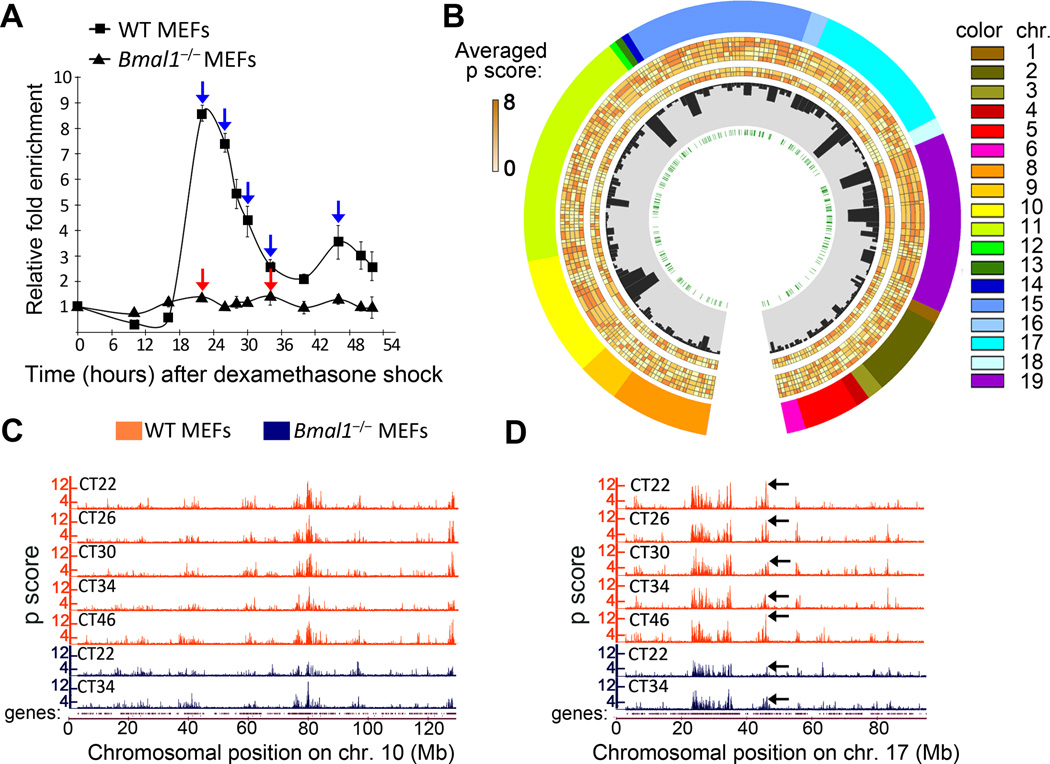
Figure 1. Characterization of genomic long-range interactions during the circadian cycle.
A, Dbp expression profile in wild type (WT) and Bmal1−/− MEFs after dexamethasone synchronization was analyzed by quantitative RT-PCR. Time 0 value was set to 1. Data were normalized to β-actin, and are represented as average and s.e.m. of three independent biological replicates. Blue and red arrows indicate the circadian times (CT) in which wild type (WT) and Bmal1−/− cells were harvested for 4C analysis. B, Circos plot representing the Dbp interactome. The layers indicate, from the outside to the inside: chromosome, number is indicated as a color code and length is proportional to the actual length of the interacting regions; averaged p scores for each genomic region shown as a color scale; histogram bars representing the gene content for each region; E-box elements location. The averaged p scores correspond to each 4C experiment, from the outside to the inside: WT CT22, WT CT26, WT CT30, WT CT34 and WT CT46, Bmal1−/− CT22 and Bmal1−/− CT34. C and D, microarray profiles showing the interaction frequencies (p score from the 4C data) between Dbp and mouse chromosomes 10 (C) and 17 (D). Orange and blue plots represent the data for wild type (WT) and Bmal1−/− MEFs respectively. The corresponding circadian time (CT) is indicated for each lane. The datasets are highly correlated, but major differences in the interaction frequencies become apparent (black arrows on plots in D). The genomic position in mm8 coordinates is indicated on the horizontal axis. The profiles for the rest of the chromosomes are provided in Supplementary Figure 3.
To explore the interaction frequencies in trans, we applied a running mean procedure using a window size on the mouse genome of 100 Kb centered at each probe of the microarray. This analysis shows a precise distribution and nonrandom interaction patterns for the Dbp locus that are coherent with interaction patterns previously described for other loci in several cell types 35,38. The genomic distribution of Dbp contacts along the circadian cycle remained largely unaltered, delineating the genomic spatial environment of the Dbp locus (Figure 1B, C and D, and Supplementary Figures 2 and 3).
We identified 201 genomic regions that contact the Dbp gene at any time of the circadian cycle in wild type MEFs, with a mean length of ~ 130 Kb (Figure 1B and Supplementary Table 1). While some chromosomes very rarely interact with Dbp, such as chromosomes 1, 3 or 12, others display preferential contacts with our bait on different locations (Figure 1B, outer layer). For example, on chromosome 11 we found 39 loci that contact Dbp. This indicates close spatial proximity and a high intermingling frequency between territories from chromosomes 7 and 11 (Refs. 39–41). Interestingly, the Dbp genomic contacts display a four-fold enrichment on gene content over randomized data. This finding is in keeping with previously described genomic contacts for active loci 31,35,38,42,43. In agreement with previous studies, the highest running mean values of the 4C data for Dbp are found at chromosomal locations with high gene density 31,35,44 (Supplementary Table 2; Supplementary Figure 3; plots for chromosomes 10, 17, 18 and 19 are shown as examples, Supplementary Figure 4A). This high enrichment for genes on specific regions interacting with Dbp (black histogram bars in the third layer in Figure 1B) reflects the spatial distribution of chromatin domains, where active and gene-dense domains tend to colocalize 26,27,45. Similar results were obtained when analyzing a parallel 4C experiment, in which a different MEF cell line was synchronized and samples were collected at CT22 and CT34 (Supplementary Figure 5).
We sought to determine the dynamics of the interaction of Dbp contacts during the circadian cycle. Our running mean analysis shows that the genomic locations of Dbp contacts remain overall similar along the circadian cycle. Remarkably, the interaction frequencies of Dbp with specific loci do vary in various degrees, depending on the locus and on the circadian time (Figure 1D; example shown by black arrows on colored genomic plots).
To determine the overall likelihood of interaction of the Dbp locus with a given genomic region, we calculated an averaged p score for every one of the 201 described contacts at each CT. The averaged p score was calculated considering the p scores of neighboring probes (see Methods). Then, the Dbp genomic contacts were classified according to their averaged p score at each CT. Interestingly, specific contacts that follow a cyclic pattern of interaction mirror Dbp circadian gene expression. These genomic regions efficiently contact Dbp at CT22, CT26 and CT48, corresponding to times at which Dbp shows highest expression. The interaction is virtually undetectable at CT34, a time of lowest Dbp expression (Figure 2A). We identified 29 genomic regions that meet these criteria; these constitute the Dbp circadian interactome (Figure 2B; Supplementary Figure 4, and Supplementary Table 3).
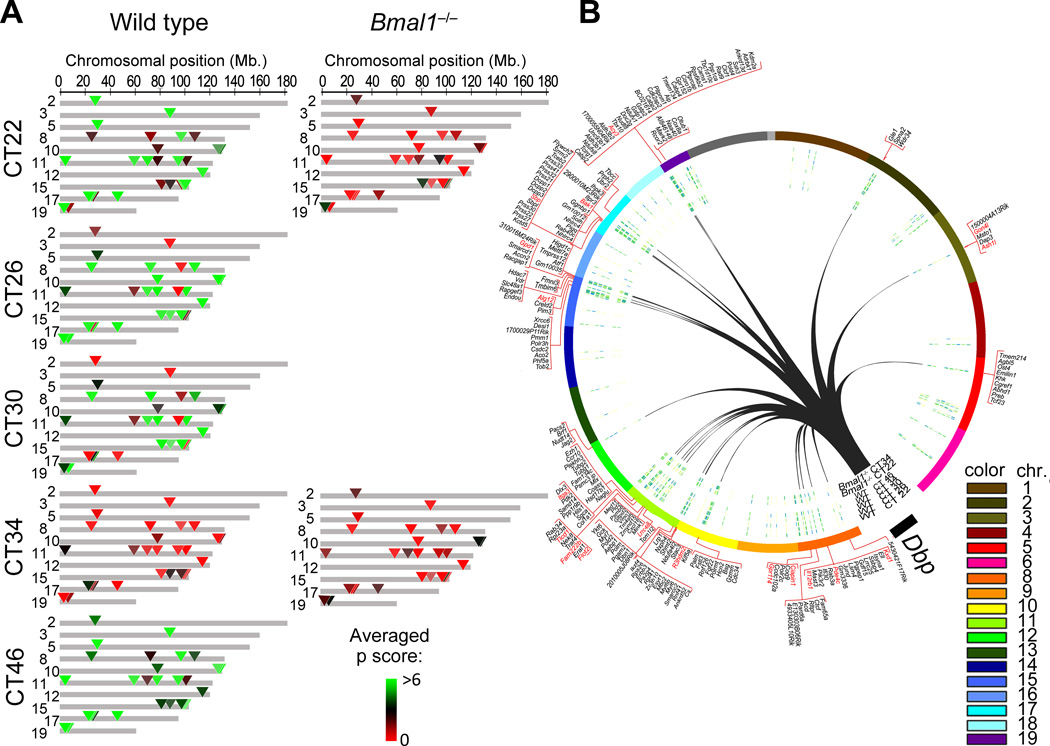
Figure 2. Genomic location of Dbp long-range contacts that follow a BMAL1-dependent circadian pattern of interaction.
A, genomic map of the Dbp circadian interactome at the indicated circadian times (CT) after dexamethasone synchronization in wild type (WT) and Bmal1−/− MEFs. Averaged p scores for each region are indicated in green-red color scale according to the intensity of the interaction which is proportional to the probe signal (4C over genomic DNA). Colored triangles indicate the positions of the Dbp circadian contacts. Gray areas do not show circadian contact. The genomic position in mm8 coordinates is indicated on the top horizontal axis. Chromosomes that do not present circadian interaction with Dbp are not represented in this plot. B, Circos plot representing the genome-wide view of Dbp circadian interactions (black lines) with the corresponding chromosomes in trans. The gene content corresponding to each contact region is indicated in the outer layer of the plot. In red color are the names of those genes that present circadian mRNA accumulation after dexamethasone synchronization as defined by the gene expression analysis (JTK p<0.01).
Bmal1 is essential for a specific circadian interactome
Next, we sought to determine whether the molecular mechanism involved in establishing the Dbp circadian interactome implicates the clock machinery. To do so, we used Bmal1−/− MEFs (Supplementary Figure 1A). BMAL1 is a basic helix-loop-helix (b-HLH) transcription factor that heterodimerizes with another b-HLH circadian transcription factor, CLOCK. These dimers bind in a precise time-of-the-day manner to E-boxes elements within circadian gene promoters to drive cyclic expression of genes 46,47. Ablation of BMAL1 drastically disrupts circadian gene expression (Figure 1A)48. A 4C analysis was performed for Bmal1−/− MEFs at CT22 and CT34 after DEX stimulation. CT22 and CT34 time points correspond to the peak and the trough of Dbp expression (Figure 1A, red arrows). As expected, Dbp contacts in Bmal1−/− MEFs appear overall similar to WT MEFs, showing a highly correlated profile of peaks and troughs (Figure 1C and 1D; Supplementary Figures 2 and S3; compare WT with KO plots). However, there is a markedly reduced contact frequency within the Dbp circadian interactome in Bmal1−/− MEFs (Figure 2A; Supplementary Table 3). Meaningfully, lack of BMAL1 is associated to a loss of circadian oscillation in the interaction of Dbp with the 29 selected genomic regions (Figure 2A, Supplementary Table 3). The profiles of the Dbp circadian contacts at both CT22 and at CT34 in Bmal1−/− MEFs show low interaction frequencies, being thereby highly similar to CT34 profile in wild type MEFs (Figure 2A). These findings indicate that the circadian system contributes to the establishment of a specific subnuclear genomic environment around the Dbp gene.
To gain insights into the molecular mechanisms that contribute to the specific circadian genomic architecture of the Dbp locus, we used MotifMap49 to identify the transcription factor binding sites on the promoters of the genes that associate with Dbp in a circadian fashion (Figure 2B; Supplementary Table 4). Among these promoters, we found a 2.5- fold enrichment on promoters containing E-boxes (p < 0.001, Fisher exact test) (Supplementary Table 4). E-boxes are highly conserved DNA elements that bind CLOCK:BMAL1 and involved in driving circadian gene expression32,50. We speculate that the Dbp gene is present within a subnuclear environment that is gradually modified during the circadian cycle, and that the circadian molecular machinery is implicated in establishing this pattern.
Visualizing Dbp specific circadian interactions
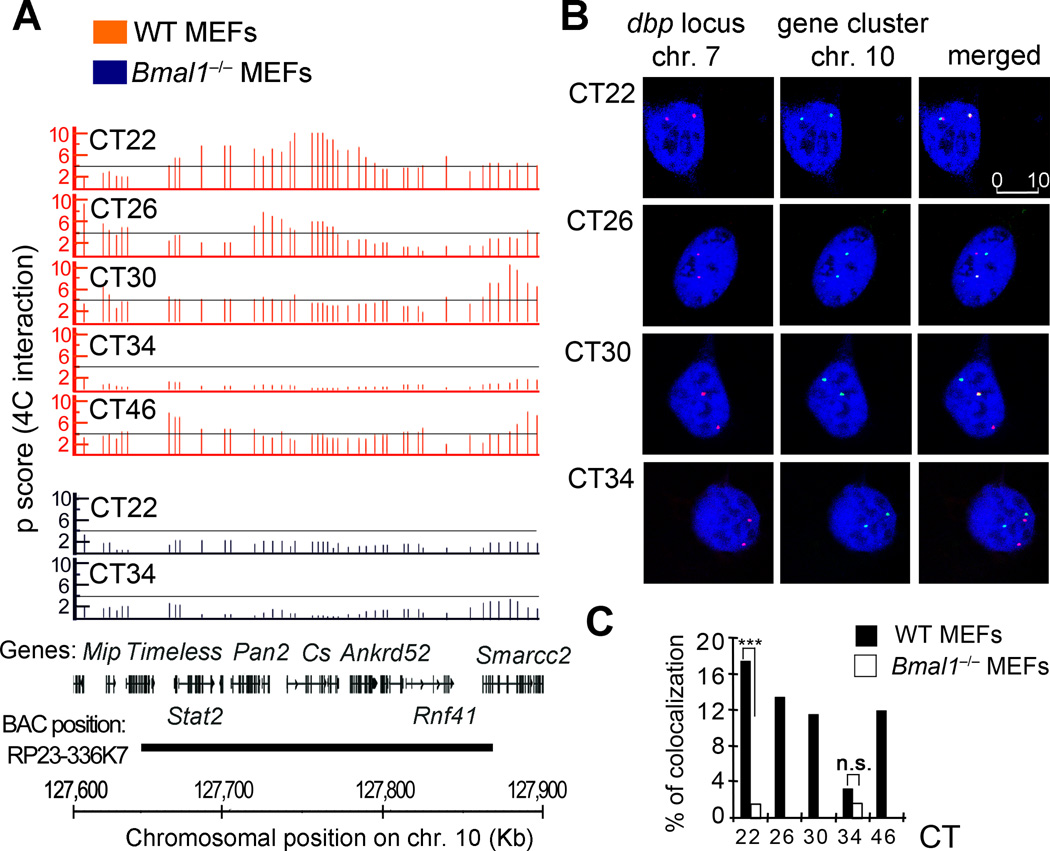
To confirm the cyclic variations in the subnuclear localization of the Dbp locus observed by 4C, we performed DNA fluorescence in situ hybridization (DNA-FISH). As example, we report the interchromosomal contact of Dbp with a high-gene content region on chromosome 10 that presents a very robust circadian profile of interaction peaking at CT22 (Figure 3A; Supplementary Table 5). The interaction frequency progressively decreases during the circadian cycle, and becomes very low at CT34 to then increase again at CT46 (Figure 3A). These cyclic interactions are abolished in Bmal1 −/− MEFs, where the frequency of interaction is low, and basically equivalent at both CT22 and CT34 (Figure 3A; Supplementary Table 5). Synchronized wild type and Bmal1 −/− MEFs were treated for FISH analysis at selected circadian times (Figure 3B). DNA-FISH signals from 280–350 cells were analyzed and monitored for fluorescence colocalization of red and green signals, whose overlapping generates a visible yellow signal. We found a marked correlation between the interaction probability from the 4C signal and the colocalization frequency revealed by DNA FISH (Figure 3). There is a significant increase of colocalization signals from the two loci at CT22 in WT MEFs when compared with the Bmal1 −/− cells at the same circadian time (Figure 3C, WT 17.4%; Bmal1−/− 1.5%, p<0.001, Fisher exact test). Wild type MEFs show a gradual decrease in colocalization events as the circadian cycle progresses. At CT34, a much smaller proportion of nuclei show colocalization signals (WT 3.2%; Bmal1−/− 1.4% of the MEFs, n.s., Fisher exact test). Colocalization is visualized again twelve hours later (Figure 3C). The range of colocalization frequencies is optimal for interacting loci analyzed by FISH techniques 44. The colocalization requires a functional clock, as demonstrated by the analyses of Bmal1−/− MEFs. Thus, the circadian machinery contributes to the adequate Dbp genomic location in a precise subnuclear environment.
Figure 3. FISH validation of 4C data.
A, 4C microarray profiles for a selected region on chromosome 10, showing the running mean enrichments of 4C signal over genomic signal for 100-kilobases windows (p scores). Threshold (p score = 4) is indicated as a grey line. The corresponding circadian time (CT) after dexamethasone synchronization, is indicated on the top right for each plot. Orange plots represent data for wild type (WT) MEFs, and the data corresponding to Bmal1−/− MEFs is shown in the blue plots. The gene position and the genomic location in mm8 coordinates for all the plots are indicated at the bottom of the figure. The region covered by the BAC used as a probe for this experiment is also indicated. B, representative double label DNA FISH for Dbp locus on chromosome 7 (red) and the selected region on chromosome 10 (green). DNA was counterstained with DRAQ5 (blue). Representative picture shots correspond to wild type MEFs synchronized with dexamethasone, and fixed for FISH analysis at the indicated hours after synchronization (circadian time, CT). 280 to 350 nuclei were analyzed for each time point and genotype using three biological replicates. The scale bar in µm is indicated in the top right panel. C, bar chart showing the interchromosomal interaction frequencies between Dbp and the selected gene cluster on chromosome 10. Dexamethasone synchronized wild type (WT) and Bmal1−/− MEFs were fixed at the indicated CTs for further DNA FISH analysis. Data is represented as a percentage of colocalization based on overlapping events of the green (chromosome 10 locus) and red (Dbp locus on chromosome 7) FISH probes from a total of 280–350 cell nuclei at each condition from three biological replicates. ***P <0.001; n.s. non-significant (P>0.05), Two tailed Fisher exact test.
Circadian contacts in functionally specific network of genes
Higher-order genome organization plays a major role in the efficient response to stimuli, in effective signaling pathways and in the coordination of lineage specific differentiation44,51,52. Our observations prompted us to question whether the genes within the Dbp circadian interactome could be functionally related. There is a lack of comprehensive information on circadian gene expression profiles in MEFs. To overcome this hurdle, we performed a high-resolution circadian gene expression microarray analysis on MEFs. Cells were synchronized with DEX and total mRNA was collected every four hours during the 24 hours circadian cycle, starting at CT22. Out of ~ 28,000 transcripts analyzed in the microarrays, a total of 1,189 (~ 4 %) transcripts displayed significantly rhythmic profiles (JTK p value <0.01; FDR < 5%; Supplementary Table 6, Supplementary Figure 4A). Hierarchical cluster analysis of the anchored data was used to order the genes by phase (Figure 4A). The mRNAs from Dbp and other circadian components (per2, per3, arntl, npas2, cry1, cry2, nr1d1, nr1d2) show a very robust circadian oscillatory pattern (Supplementary Figures 6A and 6B). These data confirmed that, as previously described, the transcriptional program in wild type MEFs oscillates systematically and gradually during the circadian cycle.
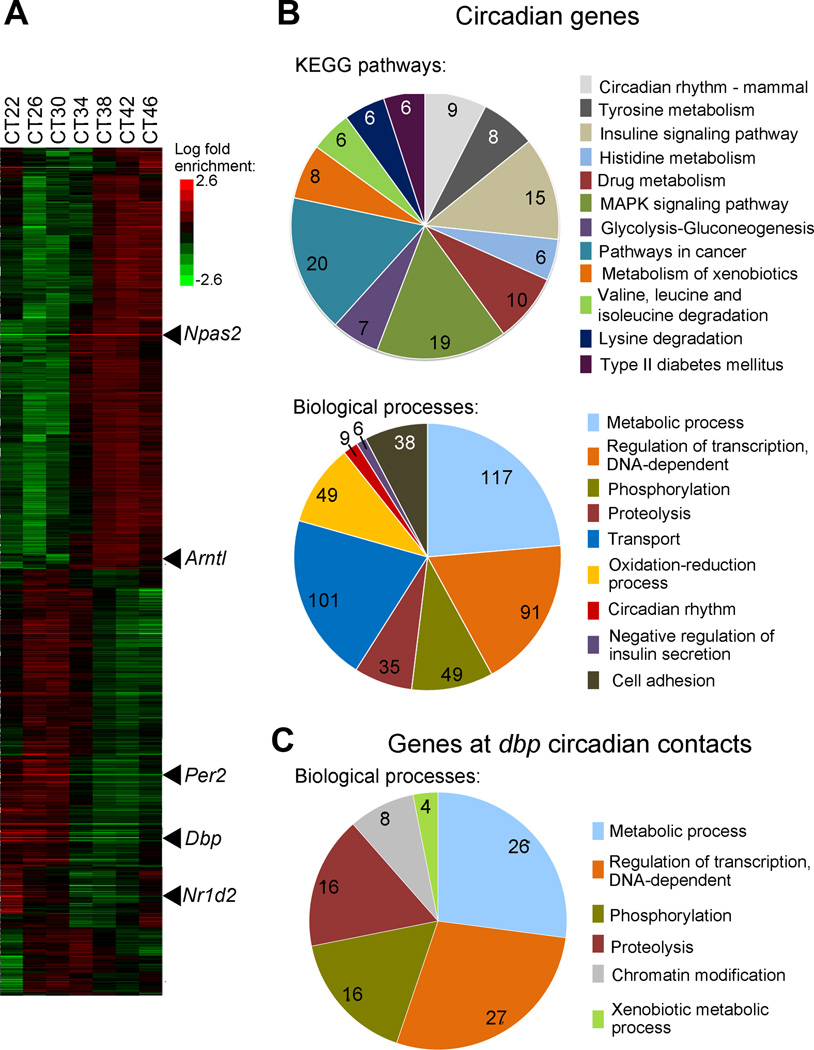
Figure 4. Circadian gene expression profiles in dexamethasone synchronized MEFs.
A, heat map view of global hierarchical cluster analysis of genes expressed in wild type MEFs on a circadian basis. Each gene is represented as a horizontal line, ordered vertically by phase determined by Cluster 3.0. mRNA was extracted from wild type MEFs every four hours after dexamethasone synchronization, and microarray analyses were done per triplicate using GeneChip Mouse Gene 2.0 ST Array. Some of the genes that constitute the circadian machinery are indicated as examples. CT, circadian time. B and C, pie charts represent functional categories of circadian genes in wild-type MEFs (B) and of the genes that are present on the 4C genomic regions that interact with Dbp on a circadian basis (C). The number of genes sharing common biological processes is presented. The key shows the functional categories used to classify the genes. P values for each functional category are indicated in Supplementary Table 7
The analysis of statistically enriched biological annotations in the identified circadian genes with respect to the mouse genome (Figure 4A and Supplementary Table 7) revealed a significant enrichment of genes involved in metabolic processes (p= 7.94E-14, Hypergeometric test corrected by Benjamini and Hochberg posttest). These include lipid, carbohydrate and nucleotide metabolism. We identified many genes encoding transcriptional regulators. Amongst them, 8 participate in chromatin remodeling processes, and 36 are known transcription factors. A significant enrichment in proteins involved in oxidation-reduction processes is apparent (p= 1.49E-9, Hypergeometric test corrected by Benjamini and Hochberg posttest), most of them consisting of proteins or enzymes that bind metabolites (NAD+, NADH, FAD or Haem) which regulate their catalytic activity. Several biological pathways appear significantly enriched in our dataset, such as mammalian circadian rhythms, amino acid metabolic pathways, insulin signaling and xenobiotic metabolism (p=1.54E-06, 1.29E-3 and 1.14E-3 respectively, Hypergeometric test corrected by Benjamini and Hochberg posttest) (Figure 4B; Supplementary Table 7). Our findings are consistent with previous reports describing circadian genes and biological pathways 13,53–55.
These data prompted us to investigate if common features are shared amongst the circadian transcriptome and the genetic content of the Dbp circadian contacts. There are 256 genes in the Dbp circadian interactome (Figure 2B, outer layer; Supplementary Table 3). They were analyzed to decipher their gene ontology enrichments (Figure 4C, Supplementary Table 7). When comparing the biological functions in both groups of genes, we found a striking parallel between the genes expressed in a circadian manner in wild type MEFs and those contained in the Dbp circadian interactome (Figure 4B and 4C. Supplementary Table 7). The two most enriched ontological groups are those related to metabolic processes and regulation of transcription (Supplementary Table 7). Both gene groups are enriched for phosphorylation and proteolysis biological processes. Moreover, amongst the genes found in Dbp circadian contacts, there is a significant enrichment for genes involved in xenobiotic metabolism (p=0.02; Hypergeometric test corrected by Benjamini and Hochberg posttest) (Figure 4C; Supplementary Table 7), a process that is under circadian control 54,56. DBP itself is a circadian-controlled transcription factor that confers rhythmicity to downstream target genes through binding to a D-box consensus promoter sequences. Interestingly, DBP has been shown to specifically modulate xenobiotic metabolism 57. In this context, the rhythmicity of the transcriptome and a time-controlled spatial nuclear organization could generate specialized environments involved in cyclic nuclear functions35,44.
Features of genes within the Dbp circadian interactome
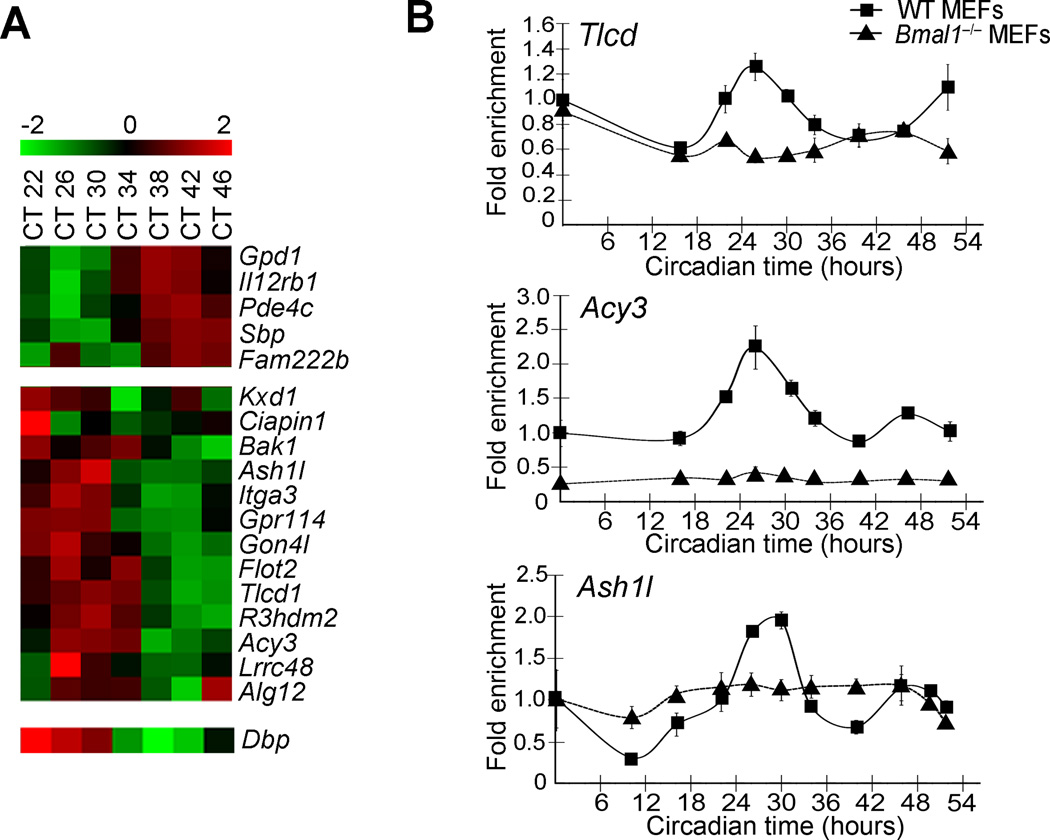
We investigated the time-specific expression of the genes within the Dbp circadian interactome. By comparing our MEFs circadian array data with the genes that appear to have circadian contacts, we found 18 genes that cycle in a circadian manner (JTK p value <0.01) (Figure 2B, red colored genes; Figure 5A, Supplementary Figure 4A and Supplementary Table 8). They can be classified in two major groups of 5 and 13 genes according to their expression phase (Figure 5A). The rhythmicity of selected genes was confirmed by RT-PCR and, as shown on Figure 5B, their mRNA oscillates in a circadian manner and further validates our microarray data.
Figure 5. Circadian expression of Dbp and genes located in spatial proximity.
A, Heat map showing the log2 expression values of the circadian genes that are found in the Dbp circadian interactome. Selected genes were plotted according to their phase. CT, circadian time. B, Quantitative Real time PCR of selected transcripts confirms the microarray data. Total RNA was collected before dexamethasone synchronization (CT0) and at 16, 22, 26, 30, 34, 40, 46 and 52 h after dexamethasone induction from wild type (WT) and Bmal1−/− MEFs. Data were normalized to β-actin, and are represented as average and s.e.m. of three independent biological replicates.
A motif discovery analysis on the promoters of these 18 genes revealed significant enrichment of recognition motifs for RORα and E-box elements (Supplementary Figure 6C), known to be centrally implicated in circadian gene expression58. A comparative analysis with available BMAL1 ChIP-seq databases13,47 indicates that the promoters of many genes located within the Dbp circadian interactome bind BMAL1 (Supplementary Figure 6D). Moreover, the motif analysis indicates that the promoters of the circadian genes within the Dbp interactome also may contain D-boxes (Supplementary Figure 6E). These findings, which are consistent with our microarray analysis, suggest the possibility that the clock machinery itself could be implicated in shaping nuclear genomic architecture.
DISCUSSION
The coordinated expression of circadian genes represents a remarkable paradigm of transcriptional control6. Accumulating evidence has recently shaped the notion that distinct changes in chromatin remodeling may be driven by the circadian clock to insure co-regulation of clock-controlled genes. The combined effects exerted by a variety of chromatin remodelers are assumed to lead to circadian activation and silencing of specific genes. The recent advances in the field of nuclear architecture suggest that the chromosome organization plays an active role in all genomic functions. In this context, understanding how the nuclear landscape is modified in a circadian time-specific manner can lead to unforeseen insights into the molecular mechanisms that sculpt circadian rhythms.
Our study reveals that the circadian clock is implicated in shaping temporal and spatial cycles in chromosomal organization. These variations in nuclear organization could provide a genomic frame to assist circadian gene expression of Dbp. Our data suggest that the genomic environment of Dbp locus around the circadian cycle remains largely constant. However, several large chromatin domains change their frequency of interaction in trans with Dbp, paralleling the progression of the circadian cycle and the transcriptional state of the gene (Figure 2). We present genetic evidence that BMAL1 protein plays a critical role in establishing a circadian time-specific Dbp interactome (Figures 2 and 3).
The outcomes of our comprehensive analysis have direct functional implications. In this respect, we have described a significant spatial clustering of genes and circadian-related DNA elements around the Dbp locus. These results point to the existence of subnuclear environments enriched with CLOCK-specific response elements. This finding is in agreement with previous reports showing that spatial congregation of DNaseI hypersensitive sites (DHS) is observed within DNA sequences that establish trans contacts 27,35. The role of specific transcriptional regulators in establishing the interactome is emerging as a new avenue for understanding the modulation and functions of genome topology 59,60. Here we provide evidence supporting that the circadian clock, including the BMAL1 transcription factor, is involved in shaping the nuclear architecture during the circadian cycle. This is further supported by our study on BMAL1-deficient cells, where the changes in the interactome around the circadian cycle are not present. Future studies are necessary to uncover the precise contribution and the hierarchical organization of all clock regulators in determining circadian genome topology.
Importantly, the analysis of the Dbp circadian interactome reveals a group of circadian genes whose transcript levels oscillate in a BMAL1-dependent fashion. While the enrichment of functionally related genes in a time-specific manner favors a scenario in which particular genomic environments assist the coordinated transcription of a program of gene expression 19,60, additional experiments will be needed to prove this point. Indeed, we cannot rule out the possibility that spatial clustering of functionally related genes appears a consequence of specific transcriptional regulators delineating the nuclear landscape during the circadian cycle. In this respect, it is possible that our study represents a paradigm for the function-structure-function model29, in which the genome activity drives the genome topology, which in turn impinges on the functions of the genome. In this model, the stochastic nature of gene expression becomes modulated by the genomic environment29 (Figure 6). In this scenario, the circadian expression of Dbp is dictated to some extent by the genomic environment (Figure 6), and gene positioning could be a key modulatory factor. However, further experiments are necessary to elucidate to what extent the genome topology controls circadian gene expression and vice-versa. The development of new technologies that allow direct manipulation of specific genomic loci in a subnuclear scale will help us to gain more insights into this question. Finally, it could be hypothesized that the availability of specific metabolites or other events related to the cell cycle may also contribute to establish the described the circadian interactome. The cyclic changes observed here could also be implicated in other nuclear, non-transcriptional functions.
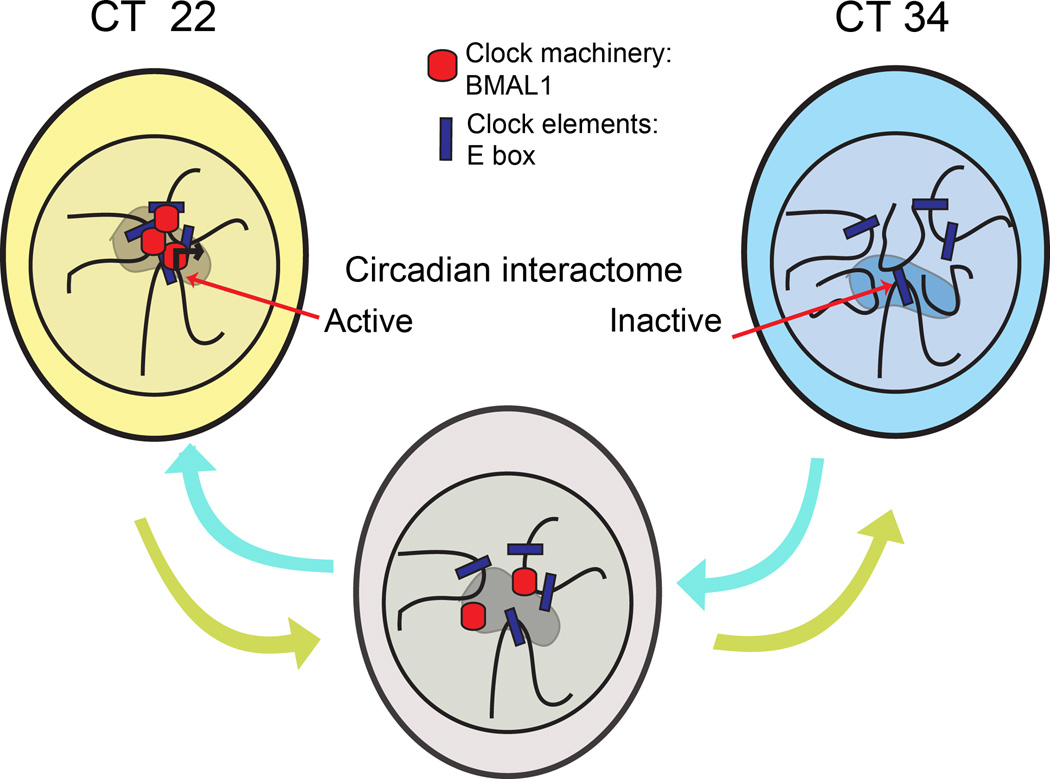
Figure 6. A schematic model of the cyclic events in chromosomal organization along the circadian cycle.
Hypothetical model of cyclic long-range chromosomal interactions dictated by the circadian clock. During the circadian time (CT) which corresponds with high Dbp gene expression (CT22), a specific genomic environment is associated with gene transcription (represented as a shaded area in the nucleus of the cells). Genes participating in this process constitute the circadian interactome. As the circadian cycle progresses, the clock machinery is disassembled and circadian gene expression decreases. When Dbp circadian transcription is at its trough (CT34), the clock machinery is already uncoupled from E-boxes, and this event correlates with a different genomic environment around Dbp locus (see Discussion).
While to date there is a lack of solid biochemical approaches that would allow deciphering the molecular mechanisms implicated in the organization of the circadian interactome, the conceptual advance derived from this research involves the notion that the circadian clock is able to operate on the physical organization of the nuclear landscape. Our study opens new avenues for understanding the global coordination of circadian gene expression. We reason that the circadian genome topology could be further studied in the context of circadian-related pathological conditions.
ONLINE METHODS
MEFs culture
MEFs cells were established from either wild type mice or Bmal1-null mutant littermates 48, as routinely done in our laboratory 61,62. MEFs were maintained in DMEM supplemented with heat inactivated 10% FBS (GibcoBRL), non-essential aminoacids (Gibco) and antibiotics. Cells were cultured at 37 °C in 5% CO2. Absence of BMAL1 protein was confirmed by western blot using α-BMAL1 antibody (Abcam Ab3350) at 1:2,500 dilution in TBST-5% nonfat milk. α-Actin antibody (Abcam Ab3280) at 1:10,000 dilution in PBST was used as a loading control.
RNA extraction and GeneChip analyses
RNA isolation for microarray was done as previously described 12,63. MEFs from wild-type mice were plated on 6-well dishes and grown until confluence. For synchronization, cells were treated with 100nM dexamethasone in DMEM for 1 hour64. The hormone was washed out twice with PBS, and normal medium was added to the cells. MEFs were harvested for RNA isolation at the circadian times (CT) of interest, considering the time of treatment CT=0. For each time point, we prepared samples to assay in triplicate. Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer instructions. RNA was further cleaned up using RNeasy Mini Kit (Quiagen). RNA quality control was assayed using Agilent 2100 Bioanalyzer platform, and quantified with NanoDrop spectrophotometer. Probe synthesis and chip hybridization was performed at the UCI DNA Microarray Core Facility. Briefly, 100ng of total RNA per sample were used as a template to obtain cDNA using the GeneChip cDNA synthesis Kit (Affymetrix). Mouse Gene 2.0 ST arrays (Affymetrix) were used to characterize the gene expression levels during the circadian cycle in WT MEFs. Data were normalized, and for each probe set, the measurements of the three microarrays were averaged. For analysis of rhythmic transcripts, the nonparametric test, JTK-cycle, was used incorporating a window of 20–28 h for the determination of circadian periodicity 65. The algorithm used by the JTK-cycle analysis is a combination of Jonckheere-Terpstra test for monotonic ordering and Kendall’s τ test for association of measured quantities. It is optimized for detection of rhythmicity. Transcripts were considered circadian if JTK p<0.01. For this significance, false discovery rate (FDR) was calculated by permutations of the data, and corresponds to 5%. Circadian genes were clustered and heat maps were visualized using the softwares Cluster 3.0, Java Treeview66,67 and MultiExperiment Viewer68,69. Briefly, Log2 transformed expression values at the analyzed CTs for the selected circadian genes were centered at their mean, so that the mean value for each gene is set to 0. Hierarchical clustering was then performed and visualized as a heatmap.
Ontological Analyses
automated extraction of concurrent gene ontology annotations was performed with GeneCodis program 70,71. The gene ontology category “Biological processes” was selected, and is extracted from http://www.geneontology.com. Biological pathways are extracted from http://www.genome.jp/kegg/ as “KEGG pathways”, from the Kyoto Encyclopedia for Genes and Genomes. The p values calculations with GeneCodis are performed using the hypergeometric distribution statistical test70,71. The p value was then corrected by an implementation of the False Discovery Rate method of Benjamini and Hochberg as described70,71.
Quantitative real-time RT-PCR
cDNA was obtained by retrotranscription of 1ug of total mRNA using iScript cDNA synthesis kit (Biorad) following the manufacturer’s instructions. Real-time RT-PCR was done using the Chromo4 real-time detection system (Bio-Rad). The PCR primers for Dbp mRNA were described elsewhere14. For a 20 µl PCR, 50 ng of cDNA template was mixed with the primers to final concentrations of 200 nM and mixed with 10 µl of iQ SYBR Green Supermix (Bio-Rad). The reactions were done in triplicates using the following conditions: 3 min at 95 °C, followed by 40 cycles of 30 s at 95 °C and 1 min at 60 °C.
Chromosome conformation capture on chip (4C)
4C assays were performed as described previously31,35 with some modifications. For each experiment, 20 million cells were synchronized with dexamethasone. Crosslinking was done in a final concentration of 2% formaldehyde, at 37°C for 10 min. The reaction was quenched by the addition of glycine (final concentration 0.125 M). Cells were washed with cold PBS and lysed for 1 hour at 4 °C with nuclear extraction buffer (10mM Tris-HCl, pH8.0, 10 mM NaCl, 0.2% NP-40, 1X complete protease inhibitors (Roche)). Cells were collected by centrifugation, resuspended in 1 ml of restriction buffer (New England Biolabs Buffer 2) and kept frozen at −80°C until all samples were harvested and processed. Once all the samples were ready, a final concentration of 0.3% SDS was added to 10 million cells, and incubated at 37°C during one hour. Triton X-100 was then added to a final concentration of 1.8%, and samples were incubated during 1 hour at 37 °C. Crosslinked DNA was digested with 500 U of HindIII (New England Biolabs) at 37°C overnight. The enzyme was heat inactivated (65°C for 30 min) and the reaction was resuspended in a total volume of 7 ml with 1X ligation buffer and 100 U of T4 DNA Ligase (Roche), followed by incubation at 16°C overnight. Samples were treated with 500 µg of Proteinase K (Ambion) and crosslinking was reversed at 65°C overnight. DNA was phenol extracted and ethanol precipitated. 50 µg of this 3C template were digested overnight with 50 U of Csp6I enzyme (Fermentas). The enzyme was heat inactivated and the DNA was phenol extracted, ethanol precipitated and subsequently ligated at low concentration to promote circularization, that is, intramolecular ligation events, in a final volume of 7 ml with 100 U of T4 DNA Ligase (Roche). Ligation products were phenol extracted and ethanol precipitated using glycogen as a carrier.
PCR reactions were performed using the Expand Long Template PCR system (Roche). We used the maximum amount of template that still shows a linear amplification by testing serial dilutions of each sample and quantifying the PCR products in agarose gels, using QuantityOne software. Finally, PCRs were done in a PTC-100 thermal cycler (Bio-Rad) using the following conditions: 2 min at 95 °C, followed by 30 cycles of 15 s at 94 °C, 1 min at 60 °C and 3 min at 68 °C; and a final extension step of 7 min at 68 °C was included. Dbp bait was amplified with the following inverse primers: Dbp4CHindIII: 5’-TCACAGGGTTTATGCCTGACAGGAT-3’; Dbp4CCsp6I: 5’-AGCACTGGTTTTTGCCTGGCGGTC-3’. The position of the bait in Dbp locus and the restriction fragments are indicated in Supplementary Figure 1A. 16 PCR reactions were pooled and purified using the QUIAquick nucleotide removal kit (Quiagen). 4C template DNA was hybridized to custom-made microarrays designed as described previously35, using the mm8 assembly of the mouse genome. The hybridization of DNA samples for ChIP-Chip analysis was performed by following manufacturer’s intructions with modificatins. Briefly, 20ug of the Cy5-labeled 4C sample and 20ug of the Cy3-labeled reference sample (genomic DNA) were mixed, dried down, and resuspended in 20 ul of NimbleGen Hybridization Buffer (NimbleGen Systems). After denaturation, hybridization was carried out in a MAUI Hybridization System (BioMicro Systems) for 24 h at 42°C. The arrays were washed using NimbleGen Wash Buffer System (NimbleGen Systems), dried by centrifugation and scanned in a two-color scanning system. Images were processed using ImageJ and Niblescan softwares as reported in the user’s guide.
The sequences located on chromosome 7 contacted the Dbp locus very frequently and they were highly represented in the 4C DNA library. Subsequently, under these conditions, the labeling and hybridization process saturated the probes from chromosome 7 in the microarray, thus generating non-quantitative data.
4C data analysis for quantitative interactions
Statistical analysis of the 4C microarray data was performed using the R environment implemented with ACME package (http://www.bioconductor.org/packages/release/bioc/html/ACME.html), as reported previously35. Briefly, the ratio sample-to-genomic DNA was calculated for each probe. Normalized data for each probe was log2 transformed. Using a procedure consisting on a running window Chi-squared test of 100-kb centered at the probe, a p-value was generated for each probe. The data was log10 transformed, thus a −log10(p-value) was assigned to each probe, as a measure of its interacting frequency. The −log 10 (p-value) is called p-score along the text. Probes with a −log10(p-value) or p-score greater than 4 in at least one of the time points were selected positive, which corresponded to FDR of ~0.3%.
The 100-kb flanking region around the probes within the top 0.1% of the p-scores was extracted and considered significant. Overlapping regions were further merged to get a set of 201 non-overlapping regions with a mean length of 130-kb. When several regions are merged, their −log10(p-value) or p-scores are averaged, a procedure which generates the averaged p-score. This procedure was carried out at different window lengths and p-values cut-offs, but there was no qualitative change in the results. Hence finally the procedure produced 201 non-overlapping regions with a time series profile of their interactions with the Dbp bait as measured by their −log10(p-value) at each time point. Using K-means clustering with Pearson correlation as the similarity metric, the interaction profiles were further clustered into 12 groups 72. We selected the genomic regions of two of the clusters, which presented a difference of (averaged −log10(p-value) at CT22 or CT26) – (averaged −log10(p-value at CT34) > 4. The selected regions were visually inspected in the K-means plots, which confirm their cyclic pattern over the investigated time points. The genomic locations were viewed with the Integrated Genome Browser (IGB) software (http://bioviz.org/igb/). FDR was determined as previously described35. Briefly, permutations of the data generated randomized 4C values for all the genomic positions represented as probes in the microarray. A Chi-squared test was performed on this data as described, which allows the FDR calculation by dividing the number of positive probes by the actual number of positive probes from the 4C microarray. All the genome analyses were finally converted to mm9 genomic coordinates and illustrated in the Supplementary Tables.
DNA FISH
For 3D FISH, MEFs were grown on UV-sterilized 22 × 22 mm coverslips placed onto 6-well plates. Cells were synchronized with dexamethasone, and grown until the desired time point. Fixation was performed in 4% paraformaldehyde for 10 min. Cells were permeabilized in 0.5 % saponin (Sigma)/0.5 % Triton X-100/ PBS 1X for 20 min at room temperature. After two washes with 2X SSC pH 7, cell were stored in 50% formamide/2X SSC buffer pH 7 during 2 hours. We used bacterial artificial chromosomes (BACs) as templates for the probes, obtained from BACPAC resources center. BACs IDs are: for Dbp, RP23–36B116; for region in chromosome 10, RP23–100C9 and RP23–336K7. BAC DNA was isolated and verified by PCR. 1µg of BAC DNA was labeled by nick translation with either biotin (Roche; Biotin-Nick Translation Mix) or digoxigenin (Roche, DIG-Nick Translation Mix), following the manufacturer’s instructions. For each slide, 600 ng of probe were ethanol precipitated in the presence of 8µg of Mouse COT1 DNA (Invitrogen) and 40 µg of tRNA. The pellet was resuspended in 5 µl of formamide, and 5 µl of 2X hybridization buffer (20% dextran sulphate/4X SSC, /2% Tween 20) was added. The probe was denatured together with the nuclei for 30 min at 80 °C, and left to hybridize during two overnights at 37 °C in a humidified chamber. The nonhybridized probe was washed out with successive washes consisting on: 3 washes with 50% formamide/2X SSC during 5 min at 45 °C, 3 washes with 1X SSC during 5 min at 60 °C and one with 0.05% tween-20/4XSSC at room temperature for 5 min. A blocking step was included with blocking solution (3% BSA/0.05% tween-20/4X SSC) during 30 min. Labeled probes were detected with 1:200 dilution in blocking solution of fluorescein-avidin (Vector labs) and Rhodamin anti-DIG (Roche), during 1 hour at 37°C in a humid chamber. The excess of fluorescent reagents were washed out by 3 washes of 5 min at 45 °C with 0.05% tween/4X SSC. DNA was counterstained with DRAQ5™ (Biostatus), according to manufacturer’s protocol. Slides were mounted in Vectashield HardSet (Vector labs) and stored at 4°C until visualization.
Microscopy and image analysis
We examined DNA-FISH signals on a Leica TSC SP5 confocal microscope with 60 × 1.4 immersion oil objective lens, controlled by a Leica LAS System. We collected confocal stacks at randomly chosen fields and included all cells with clear signals for the four alleles in the analysis. We captured confocal stacks for these cells using green and red channels to detect both Dbp and the genomic locus on chromosome 10. A minimum of 250 cells were analyzed and scored for colocalization events by a person blind to the experimental sampling.
Promoter analysis
Genes overlapping 4C regions were identified by using all known RefSeq 73 transcripts for the mouse genome. All gene promoters that overlap regions from the 4C data were further analyzed for transcription factor binding sites using the MotifMap database 49,74. Briefly, in MotifMap the mouse genome is searched for binding sites using position specific weight matrices from TRANSFAC75 and JASPAR76. The sites are then filtered with a combination of motif matching score (Z-score) and conservation score (Bayesian Branch Length Score – BBLS). For this study, only sites with BBLS greater than 1 were retained. Using all promoters (defined as 8 KB upstream and 3 KB downstream of the transcription start sites) in the mouse genome as control (mm9), a Fisher test was performed to find transcription factor binding sites that are specifically enriched in the promoters that overlap the 4C regions.
Motif discovery analysis was performed using the algorithm MEME 4.9.0, available at http://meme.nbcr.net/meme/cgi-bin/meme.cgi. The selected promoters were used as inputs in FASTA format, and output files were examined to retrieve matching motifs for known transcription factors. To do so, the discovered motifs were queried against the database JASPAR76 using Tomtom.
Circos plots
Circos plots were constructed using the Circos software 77. Data from the 4C experiment, gene-overlap, and transcription factor binding site analysis were used as input.
Supplementary Material
Acknowledgements
We thank R.L. Schiltz and T.A. Johnson (National Cancer Institute, National Institute of Health, Bethesda, Maryland, USA) for assisting with cell culture; R. Orozco-Solis, K. Eckel-Mahan, S. Sahar (Center for Epigenetics and Metabolism, University of California Irvine, Irvine, California, USA) and M. Groudine (Fred Hutchinson Cancer Research Center, Seattle, Washington, USA) for critical reading of the manuscript; S. Dilag (Center for Epigenetics and Metabolism, University of California Irvine, Irvine, California, USA) for technical support; X. Kong (Department of Biological Chemistry, University of California Irvine, Irvine, California, USA) for sharing FISH expertise and reagents; and all the members of P.S.-C., G.L.H. and P.B. laboratories for discussions. This work was in part supported by the following grants: European Molecular Biology Organization (EMBO) long term fellowship ALTF 411-2009 (to L.A.-A.) US National Institutes of Health (NIH) Grants R01-GM081634 and AG033888 (to P.S.-C.), and Sirtris Pharmaceuticals SP-48984 (to P.S.-C.). The work of V.R.P. and P.B. is supported by the following grants: National Science Foundation IIS-0513376, NIH Grants LM010235-01A1 and 5T15LM007743 (to P.B.).
Footnotes
Accession codes: The microarray source data has been already uploaded to the GEO database. The accession codes will be reported as soon as they come available, with the corrections to the galley proofs at the latest.
Author Contributions: L. A-A., O.H., G.L.H. and P. S-C. conceived and designed research. L.A-A. and O.H. performed 4C experiments. L.A-A. performed FISH and gene expression experimental work. L.A-A., O.H., V.R.P. and P.B. performed bioinformatical analyses. V.R.P. performed promoter analysis using MotifMap. L.A-A. and O.H. analyzed and interpreted the data. L.A-A. and P. S-C. wrote the manuscript.
REFERENCES
- 1.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 2.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 3.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annual review of genetics. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 4.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annual review of neuroscience. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Doherty CJ, Kay SA. Circadian control of global gene expression patterns. Annual review of genetics. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguilar-Arnal L, Sassone-Corsi P. The circadian epigenome: how metabolism talks to chromatin remodeling. Current opinion in cell biology. 2013 doi: 10.1016/j.ceb.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Molecular cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 10.DiTacchio L, et al. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etchegaray JP, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. The Journal of biological chemistry. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 12.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nature structural & molecular biology. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 16.Vollmers C, et al. Circadian Oscillations of Protein-Coding and Regulatory RNAs in a Highly Dynamic Mammalian Liver Epigenome. Cell Metabolism. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Martelot G, et al. Genome-Wide RNA Polymerase II Profiles and RNA Accumulation Reveal Kinetics of Transcription and Associated Epigenetic Changes During Diurnal Cycles. PLoS biology. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakim O, Sung MH, Hager GL. 3D shortcuts to gene regulation. Current opinion in cell biology. 2010;22:305–313. doi: 10.1016/j.ceb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajapakse I, Groudine M. On emerging nuclear order. The Journal of cell biology. 2011;192:711–721. doi: 10.1083/jcb.201010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giles KE, Gowher H, Ghirlando R, Jin C, Felsenfeld G. Chromatin boundaries, insulators, and long-range interactions in the nucleus. Cold Spring Harbor symposia on quantitative biology. 2010;75:79–85. doi: 10.1101/sqb.2010.75.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Current opinion in genetics & development. 2012;22:110–114. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Seminars in cell & developmental biology. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature reviews Genetics. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 25.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavalli G, Misteli T. Functional implications of genome topology. Nature structural & molecular biology. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlsson R. & Gondor, A The 4C technique: the 'Rosetta stone' for genome biology in 3D? Current opinion in cell biology. 2007;19:321–325. doi: 10.1016/j.ceb.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature genetics. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 32.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nature genetics. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 33.Stratmann M, Suter DM, Molina N, Naef F, Schibler U. Circadian Dbp Transcription Relies on Highly Dynamic BMAL1-CLOCK Interaction with E Boxes and Requires the Proteasome. Molecular cell. 2012;48:277–287. doi: 10.1016/j.molcel.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Wuarin J, Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63:1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- 35.Hakim O, et al. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome research. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cremer T, Cremer M. Chromosome territories. Cold Spring Harbor perspectives in biology 2, a003889. 2010 doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 40.Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. The Journal of cell biology. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature genetics. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 42.Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Current opinion in cell biology. 2012;24:359–366. doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shopland LS, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. The Journal of cell biology. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilbert N, et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS biology. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daily K, Patel VR, Rigor P, Xie X, Baldi P. MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC bioinformatics. 2011;12:495. doi: 10.1186/1471-2105-12-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. Journal of biological rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- 51.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosak ST, et al. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS biology. 2007;5:e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 56.Claudel T, Cretenet G, Saumet A, Gachon F. Crosstalk between xenobiotics metabolism and circadian clock. FEBS letters. 2007;581:3626–3633. doi: 10.1016/j.febslet.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metabolism. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 58.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nature genetics. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 59.Hakim O, et al. Spatial congregation of STAT binding directs selective nuclear architecture during T cell functional differentiation. Genome research. 2012 doi: 10.1101/gr.147652.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Current opinion in genetics & development. 2010;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 61.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 62.Pando MP, Morse D, Cermakian N, Sassone-Corsi P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–117. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- 63.Grimaldi B, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell metabolism. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 65.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of biological rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 68.Saeed AI, et al. TM4 microarray software suite. Methods in enzymology. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 69.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 70.Nogales-Cadenas R, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic acids research. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome biology. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baldi P, Brunak S. Bioinformatics: the Machine Learning Approach. Vol. 476. MIT Press; 2001. [Google Scholar]
- 73.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic acids research. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie X, Rigor P, Baldi P. MotifMap: a human genome-wide map of candidate regulatory motif sites. Bioinformatics. 2009;25:167–174. doi: 10.1093/bioinformatics/btn605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matys V, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic acids research. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Portales-Casamar E, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic acids research. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.