Abstract
Background
The RSK2 gene is responsible for Coffin-Lowry syndrome, an X-linked dominant genetic disorder causing mental retardation, skeletal growth delays, with craniofacial and digital abnormalities typically associated with this syndrome. Craniofacial and dental anomalies encountered in this rare disease have been poorly characterized.
Methodology/Principal Findings
We examined, using X-Ray microtomographic analysis, the variable craniofacial dysmorphism and dental anomalies present in Rsk2 knockout mice, a model of Coffin-Lowry syndrome, as well as in triple Rsk1,2,3 knockout mutants. We report Rsk mutation produces surpernumerary teeth midline/mesial to the first molar. This highly penetrant phenotype recapitulates more ancestral tooth structures lost with evolution. Most likely this leads to a reduction of the maxillary diastema. Abnormalities of molar shape were generally restricted to the mesial part of both upper and lower first molars (M1). Expression analysis of the four Rsk genes (Rsk1, 2, 3 and 4) was performed at various stages of odontogenesis in wild-type (WT) mice. Rsk2 is expressed in the mesenchymal, neural crest-derived compartment, correlating with proliferative areas of the developing teeth. This is consistent with RSK2 functioning in cell cycle control and growth regulation, functions potentially responsible for severe dental phenotypes. To uncover molecular pathways involved in the etiology of these defects, we performed a comparative transcriptomic (DNA microarray) analysis of mandibular wild-type versus Rsk2-/Y molars. We further demonstrated a misregulation of several critical genes, using a Rsk2 shRNA knock-down strategy in molar tooth germs cultured in vitro.
Conclusions
This study reveals RSK2 regulates craniofacial development including tooth development and patterning via novel transcriptional targets.
Introduction
The ribosomal S6 family of serine/threonine kinases is composed of 4 highly related members in mammals: RSK1 (human chromosome 3), RSK2 (RPS6KA3, Xp22.2-p22.1), RSK3 (chromosome 6) and RSK4 (Xq21), which are 75% homologous and are implicated in several important cellular events including proliferation, differentiation, cellular stress response and apoptosis. Mutations in RSK2 cause X-linked Coffin-Lowry syndrome (OMIM #303600) characterized by psychomotor and growth retardation, with typical facial and digital abnormalities and progressive skeletal malformations like delayed bone development, spinal kyphosis/scoliosis, and sternum/rib protrusins (pectus carinatum) or depression (excavatum) [1], [2], [3]. Orodental findings include a high narrow palate, a midline lingual furrow, malocclusion, hypodontia, and peg shaped incisors (Fig. 1). Premature loss of the primary dentition was also observed, and wide spaced teeth and large medial incisors were reported [1], [4], [5].
Figure 1. Dental anomalies encountered in Coffin-Lowry syndrome.
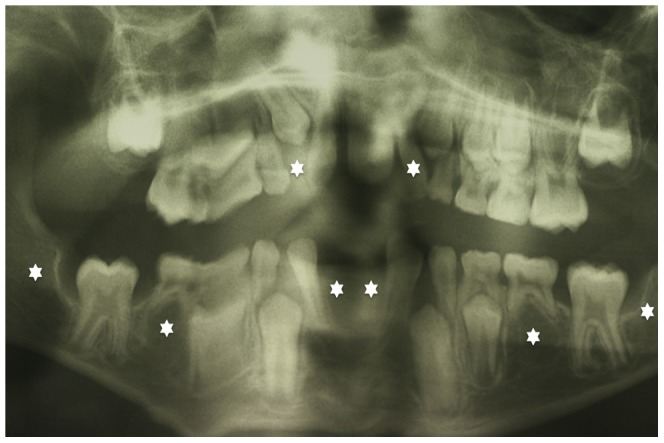
This panoramic radiograph shows oligodontia (more than six permanent missing teeth (white stars: 12, 22, 41, 31, 45, 35, 47, 37). The deformations observed on this panoramic radiograph are linked to movements during the X-Ray acquisition process and difficulties for handicapped patient to remain still during the procedure.
RSKs are Ser/Thr protein kinases that act at the distal end of the Mitogen-Activated Protein Kinase/Extracellular signal-Regulated Kinase (MAPK/ERK) signalling pathway. The various RSK proteins are widely expressed, with many cell types expressing several members. RSKs are directly phosphorylated and activated by ERK1/2 in response to several growth factors [6]. In the cytosol, RSK proteins have been shown to phosphorylate substrates including GSK3, L1CAM, the Ras GEF-Sos, IkB, the p34cdc2-inhibitory kinase Myt1, the translation factors eEF2 and eIF4B, and the pro-apoptotic protein BAD [7], [8]. Moreover, upon activation a fraction of the cytosolic RSKs translocate to the nucleus where they are thought to regulate gene expression through phosphorylation of transcription factors such as CREB1, ERa, Nurr77 and SRF, as well as histones [7]. The contribution of each RSK family member to the in vivo activation of most of these substrates is currently not well defined.
RSK orthologues have been identified in mouse, rat, chicken, Xenopus and Drosophila [9]. Rsk2 knockout mice display spatial learning and memory impairment [10] and develop a progressive osteopenia due to impaired osteoblast function [11]. Lack of phosphorylation of the transcription factor ATF4 is responsible for the skeletal abnormalities in Rsk2 knockout mice [10]. Murine Rsk2 is highly expressed during somitogenesis, suggesting skeletal and muscle growth and/or patterning roles, potentially leading to the numerous skeletal abnormalities encountered in the human syndrome [12]. It is interesting to note that Rsk2 shows specific developmental patterns of expression in the maxillary and the mandibular components of the first branchial arch [12], and that craniofacial and dental anomalies are present in the clinical synopsis of Coffin-Lowry syndrome.
The mouse dentition is a powerful and useful model to study the mechanisms leading to human dental anomalies, despite some intrinsic differences. The mouse has a monophyodont dentition that encompasses only four permanently growing incisors and 12 molars separated by a diastema [13]. Most mutant mouse models for human syndromic and non-syndromic genetic disorders displaying dental defects mimicking human pathologic phenotypes [14], although some discrepancies have been reported [15]. In this study, we describe the craniofacial and orodental phenotype of Rsk2-/Y knockout mice, as well as triple Rsk1,2,3-/- mutants. By employing several additional approaches (expression analysis by in situ hybridization, comparative transcriptomic analysis, shRNA knock-down in tooth germ explants) we establish RSK2 regulates defined processes in odontogenesis and craniofacial growth.
Results
Rsk2-/Y craniofacial phenotype
The craniofacial phenotype was assessed by X-Ray microtomographic imaging and analyzed through euclidean distances measurement and comparison. This phenotype was variable, ranging from normality (mutant mice 702, 731) to obvious craniofacial dysmorphology (mutant mice 150, 700) (Fig. 2). Some mutant mice show an intermediate phenotype, with skulls smaller in length but not in width.
Figure 2. Craniofacial phenotype of Rsk2-/Y mice assessed by X-Ray microtomography.
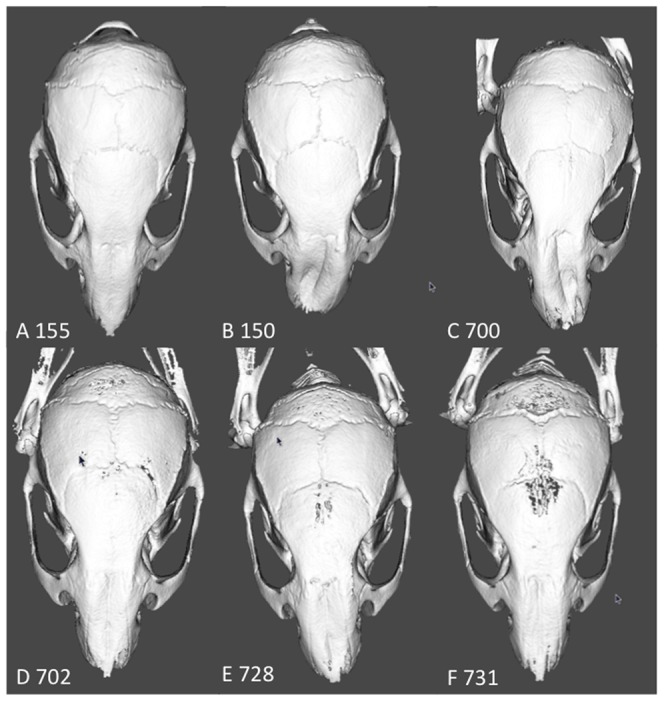
The craniofacial phenotype is highly variable, ranging from an almost normal-shaped cranium (mutant 155), although smaller in length, to a very dysmorphic appearance with a lateral nasal deviation. For mutant identification see Table 1.
An overall skull length reduction from the nasal to the occipital bone was a collective measure of the sum defects in the nasal, frontal, parietal, interparietal/occipital bones. The frontal bone always exhibited the greatest reductions in length of all skull bones. The interparietal/occipital bones were the least affected (distances 1–4, 1–5, 2–3, 2–4, 2–5 being more reduced than 1–2 or 1–3: see Table 1; measurements were performed according to anatomical landmarks detailed in Fig. S1). A nasal deviation was observed in some mutant mice (Fig. 2B,C,E; Table 1). Craniofacial microtomographic analysis of Rsk1,2,3-/- mice was not performed, due to rare availability of these triple mutants. Analysis thus focused on Rsk2-/Y mice, a defined genetic model of human Coffin-Lowry syndrome.
Table 1. Distances between skull landmarks on wild-type and Rsk2-/Y mutant mice.
| Distances | Mean WT | STD WT | Mean Rsk2 mutants | STD Rsk2 mutants | p-value | |
| Between 1 and 2 | 7.10 | 0.59 | 6.84 | 0.35 | 0.03 | |
| Between 1 and 3 | 15.05 | 0.46 | 13.92 | 0.59 | 0.01 | |
| Between 1 and 4 | 18.59 | 0.41 | 17.24 | 0.57 | 0.005 | |
| Between 1 and 5 | 21.87 | 0.49 | 20.48 | 0.48 | 0.005 | |
| Between 2 and 3 | 8.07 | 0.45 | 7.25 | 0.47 | 0.005 | |
| Between 2 and 4 | 11.72 | 0.38 | 10.69 | 0.45 | 0.005 | |
| Between 2 and 5 | 15.21 | 0.35 | 14.13 | 0.31 | 0.005 | |
| Between L and R 6 | 12.08 | 0.17 | 12.16 | 0.13 | 0.47 | |
| Between L and R 7 | 6.42 | 0.21 | 6.45 | 0.27 | 0.68 | |
| Between L and R 8 | 4.77 | 0.11 | 4.79 | 0.73 | 0.22 | |
| Between L and R 9 | 6.27 | 0.25 | 6.13 | 0.12 | 0.06 | |
| Between L and R 10 | 0.74 | 0.06 | 0.71 | 0.08 | 0.47 | |
| Between L and R 11 | 1.05 | 0.05 | 1.13 | 0.11 | 0.29 | |
| Between L and R 12 | 10.08 | 0.37 | 10.22 | 0.28 | 0.29 | |
| Between L and R 15 | 4.53 | 0.19 | 4.72 | 0.51 | 0.37 | |
| Between L and R 16 | 2.29 | 0.22 | 2.23 | 0.23 | 0.57 | |
| Between 1 and 6R | 15.24 | 0.34 | 14.39 | 0.52 | 0.008 | |
| Between 1 and 6L | 15.20 | 0.32 | 14.47 | 0.31 | 0.008 | |
| Between 1 and 7R | 7.92 | 0.28 | 7.18 | 0.58 | 0.008 | |
| Between 1 and 7L | 7.89 | 0.22 | 7.24 | 0.34 | 0.005 | |
| Between 1 and 8R | 6.90 | 0.24 | 6.29 | 0.46 | 0.88 | |
| Between 1 and 8L | 6.90 | 0.25 | 6.35 | 0.29 | 0.68 | |
| Between 1 and 9R | 20.65 | 0.47 | 19.55 | 0.46 | 0.008 | |
| Between 1 and 9L | 20.62 | 0.48 | 19.56 | 0.32 | 0.008 | |
In bold are distances significantly different between wild- type (WT) and mutants (all these distances were shorter in mutants). Anatomical landmarks are explained in Fig. S1 (supplementary information). All distances are in millimeters. STD: standard deviation; L: left; R: right.
Rsk2-/Y and Rsk1,2,3-/- dental phenotype
Phenotype of molar tooth rows
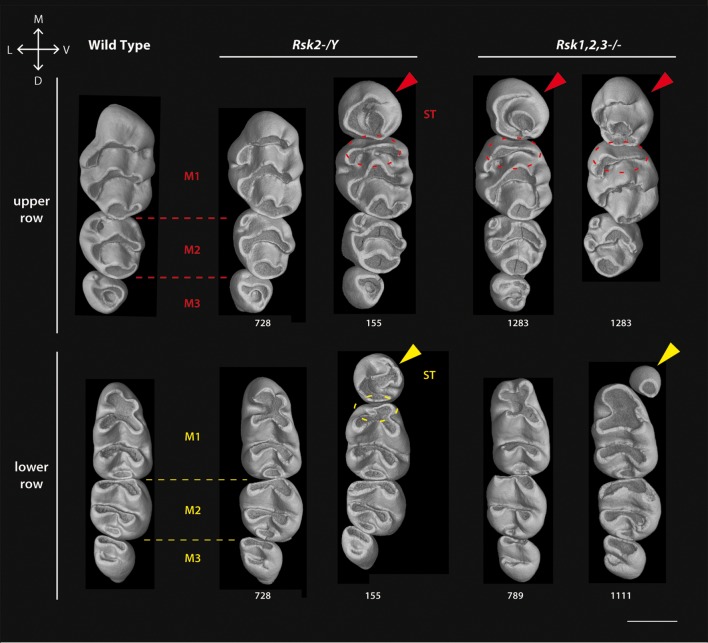
We analyzed 15 Rsk2-/Y mice and 9 Rsk1,2,3-/- mutant mice (see Table 2 and Fig. 3). Supernumerary teeth (ST) occurred in all specimens except for two Rsk2-/Y mice (702, 731). ST were aligned with the molar tooth row and located just in front and at the contact of the first molars (M1). The phenotype penetrance was higher in the upper dentition (65% in Rsk2-/Y mice, 83% in Rsk1,2,3-/- mice) than in the lower dentition (31% and 11%, respectively). Shape and size of the supernumerary teeth were variable and ranged from a monocuspid tooth to a well-shaped molar-like tooth. ST were always smaller in size than the adjacent first molars, and they were sometimes, especially in the Rsk1,2,3-/- upper rows, as big as the M2. Inactivation of Rsk2 alone was sufficient to generate the phenotype. When a ST was present, the mesial part of the first molar displayed altered crown morphology (Fig. 3). Even when the phenotype looked almost normal, a significant reduction in M1-M3 length was observed (Table 3). When ST occurred, the length mean value was always reduced for both M1 and M2 (Table 3). ST were always longer than M3. Molar width remained stable for M1, but was reduced for M2 and M3, except when only 3 molars were present in the mandible (Table 4). Shape abnormalities mainly occurred in mesial parts of the first lower (M1) and first upper (M1) molars in Rsk2-/Y mice. In upper M1, the central cusp of the mesial crest tended to be reduced, and the crest itself was more flattened (see red dotted circles in Fig. 3). Accordingly the mesial crest of the lower M1 also had an abnormal shape, because this crest tended to be strongly reduced when a ST occurred (see yellow dotted circles in Fig. 3). In triple mutants without ST (e.g. 789), the lower M1 displayed a mesial crest with vestibular cusp bigger than in WT.
Table 2. Presence (+) of a supernumerary tooth (mesial to the first molar) in Rsk mutant mice.
| Mouse | Genotype | Upper Right | Upper Left | Lower Right | Lower Left |
| 11 | Rsk2-/Y | + (4) | + (4) | + (4) | N |
| 25 | Rsk2-/Y | + (4) | + (4) | N | + (4) |
| 37 | Rsk2-/Y | + (4) | + (4) | + (4) | N |
| 39 | Rsk2-/Y | + (4) | + (4) | N | N |
| 46 | Rsk2-/Y | + (4) | N | N | N |
| 61 | Rsk2-/Y | + (4) | + (4) | N | N |
| 62 | Rsk2-/Y | N | + (4) | N | N |
| 150 | Rsk2-/Y | + (4) | + (4) | + (4) | + (4) |
| 155 | Rsk2-/Y | + (4) | + (4) | + (4) | + (4) |
| 184 | Rsk2-/Y | + (4) | + (4) | + (4) | + (4) |
| 185 | Rsk2-/Y | + (4) | + (4) | N | N |
| 700 | Rsk2-/Y | N | N | N | + (4) |
| 702 | Rsk2-/Y | N | N | N | N |
| 728 | Rsk2-/Y | + (4) | N | N | N |
| 731 | Rsk2-/Y | N | N | N | N |
| 789 | Rsk1,2,3-/- | + (4) | + (4) | N | N |
| 793 | Rsk1,2,3-/- | + (4) | N | N | N |
| 860 | Rsk1,2,3-/- | + (4) | N | + (4) | N |
| 883 | Rsk1,2,3-/- | + (4) | + (3) | N | N |
| 1105 | Rsk1,2,3-/- | + (4) | N | N | N |
| 1111 | Rsk1,2,3-/- | + (3) | + (3) | + (4) | N |
| 1283 | Rsk1,2,3-/- | + (3) | + (4) | N | N |
| 1139 | Rsk1,2,3-/- | + (3) | + (4) | N | N |
| 1159 | Rsk1,2,3-/- | + (4) | + (3) | N | N |
N: normal pattern of 3 molars M1, M2, M3. (3) means the presence of only 3 molars but the size of the second one is bigger (most probably there is a supernumerary tooth ST in front of M1, and M3 is missing).
Figure 3. Variation of molar shape, size and number in Rsk2-/Y and Rsk1,2,3-/- mice analyzed by X-Ray microtomography.
All molar rows are oriented in the same manner (top corresponds to mesial and left to lingual side). At the left are wild-type molars, other rows are mutants as indicated. Arrowheads point to the supernumerary teeth ST; dotted ellipses show the reduction of the mesial-most affected cusp. Scale bar: 0.7 µm.
Table 3. Comparison of each molar length, of the total molar field length, and the total length of M1+ M2, between wild-type (WT) mice and RSk2-/Y mutant mice with 3 or 4 molars.
| Mandibular molars WT | Mandibular molars Mutant 3M | Mandibular molars Mutant 4M | Maxillary molars WT | Maxillary molars Mutant 3M | Maxillary molars Mutant 4M | |
| Mean ST | 0.60 | 0.74 | ||||
| STD | 0.01 | 0.02 | ||||
| Mean M1 | 1.44 | 1.37 | 1.14 | 1.56 | 1.38 | 1.14 |
| STD | 0.03 | 0.11 | 0.05 | 0.07 | 0.10 | 0.09 |
| p-value | 0.23 | 0.0004 | 0.01 | 0.001 | ||
| Mean M2 | 0.89 | 0.85 | 0.79 | 0.92 | 0.85 | 0.82 |
| STD | 0.03 | 0.02 | 0.03 | 0.02 | 0.04 | 0.05 |
| p-value | 0.003 | 0.004 | 0.02 | 0.01 | ||
| Mean M3 | 0.59 | 0.53 | 0.49 | 0.60 | 0.63 | 0.63 |
| STD | 0.02 | 0.03 | 0.04 | 0.03 | 0.03 | 0.05 |
| p-value | 0.0002 | 0.0004 | 0.96 | 0.15 | ||
| Total | 2.93 | 2.75 | 2.88 | 3.08 | 2.85 | 3.31 |
| STD | 0.04 | 0.15 | 0.28 | 0.10 | 0.13 | 0.06 |
| p-value | 0.0008 | 0.36 | 0.01 | 0.001 | ||
| M1+ M2 | 2.33 | 2.10 | 2.15 | 2.48 | 2.30 | 1.96 |
| STD | 0.04 | 0.16 | 0.21 | 0.08 | 0.14 | 0.13 |
| p-value | 0.03 | 0.004 | 0.002 | 0.001 |
M1 size is reduced in mutants with 4 molars both in mandible and maxilla, but only in the maxilla of mutants showing 3 molars. M2 is reduced both in maxilla and mandible whatever the molar mutant phenotype. M3 mesiodistal dimension is reduced only in the mandible, irrespective of the presence of 3 or 4 molars. The total size of the molar field is reduced in the mandible and maxilla in mutant with 3 molars. The total size of the molar field is significantly increased in the maxilla in mutants with 4 molars. The total size of M1+M2 was always reduced when compared to the wild type value. All distances are in millimeters. Significant p-values are shown in bold.
M1 : molar 1; M2 : molar 2; M3 : molar 3; ST : supplementary tooth in front of M1; STD : standard deviation; Mutant 3M : mutant with 3 molars; Mutant 4M : mutant with 4 molars; Total : total length of all molars in the molar field.
Table 4. Comparison of each molar width (vestibulolingual dimension) between wild-type and Rsk2-/Y mutant mice with 3 or 4 molars.
| Mandibular molars WT | Mandibular molars Mutants 3M | Mandibular molars Mutants 4M | Maxillary molars WT | Maxillary molars Mutants 3M | Maxillary molars Mutants 4M | |
| Mean ST | 0.59 | 0.76 | ||||
| STD | 0.09 | 0.04 | ||||
| Mean M1 | 0.79 | 0.77 | 0.82 | 1.04 | 1.01 | 1.06 |
| STD | 0.02 | 0.04 | 0.03 | 0.05 | 0.05 | 0.02 |
| p-value | 0.19 | 0.12 | 0.19 | 0.56 | ||
| Mean M2 | 0.85 | 0.85 | 0.81 | 0.90 | 0.86 | 0.86 |
| STD | 0.02 | 0.02 | 0.03 | 0.02 | 0.05 | 0.03 |
| p-value | 0.89 | 0.02 | 0.03 | 0.02 | ||
| Mean M3 | 0.60 | 0.57 | 0.58 | 0.61 | 0.58 | 0.54 |
| STD | 0.02 | 0.02 | 0.05 | 0.03 | 0.02 | 0.04 |
| p-value | 0.006 | 0.06 | 0.02 | 0.01 |
In mutants the width of M1 was not modified. The width of the maxillary M2 and M3 was reduced. In the mandible the width of M2 was lower only when 4 molars were present, and the width of M3 was lower only in mutants with 3 molars (all distances in millimeters; significant p-values in bold).
M1 : molar 1; M2 : molar 2; M3 : molar 3; ST : supplementary tooth in front of M1; STD : standard deviation; Mutant 3M : mutant with 3 molars; Mutant 4M : mutant with 4 molars.
The length of the molar row was significantly reduced in both the lower and upper jaws when no ST was present. When a ST occurred, the length of the molar row was almost normal in the mandible, but was significantly higher in the maxilla when compared to WT (Table 3). Some Rsk1,2,3-/- mouse specimens displayed 3 upper postcanine teeth, as in the WT, but the dental row encompassed maxillary ST-M1-M2, and lacked the M3. Consequently, the second tooth (M1) was bigger than neighbouring teeth ST and M2 (Fig. 3, specimen 1283).
Incisor phenotype
No statistically significant differences were revealed between right and left incisors in both groups (p>0.05). As a consequence, results from right and left incisors were merged (Table 5). No statistical difference was observed between WT and Rsk2-/Y (p>0.05). No obvious incisor anomaly was detected. Analysis of the incisor phenotype in Rsk1,2,3-/- mice was not performed due to a reduced number of available animals, visual inspection however did not revealed any incisor dental defects. The existence of incisor anomalies in Rsk1,2,3-/- cannot be ruled out.
Table 5. Morphological analysis of mandibular incisors in WT and Rsk2-/Y mice.
| Li (mm) | hi (mm) | Hi (mm) | li (mm) | ti (mm) | lpa (mm2) | ||
| WT | Mean | 11.33 | 1.11 | 2.96 | 9.38 | 0.65 | 10.36 |
| STD | 2.34 | 0.07 | 0.20 | 1.56 | 0.02 | 1.07 | |
| Rsk2-/Y | Mean | 11.29 | 1.14 | 2.97 | 9.05 | 0.65 | 10.18 |
| STD | 1.66 | 0.05 | 0.21 | 1.65 | 0.03 | 1.33 |
No statistically significant differences were revealed between right and left incisors in both groups (p>0.05). As a consequence, results from right and left incisors were merged. No statistical difference was observed between WT and Rsk2-/Y (p>0.05).
(Li) length of the longest and external bow, (hi) height of median part of the incisor, (Hi) total height of the incisor, (li) horizontal length of the incisor joining the tip to the distal extremity of the root, (lpa) projected area in lateral view of the incisor, (ti) thickness the median part of the incisor. For more explanations on the measured distances, see Figure S3. STD : standard deviation.
Abnormal molar root development in Rsk2-/Y mice
Both the number and shape of molar roots also reflect the tooth identity. In WT mice the mandibular molars M1 and M2 have 2 roots, whereas M3 has only one root. The maxillary molars M1 and M2 have 3 roots. Variations in the root number of M3 occur both within and among various WT strains. The upper M3 molar usually has 2 or 3 roots, although sometimes it has only 1 root. Thus, the total number of molar roots is 5 per hemi-mandible, whereas it can vary from 7 to 9 per hemi-maxillary. In Rsk2-/Y mice, hemi-mandibles harbouring a ST had a total root number of 6, with the ST always having a single root. In the maxillary quadrant, the total root number varied from 8 to 9 when a ST was present. ST could have 1 or 2 roots, M1 had 2 roots, but the M2 root number remained stable with 3 roots (Table S1 in File S1). The presence of maxillary M1 with only 2 roots probably correlates with the occurrence of ST, often associated to a compression and a reduction of the mesial part of M1.
Rsk2-/Y and the diastema
The maxillary diastema was significantly reduced in Rsk2-/Y mice harbouring ST (Table 6). This was not the case for the mandibulary diastema, whatever the number of teeth.
Table 6. Upper and lower diastema size in wild-type and RSk2-/Y mutant mice with 3 or 4 molars.
| Mean size (mm) | STD | p-value | |
| Mandibular diastema Wild-type | 2.99 | 0.09 | |
| Mandibular diastema Mutants with 3 molars | 2.83 | 0.21 | 0.22 |
| Mandibular diastema Mutants with 4 molars | 2.71 | 0.1 | 0.26 |
| Maxillary diastema Wild-type | 5.45 | 0.18 | |
| Maxillary diastema Mutants with 3 molars | 5.33 | 0.21 | 0.49 |
| Maxillary diastema Mutants with 4 molars | 5.01 | 0.21 | 0.0019 |
Only the upper (maxillary) diastema from mutants with 4 molars was significantly smaller compared to wild-type (bold). STD : standard deviation.
Expression patterns of Rsk genes during mouse odontogenesis
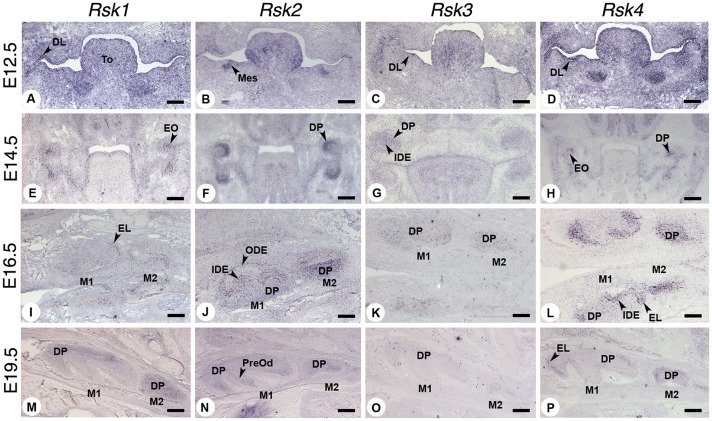
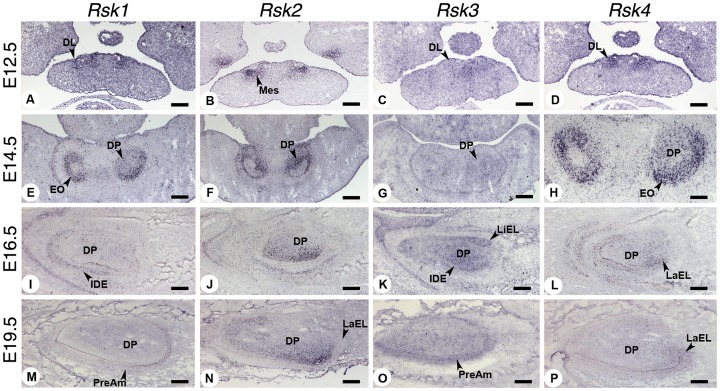
To investigate whether the dental anomalies observed in the Rsk-deficient mice may correlate with distinct patterns of expression of Rsk genes during odontogenesis, we performed an in situ hybridization analysis of Rsk1, Rsk2, Rsk3 and Rsk4 mRNA transcripts at various stages of tooth development. All four genes were found to be expressed in specific areas of the tooth anlagen, as illustrated in Fig. 4 for the developing first molars, and Fig. 5 for the mandibular incisors. Their expression features are summarized in Table 7.
Figure 4. Expression patterns of Rsk genes during mouse molar odontogenesis at E12.5 (dental lamina), E14.5 (cap), E16.5 (bell) and E19.5 (late bell) stages, analyzed by in situ hybridization.
Abbreviations: DL dental lamina, DP dental papilla, EL epithelial loop, EO enamel organ, IDE inner dental epithelium, M1 first molar, M2 second molar, Mes ectomesenchyme, ODE outer dental epithelium, PreOd preodontoblasts, To tongue. Scale bars 140 µm (A,B,C,D,I,J,K and L) and 250 µm (E,F,G,H,M,N,O and P).
Figure 5. Expression patterns of Rsk genes during incisor odontogenesis at E12.5 (dental lamina), E14.5 (cap), E16.5 (bell) and E19.5 (late bell) stages analyzed by in situ hybridization.
Abbreviations: DL dental lamina, DP dental papilla, EL epithelial loop, EO enamel organ, IDE inner dental epithelium, LaEL labial epithelial loop, LiEL lingual epithelial loop, Mes ectomesenchyme, PreAm preameloblasts. Scale bars 140 µm (A,B and C), 200 µm (D,E, F,G,I, J, K and L), 250 µm (M,N,O and P) and 50 µm (H).
Table 7. Rsks expression patterns during odontogenesis.
| Rsk1 | Rsk2 | Rsk3 | Rsk4 | |||||||||||||
| Inc | M1 | M2 | M3 | Inc | M1 | M2 | M3 | Inc | M1 | M2 | M3 | Inc | M1 | M2 | M3 | |
| E12.5 | ||||||||||||||||
| Oral epithelium | + | + | + | + | ||||||||||||
| Dental lamina | + | + | + faint | + | + | + | ||||||||||
| Mesenchyme | + | + | + | + | ||||||||||||
| E14.5 | ||||||||||||||||
| Enamel organ | + | + | + | + | ||||||||||||
| IDE | + | + | + | + | ||||||||||||
| Enamel knot | ||||||||||||||||
| Dental Papilla | + π faint | + | + | + | + | + | + | |||||||||
| Dental sac | + | |||||||||||||||
| E16.5 | ||||||||||||||||
| ODE | + | + | + | |||||||||||||
| IDE | + | + | + | + | + | + | + | + | ||||||||
| Epithelial loop | + | + | + | + | + | + | + | |||||||||
| Dental Papilla | + lab | + lab | + | + | + π | + | + | + | + | + | ||||||
| E19.5 | ||||||||||||||||
| ODE | ||||||||||||||||
| SR | ||||||||||||||||
| SI | ||||||||||||||||
| IDE | + | + | + | + | ||||||||||||
| PreAm | + | + | ||||||||||||||
| Am | ||||||||||||||||
| Epithelial loop | + | + | + | + lab | + | + | ||||||||||
| Od | ||||||||||||||||
| PreOd | + | |||||||||||||||
| Dental papilla | + | + | + | + lab | + | + | + | + | + | + lab | + | + | ||||
| Dental sac | + |
+ : positive signal; lab: labial; π posterior area
IDE: inner dental epithelium; ODE: outer dental epithelium; SR: stellate reticulum; SI: stratum intermedium; PreAm: preameloblasts; Am: ameloblasts; Od: odontoblasts, PreOd: Preodontoblats
Rsk2 transcripts were mainly localized in the mesenchymal compartment or dental papilla from E12.5 (dental lamina stage) to E19.5 (late bell stage). At E16.5 (bell stage), the transcripts were scattered in the area facing the epithelial loops or at the base of the cusps; the second molar dental mesenchyme was uniformly labelled. In the incisors the transcripts were also located in the mesenchyme, and the signal was more intense in the labial area and posterior area.
The expression of Rsk1 and Rsk4 was mainly epithelial, marking the inner dental epithelium and especially the epithelial loop areas. A faint signal was also observed for Rsk4 in the posterior mesenchymal area facing the epithelial loops especially the labial loop for incisors (E16.5 and 19.5).
Rsk3 transcripts were mainly visible in the dental papilla at E14.5 (cap stage), E16.5 (bell stage) and E19.5 (Figs. 4, 5 and Table 7). At E19.5 the transcripts were concentrated in the cervical part of the papilla for the first molar and had a wider distribution in the second molar mesenchyme.
Rsk2-/Y supernumerary tooth is associated with an extra mesial placode
An extra tooth placode, and subsequent enamel knot labelled with Sonic hedgehog (Shh) as a marker, was clearly visible at E14.5, mesially to M1 in Rsk2-/Y mice when compared to WT littermates (Fig. 6). The presence of this extra placode was seen in nearly 30% of the analysed embryo and was not always simultaneously present on right and left sides of the jaw. During WT development the three molars (M1, M2 and M3) are forming from a unique molar placode. The presence of an additional dental placode mesially to the M1 placode likely corresponds to the primordium of ST developing in Rsk2-/Y mutants.
Figure 6. Whole mount in situ hybridization showing the molar and incisor placodes within the mandible at E14.5 in a wild-type mouse (A) and Rsk2-/Y mouse (B: detail of the right-side mandible) labelled with a Shh riboprobe.
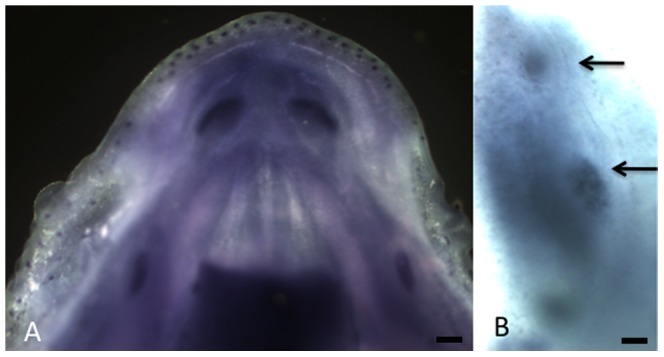
Two molar placodes are clearly visible in B (arrows) Scale bars 200 µm (A), 100 µm (B).
Rsk2-/Y transcriptome analysis
We performed a comparative transcriptomic analysis (Affymetrix GeneChips) of wild-type and Rsk2-/Y mice, analyzing RNA isolated from dissected E14.5 mandibular molar tooth germs. By using a principal component analysis (PCA), it was possible to discriminate between the molar samples according to their genotype (wild-type versus Rsk2-/Y) (Fig. 7). 504 genes were retrieved as being differentially expressed, with fold changes exceeding ±1.2 and a false discovery rate below 0.1. The fold changes were however rather low, ranking between +2.044 and −1.98, suggesting that rather minimal molecular changes, at the transcriptome level, were taking place. These changes could be also minimized by a heterogeneous sampling for Rsk2-/Y molars, with putative phenotypes ranking as previously described between normality and a 3 molars with a supernumerary tooth phenotype; this phenotype was not known at the E14.5 cap stage, when molar germs were pooled for transcriptomic analysis. The most affected (“top ten”) genes, and a selection of additional genes being up- or down-regulated, are listed in Table 8 and Table 9.
Figure 7. Principal component analysis (PCA) of transcriptomic data of wild-type versus Rsk2-/Y mandibular molars samples.
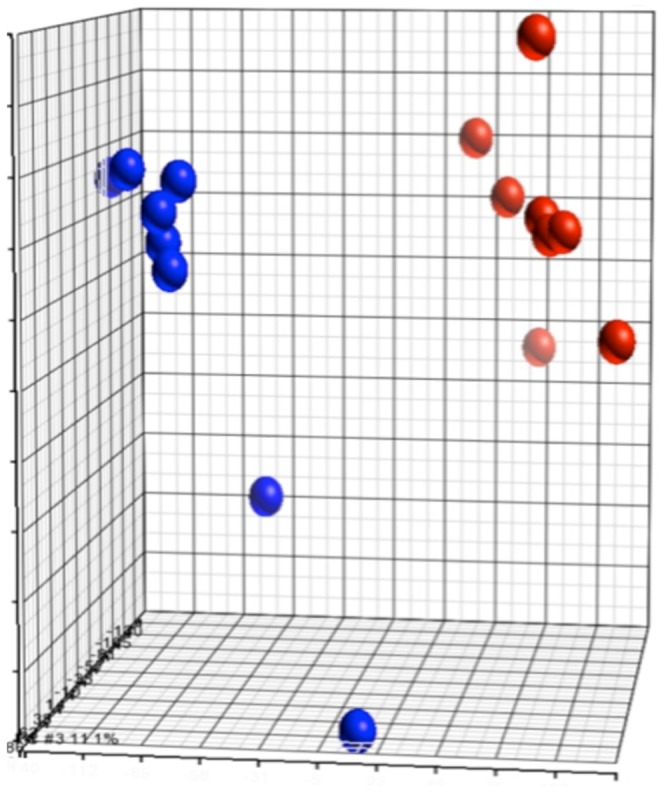
Wild-type samples are represented as red balls, whereas mutant samples are represented in blue (X, Y and Z units are arbitrary units created by the software). The units are data-dependent and are generated by the software, which gives coordinates to each sample according to three axes that relate to the weight (inertia) of the decomposition into 3 principal components. For this analysis, samples segregate in two distinct groups, showing relevant transcriptional differences between WT and Rsk2-/Y samples.
Table 8. Overview of top ten genes differentially expressed in Rsk2-/Y and wild-type developing mandibular molars.
| Top ten up-regulated genes | Fold change | P-value | Name |
| Rdh1 | 2.04458 | 5.22E-11 | retinol dehydrogenase 1 (all trans) |
| Dip2c | 1.82248 | 4.50E-11 | DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| 1700001J11Rik | 1.6761 | 2.92E-08 | - |
| Olfr319 | 1.66441 | 1.56E-08 | olfactory receptor 319 |
| Myh1 | 1.64957 | 1.91E-06 | myosin. heavy polypeptide 1 skeletal muscle |
| Acaa1b | 1.62775 | 1.64E-07 | acetyl-Coenzyme A acyltransferase 1B |
| 6430550D23Rik | 1.62394 | 3.80E-08 | - |
| Hmgb1 | 1.61691 | 4.11E-10 | high mobility group box 1 |
| 4930555K19Rik | 1.58002 | 2.16E-09 | - |
| 9630041A04Rik | 1.57948 | 3.38E-09 | - |
| BC022713 | 1.57732 | 2.80E-07 | cDNA sequence |
| Gm9968 | 1.56689 | 4.24E-12 | predicted gene 9968 |
| Gm6718 | 1.56677 | 4.74E-07 | predicted gene 6718 |
| Naip2 | 1.56543 | 3.72E-09 | NLR family. apoptosis inhibitory protein 2 |
The table is highlighting genes showing the highest degree of enrichment (positive values) or down-regulation (negative values).
Table 9. Overview of other genes differentially expressed in Rsk2-/Y and wild-type developing mandibular molars.
| Up-regulated genes | Fold change | P-value | Name |
| Mdm2 | 1.42143 | 6.43E-07 | p53 E3 ubiquitin protein ligase homolog oncoprotein |
| Neurog3 | 1.39695 | 4.46E-08 | neurogenin 3 |
| Pou6f2 | 1.38695 | 2.35E-07 | POU domain class 6 transcription factor 2 |
| Dffa | 1.38653 | 4.93E-10 | DNA fragmentation factor alpha subunit |
| Bdkrb1 | 1.38509 | 4.15E-08 | bradykinin receptor beta 1 |
| Ofd1 | 1.37103 | 4.33E-08 | oral-facial-digital syndrome 1 gene homolog; ciliogenic protein |
| Zfp759 | 1.35236 | 5.23E-07 | zinc finger protein 759 |
| Cflar | 1.32394 | 7.32E-07 | CASP8 and FADD-like apoptosis regulator |
| Aak1 | 1.23472 | 3.15E-06 | AP2 associated kinase 1 positive regulation of Notch signaling pathway |
The table is highlighting genes selected on the basis of their known involvement in pathways regulating odontogenesis.
The Rdh1 gene encoding retinol dehydrogenase (RDH) 1 was found to be the most up-regulated gene in Rsk2-/Y mutant mice. In addition to retinol dehydrogenase activity, this enzyme has strong 3alpha-hydroxy steroid dehydrogenase- and weak 17beta-hydroxy steroid dehydrogenase- activities. Rdh1 exhibits widespread and intense mRNA expression in many embryonic tissues including the neural tube, gut, and neural crest, as well as in adult mice [16]. Interestingly, retinoic acid (the end product of retinol oxidation initiated by RDH enzymes) is involved in tooth morphogenesis and differentiation, and abnormal retinoic acid levels can lead to dental anomalies [17], [18], [19], [20].
Sectm1b, encoding secreted and transmembrane protein 1B, was the most down-regulated gene in Rsk2-/Y mutant mice. This gene and protein had never been described previously as potentially involved in odontogenesis.
Among the differentially expressed genes selected (Table 9) as belonging to signalling pathways acting during tooth development (TGF-beta, FGF, Wnt, NF-kappaB) and or/cell cycle progression, Eaf2 is a proto-oncogene acting together with Wnt4 in an autoregulatory feedback loop [21]. Inactivation of this gene causes numerous tumours in mice [22]. Eaf2 was down-regulated in Rsk2-/Y mutants.
We tried unsuccessfully to confirm those results on a selection of genes by qRT-PCR, comparing three Rsk2-/Y and three WT molar RNA samples. This negative result might be explained by the heterogeneous nature of the Rsk2-/Y phenotype (as described above), the relative low level of expression changes, and may not invalidate the microarray data.
Rsk2 inactivation in vitro
In order to overcome the potential problem of Rsk2-/Y molar samples heterogeneity we decided to use an in vitro system, and inactivated Rsk2 by microinjection and electroporation of shRNA in E14.5 molar tooth germs. In vivo electroporation in the mandible, as previously described by [23] was not suitable for our study, because of the lack of precision on the targeted zone and our aim to inactivate the gene directly within the tooth anlagen.
We optimized the experimental settings to inject and electroporate Rsk2 shRNA in cap stage molars explanted and cultured ex vivo. Control explants were electroporated with a random shRNA construct. After electroporation the explants were placed into collagen drop culture to maintain their morphology [24] and kept in a defined culture medium for 24 h, before being processed for qRT-PCR analysis. Tooth morphology was preserved and neither apoptosis nor cell death were induced, as shown by activated caspase3 and TUNEL assays (data not shown).
Rsk2 shRNA electroporation efficiently diminished Rsk2 expression by 75% (Fig. 8A). We then analyzed Eaf2 and Rdh1 expression, and found that Eaf2 expression was down-regulated (75% decrease) (Fig. 8B), while Rdh1 expression was up-regulated (50% increase)(Fig. 8C). These results were in accordance with the microarray data. The new combined technique allowed us not only to optimize a protocol for studying the molecular mechanisms of early tooth development, but also to set up a new approach for microarray validation.
Figure 8. Summary of experiments involving in vitro inactivation of Rsk2 and its consequences on target gene expression, as observed after microinjection in E14.5 mandibular molar explants of a Rsk2 shRNA, followed by qRT-PCR analysis 24 h after injection.
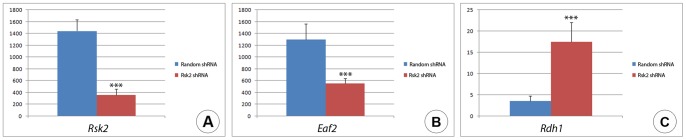
A: Rsk2 was down regulated 4.01 fold after electroporation. B: Eaf2 was significantly down-regulated, and C: Rdh1 was up-regulated after electroporation. Histograms show expression levels in molars electroporated with the random shRNA (blue) and microinjected-electroporated explants with the Rsk2 shRNA (red), with values normalized with respect to Gapdh expression. Data (mean ±SEM) were analyzed with Student t-test; ***p<0.001; **p<0.01; *p<0.05.
Discussion
A peculiar dental phenotype
Mice are commonly studied mammals for investigating the mechanisms of tooth development and evolution. The orodental phenotype of mouse models of human genetic disorders also often mimics dental anomalies encountered in human rare diseases [14]. Basal placental mammals had a dental formula including three incisors, one canine, four premolars, and three molars per dentition quadrant [13], [25]. Through evolution, almost all mammals reduced their dental formulas, but lost functional teeth sometimes remaining as rudiments that begin to develop and then abort. The dentition of the mouse comprises 4 functional teeth in each quadrant (1 incisor and 3 molars separated by a toothless space called diastema). Rudimentary tooth primordia have been reported to develop in the distal part of the diastema through the initial stages of odontogenesis of WT mice. However their development ceases before the cap stage and they regress by apoptosis [26], [27], [28]. The tooth phenotype encountered in the Rsk2-/Y mutant mouse is remarkable by the occurrence of supernumerary teeth at the positions of the fourth premolars. Fourth premolars are still present in certain rodent groups such as squirrels, and have been lost through muroid rodent evolution [13].
Rsk2 phenotype is similar to the tabby phenotype
Supernumerary teeth within the diastema were described in other mouse mutants for ectodin, Lrp4, Sprouty2/4, Wise, Polaris, and Gas1 [29], [30], [31], [32], [33], [34], [35], [36]. Dysfunctions of the FGF, Shh, Wnt, and/or NF-kappaB signalling pathways are thought to induce formation of these supernumerary teeth within the diastema, indicating that the mouse maintains genetic potentialities that could be stimulated and induce the formation of supernumerary teeth [37], [38].
Ectopic teeth in the diastema are also observed in mice misexpressing Eda (ectodysplasin A; also known as Ta; Ed1; EDA1; XLHED; tabby) and EdaA receptor (Edar) [39], [40], [41], [42]. The dental phenotype of the Rsk2-/Y mouse resembles that encountered in the tabby mouse (a model for X-linked hypohidrotic ectodermal dysplasia- an ectodermal dysplasia characterized by an absence or dystrophic development of hair, nail, sweat- lacrymal- mammary- glands and tooth; OMIM #305100). Mutations in the human X-linked gene ED1 (the Eda orthologue) are responsible for this disorder [43], [44], [45], [46]. Ontogenetic and phylogenetic data together suggest that the occurrence of functional supernumerary teeth in tabby mice diastema is caused by the persistence of the premolar rudiments that usually regress in WT mice. It has thus been proposed that this phenotype may represent an evolutionary throwback or atavism [43], [44], [45], [46].
Recently is has been shown that the number of teeth is regulated by fine-tuning of the level of receptor-tyrosine kinase signalling [32]. Known cytosolic RSK substrates include I-kappaB alpha [7], [47], [48], a member of the NF-kappaB pathway. This pathway is involved in development and in some human disorders including ectodermal dysplasias that affect ectodermal derivatives like the teeth, hair, salivary, mammary glands and skin. I-kappaB alpha has specifically been associated with an autosomal dominant form of anhidrotic ectodermal dysplasia and with T cell immunodeficiency [49]. The fact that I-kappaB alpha can be phosphorylated by RSKs, leading to increase of NF-kappaB activity, may therefore explain some of the dental anomalies encountered in the Rsk2-/Y transgenic mice and in Coffin-Lowry syndrome patients. However, the downstream signalling pathway(s) of Eda/Edar involved in tooth formation in the diastema are not yet known.
The phenotype in the light of Coffin-Lowry syndrome
The mouse model for Coffin-Lowry syndrome, obtained by inactivation of the Rsk2 gene and used in this study, was previously described by [11]. Mutant mice weigh 10% less and are 14% shorter than their wild-type littermates. This is in agreement with a role of RSK2 in organismal growth. These mice exhibit impaired learning and poor coordination, providing evidence that RSK2 plays similar roles at least for cognitive functions, in mouse and human. No previous description of the craniofacial or dental phenotype was so far reported. The craniofacial features described in Coffin-Lowry syndrome include a thick calvarium, hyperplastic supra-orbital ridges, hypertelorism, a flat malar region, and a prominent mandible. Clearly it is difficult in the light of our present observations, to relate the Rsk-knockout craniofacial phenotype to the anomalies encountered in patients. The small size of the skull could be associated to the general growth defect. However it is interesting to notice that in Coffin-Lowry patients, teeth agenesis and/or hypodontia is described. An apparent opposite phenotype between human patients and a mouse model has already been described in another rare disease, the cleido-cranial dysplasia syndrome, which is characterized by skeletal defects, numerous supernumerary teeth, and delayed tooth eruption. The mouse model of this disease (Cbfa1-KO) exhibits arrested tooth development [15]. Mostly loss of function mutations in RSK2 both in human and mouse are responsible for the phenotype. Despite striking similarities mouse and human tooth development, cross-species comparison of mouse/human dentitions show they are clearly not alike. This suggests differential output and involvement of identical or superimposed pathways. These differences may account for this apparently opposite in nature dental phenotype i.e. hypodontia in human versus supernumerary teeth in mouse.
Cell cycle progression and cell proliferation
It has been shown that tooth number, size, and shape are linked to cell proliferation and apoptosis events [50], [51], [52], [53]. It is interesting to note that Rsks genes are expressed throughout tooth development, especially in proliferating areas (this study, Figs. 4 and 5), either in the mesenchyme (Rsk2, Rsk3) or in the epithelial loops (Rsk1, Rsk4). The expression patterns described for the Rsk genes in other embryonic regions [12], [54], [55] also indicate a correlation with proliferative areas. Rsk1 is highly expressed in tissues harbouring highly proliferating cells like liver, lung, thymus, and olfactory and respiratory epithelia. Particularly intense Rsk1 expression is observed in the gut epithelium. Rsk2 is expressed at very low levels throughout mouse development in dorsal root ganglia, thyroid gland, kidney, lung, liver and skeletal muscles. Rsk3 is mainly expressed in the nervous system, but also in the thyroid gland and testis cords. RSK3 may regulate proliferation and seems linked to cellular differentiation of neuroepithelial cells during development [55]. Rsk4 ubiquitous low expression was observed throughout development. In teeth Rsk4 expression was rather specifically marking the epithelial loops.
It is well established that RSKs control cell proliferation through the regulation of mediators of the cell cycle [7]. RSK2 for example promotes cell cycle progression by phosphorylating c-Fos, a transcription factor regulating the expression of cyclin D1 during G1/S transition. RSK2 activates and phosphorylates p53 in vitro and in vivo and colocalizes with p53 in the nucleus. The RSK2-p53-histone H3 complex may likely contribute to chromatin remodelling and cell cycle regulation [56]. A list of phosphorylation substrates of the RSK isoforms was reported by [7] including c-Fos, NHE-1, Erα, p27kip1, Sos1, RanBP13, YB-1, Erp1, elF4B, rpS6, Bad, DAPK, Nur77, NFAT3, TIF-1A, ATF4, ATF1, MEF2c, Filamin A, Raptor, CCTβ, CRHSP24, Shank. Interestingly, alterations in the expression of a number of genes involved in cell cycle control (Ccna1, Hmgb1, Mdm2, Tpd52) or in cell growth (Ngf, Bdkrb1, Amhr2) is shown in our microarray analysis.
Increased proliferation and decreased apoptosis are necessary to develop supernumerary teeth, as seen by differential regulation of Sprouty genes [57]. The localisation of Rsks genes in proliferative areas, their role in the regulation of proliferation and apoptosis [58] as well as the interconnection of the involved pathways could explain that the deletion of Rsk2 gene could induce the formation of supernumerary teeth and therefore reveal ancestral stages of murine dental evolution.
RSK and the Ras/MAPK pathway
Altered ERK/MAPK signalling (with abnormally increased phosphorylation of ERK1/2) was recently reported in the hippocampus of the Rsk2-/Y mice [59]. RSK2 was also proven to exert a feedback inhibitory effect on the ERK1/2 pathway. Our microarray data did not indicate a similar trend at the level of the developing molar. Ngf was the only gene detected by our microarray analysis, known to be regulated at the transcriptional level by ERK1/2 and involved in tooth development [60].
Unravelling target genes and involved pathways
The heterogeneity of the molar samples and the variability of the Rsk2-/Y molar phenotype has certainly interfered with the analysis of the microarray data and may have prevented the confirmation of target gene expression by qRT-PCR. However, genes involved in TGF-beta receptor signalling (Amhr2), or in the FGF (Fgfbp3) and Wnt (Sfrp5) pathways, were shown to be affected by the inactivation of Rsk2. These pathways are known to be involved in tooth development [61] and, when affected, are responsible for dental anomalies both in mouse models and human [62]. The in vitro assay we have developed, involving micro-injection and electroporation of a Rsk2 shRNA in explanted molar tooth germs, proved to be a valuable system to assess quantitative changes in target gene expression.
Conclusions
Analysis of Rsk2-/Y mutant mice revealed an important role of Rsk2 in craniofacial development, especially in dentition development and patterning. The loss of function of Rsk2 allows the reappearance of supernumerary diastemal teeth considered as remnants of teeth lost over evolution. Our in situ hybridization analysis indicated that Rsk2 expression pattern may correlate with proliferative areas of the developing teeth, consistent with a biological function of RSK2 in cell-cycle control and cell growth. Transcriptome profiling analysis was difficult because of the heterogeneity and variability of the Rsk2-/Y dental phenotype, however it provided candidate genes and pathways potentially involved in this phenotype. In vitro inactivation of Rsk2 using shRNA was found to be a promising approach to address target genes and showed, in particular, an interference with an enzyme involved in the retinoid/steroid biosynthesis pathway. The fact that the regression of the fourth premolar seen in mouse is not found in all mammals, and is never observed in humans, may provide an explanation for the divergent phenotypes in Rsk2-/Y and human patients.
Materials and Methods
Patients and Ethics Statement
Patients attending the Reference centre for orodental manifestations of rare diseases, Pôle de médecine et chirurgie bucco-dentaire, Hôpitaux Universitaires de Strasbourg, or their legal representatives (parents, next of kin, caretakers or guardians) if minor and/or not in capacity to provide his or her own consent, give written informed consent for the use and publication of medical data. The picture proposed as figure 1 was uploaded in D4/phenodent database (CCTIRS positive report 11/09/2008, CNIL authorization 18/05/2009 (N°908416). A copy of the consent form can be downloaded at http://www.phenodent.org/consentement.php
Animals and Ethics Statement
Rsk2 gene targeting was previously described [11]. As in human, the mouse Rsk2 gene is located on the X chromosome. Analyses were performed on Rsk2-/Y (“knockout”) male mice, with littermate wild-type (Rsk2+/Y) males used as control animals. Triple Rsk1,2,3-/- mutant mice were obtained by breeding of single knockout mice, themselves obtained by excision of exons three and four (RSK1 knockout) or exon two (RSK2 and RSK3 knockouts) of the corresponding genes, leading to frameshift mutations [11]. The experiments were performed in accordance with the French national and European Laws and Directives Concerning Laboratory Animal Housing, Welfare and Experimentation and after approval from the CERBM-GIE: ICS/IGBMC Ethical Research Board.
X-Ray microtomography
A cohort of 6 mutant Rsk2-/Y male mice (#150, 155, 700, 702, 728, 731) and 6 wild-type (WT) littermates (#149, 154, 699, 727, 729, 730), as well as 7 Rsk1,2,3 compound mutant male mice (#789, 860, 883, 1105, 1111, 1139, 1283), were analyzed through X-ray micro-computed tomography (micro-CT) (GE eXplore Vision CT120 (General Electric, Waukesha, USA) for craniofacial exploration and Phoenix Nanotom (General Electric measurement and control, Billerica, USA) for tooth analysis). For craniofacial exploration the micro-CT was performed using 220 projections with an angle increment of 0.877 degrees (Parker mode) and one average frame per projection (70.0 kV, 32 mA, and exposition time of 16 milliseconds). 3D-reconstructions were done using a Feldkamp algorithm giving voxels of 100×100×100 µm3. For teeth a second micro-CT acquisition was performed with a smaller reconstructed voxel size (3×3×3 µm3) using 2 000 projections and three average frames per projection (100.0 kV, 70 µA, and exposure time of 500 ms). 3D-reconstructions were done using datos|x software and algorithm including beam hardening correction and ring artefact reduction.
Anatomical landmarks were identified as remarkable points (for instance cranial sutures) that could be easily recognized on each specimen. Fig. S1 (supplementary material) illustrates this step, showing on a 3D isosurface rendering the landmarks (red points) and their anatomical definitions. A similar approach was used for molar analysis (Fig. S2).
Calculation involved euclidean distances between selected pairs of points that could be compared to identify statistical significant differences between WT and mutant mice. The methodology is based upon previously described work [63], [64]. To avoid any bias in landmarks position, landmarks were placed two times by one investigator (intra-investigator reproducibility) and one time by another (inter-investigator reproducibility). As no statistical differences were observed between distances measured within or in between investigators, we decided to base our study on the average of the three measured distances. Statistical analysis comparing WT and mutant mice was performed using Mann-Whitney non parametric test. P-values lower than 0.05 were considered as significant.
Analysis of incisor morphology
All image processing tasks were done using MicroView (GE Healthcare, Waukesha, USA). Right and left mandibular incisors were isolated by manual segmentation from mandible micro-CT acquisitions of the two mouse groups (6 Rsk2-/Y and 6 WT mice). Several measurements were achieved for morphology description, on the lateral and dorsal views of a 3D isosurface rendering of incisors (threshold value = 700 Hounsfield units): the length of the longest and external bow (Li), the height of median part of the incisor (hi), the total height (Hi), the horizontal length joining the tip of the incisor and the distal extremity of the root (li), projected area in lateral view of the incisor (lpa) and the thickness of the median part of the incisor (ti) (Fig. S3). A Wilcoxon-Mann Whitney test was used to compare both groups.
In situ hybridization on cryosections
In situ hybridizations were performed according to an automated procedure described in [65], using cDNA plasmids described in Table S2 in File S1. Mouse embryos/fetuses (C57BL6) were collected at E12.5, E14.5, E16.5, and at the day of birth (referred to as E19.5). For E14.5 and older samples, the whole head was embedded in OCT 4583 medium (Tissue-TEK, Sakura) and frozen on the surface of dry ice. E12.5 embryos were fixed overnight in 4% paraformaldehyde (pH 7.5, w/v) in phosphate-buffered saline (PBS), cryo-protected by overnight incubation in 20% sucrose (w/v) in PBS, and cryo-embedded as described above. Cryosections (Leica CM3050S cryostat) at 10 µm were collected on Superfrost plus slides and stored at −80°C until hybridization. E12.5 and E14.5 samples were sectioned in a frontal plane, whereas other stages were sectioned sagitally.
Whole mount in situ hybridization
Whole mount in situ hybridization of mandibles collected at E14.5 was performed with a digoxigenin-labeled Shh riboprobe as described [66], using an Intavis InSituPro robot (for details, see http://www.empress.har.mrc.ac.uk/browser/, Gene Expression section). The Shh template plasmid was kindly provided by A. McMahon (Harvard University).
DNA microarray analysis
Tissue preparation
Embryos at E14.5 were dissected and mandibular molars were isolated. The tail was used for embryo genotyping. Individual tissue samples were frozen in liquid nitrogen and kept at -80°C until genotyping results were available and further used.
Microarray hybridization
To obtain enough RNA for hybridization on DNA microarrays, total RNA was extracted from molar tooth germs dissected manually (pooling samples from two independent wild-type or Rsk2-/Y embryos). Total RNA was extracted with the RNAeasy micro Kit (Qiagen). RNA quality was verified by analysis on the 2100 Bioanalyzer (Agilent). All samples displayed a RNA Integrity Number greater than 9.8. Eight Rsk2-/Y and 8 WT samples were hybridized and compared.
Biotinylated single strand cDNA targets were prepared, starting from 300 ng of total RNA, using the Ambion WT Expression Kit (Cat #4411974) and the Affymetrix GeneChip WT Terminal Labeling Kit (Cat #900671), according to Affymetrix recommendations. Following fragmentation and end-labeling, 1.9 µg of cDNAs were hybridized for 16 h at 45°C on GeneChip Mouse Gene 1.0 ST arrays (Affymetrix) interrogating 28,853 genes represented by approximately 27 probes spread across the full length of the gene. The chips were washed and stained in the GeneChip Fluidics Station 450 (Affymetrix) and scanned with the GeneChip Scanner 3000 7G (Affymetrix). Finally, raw data (.CEL Intensity files) were extracted from the scanned images using the Affymetrix GeneChip Command Console (AGCC) version 3.1. See also [67].
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) [68] and are accessible through GEO Series accession number GSE51034. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51034)
Microarray analysis
CEL files were further processed with the Partek software to obtain principal component analysis (PCA) and to select and consider only genes with a signal value above 5 (20th percentile of all expression values) in at least one sample. Genes were considered as differentially expressed if the false discovery rate from Benjamini and Hochberg test was under 0.1 and if the fold change was greater than 1.2 or lower than -1.2.
In vitro culture and shRNA inactivation
E14.5 tooth germs were dissected and cultured in chemically defined culture medium as previously described [19]. The tooth explant consisted of both the enamel organ (epithelial component) and the attached dental mesenchyme. A DNA solution containing 1 µg of Rsk2 shRNA (SABiosciences SureSilencing shRNA Plasmid for Mouse Rps6kA3 clone ID1– linearized pGeneclip Neomycin vector, insert sequence TGATGATACTCCAGAGGAAT) or a random shRNA (insert sequence GGAATCTCATTCGATGCATAC), 0.5% sucrose, and Fast Green (Sigma; 1/10.000) was injected into the developing molars with a Femtojet Eppendorf device (t = 0.2 s, P = 61 hPa). Two platinum needle electrodes (0.1 mm tip, Sonidel) were inserted into the tissue lying in a cover round platinum flat electrode (Sonidel). DNA was then transferred into the cells using an ECM 830 Electroporation System (BTX Harvard Apparatus), applying 1 set of 5 pulses. Electroporation settings were set to 5 V for 50 ms, spaced by 50 ms intervals. As negative control, explants were electroporated with a random shRNA sequence.
After electroporation, the explants were embedded into 14 µl rat collagen drops (BD Biosciences) containing 0.2 ng laminin (BD Biosciences), 50 µl DMEM 10x concentrated, 5 µl HEPES (1 mM), 60 µl 7.5% NaHCO3 (adapted from [24]). The collagen drops were kept on ice to prevent them from polymerizing. The collagen was then polymerized at 37°C for 10–15 min before addition of the culture medium.
All explants were subsequently cultured for 24 hours in a DMEM-HAM F12 medium supplemented with Vitamin C (18 mg/ml); L-Glutamine (200 mM); Streptomycine (1000 U/ml) and 20% fetal calf serum for optimal tooth growth. Explants were then processed for RT-qPCR analysis.
Real-time quantitative RT-PCR
RT-PCR assays were performed in duplicate, comparing 3 wild-type and 3 Rsk2-/Y RNA samples (4 teeth were used in each sample for validation of microarray data), or 6 molar samples electroporated with Rsk2 shRNA and 6 control samples (for shRNA experiments). RNA extractions were performed as previously described (ref. [67]). Oligo-dT primed cDNAs were generated using the Superscript II kit (Invitrogen) according to the manufacturer's protocol. Quantitative real-time PCR was achieved using SybrGreen and LightCycler 480 (Roche). The sequences of primers used for the various tested genes are given in Table S3 in File S1. A probe set for detection of mouse Gapdh mRNA (encoded by a housekeeping gene) was used for normalization. For each sample the ratio between signals for the gene of interest and Gapdh was calculated to normalize concentration values. To verify if genes were differentially expressed between electroporated and control samples, the average of ratios were then compared.
Statistical tests
Data, presented as means ± SEM, were compared using Student t-test (for 2 groups comparison). P-values below 0.05 were considered as significant.
Supporting Information
Description of the craniofacial anatomical landmarks used for X-Ray microtomographic analysis of Rsk mutant mice.
(TIF)
Molar landmarks used for the analysis of X-Ray tomography images.
(TIF)
Three-dimensional isosurface rendering of a right mandibular incisor from a wild-type mouse, in lateral (A) and dorsal (B) views, depicting the distances measured for the morphometric analysis.
(TIF)
Supporting Information file containing Tables S1–S3. Table S1 in file S1. Molar root numbers in WT and Rsk2-/Y mice. UR: upper right. UL: upper left. LR: lower right. LL: lower left. M1: first molar. M2: second molar. M3: third molar. ST: supernumerary tooth. Table S2 in file S1. Template plasmids used for in situ hybridization. Table S3 in file S1. Sequences of primers used for quantitative RT-PCR experiments.
(DOCX)
Acknowledgments
The authors are grateful to the patients and families attending the Reference centre for orodental manifestations of rare diseases, HUS, Strasbourg, France, and to Dr. Karen Niederreither for proofreading of the manuscript. They also wish to thank Cathy Herouard-Molina, Doulaye Dembele, and Christelle Thibault from the IGBMC Microarray and sequencing platform.
Funding Statement
This work was supported by grants (to AB-Z) from the University of Strasbourg, the Hôpitaux Universitaires de Strasbourg (API, 2009-2012, “Development of the oral cavity: from gene to clinical phenotype in Human”), IFRO (Institut Français pour la Recherche Odontologique), by institutional funds from the Centre National de la Recherche Scientifique (CNRS) and Institut National de la Santé et de la Recherche Médicale (INSERM). This project, A27 “Orodental manifestations of rare diseases”, selected by the Offensive Sciences of the Trinational Metropolitan Area Upper Rhine (TMR), is co-financed by the European Regional Development Fund (ERDF) of the European Union, in the framework of the INTERREG IV Upper Rhine programme. VL-H was the recipient of a PhD fellowship from the Ministère Français de la Recherche. MP was supported by a grant from the Agence Nationale de la Recherche (to PD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hanauer A, Young ID (2002) Coffin-Lowry syndrome: clinical and molecular features. J Med Genet 39: 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Temtamy SA, Miller JD, Hussels-Maumenee I (1975) The Coffin-Lowry syndrome: an inherited faciodigital mental retardation syndrome. J Pediatr 86: 724–731. [DOI] [PubMed] [Google Scholar]
- 3. Temtamy SA, Miller JD, Dorst JP, Hussels-Maumenee I, Salinas C, et al. (1975) The Coffin-Lowry syndrome: a simply inherited trait comprising mental retardation, faciodigital anomalies and skeletal involvement. Birth Defects Orig Artic Ser 11: 133–152. [PubMed] [Google Scholar]
- 4. Igari K, Hozumi Y, Monma Y, Mayanagi H (2006) A case of Coffin-Lowry syndrome with premature exfoliation of primary teeth. Int J Paediatr Dent 16: 213–217. [DOI] [PubMed] [Google Scholar]
- 5. Day P, Cole B, Welbury R (2000) Coffin-Lowry syndrome and premature tooth loss: a case report. ASDC J Dent Child 67: 148–150. [PubMed] [Google Scholar]
- 6. Frodin M, Gammeltoft S (1999) Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol 151: 65–77. [DOI] [PubMed] [Google Scholar]
- 7. Romeo Y, Zhang X, Roux PP (2012) Regulation and function of the RSK family of protein kinases. Biochem J 441: 553–569. [DOI] [PubMed] [Google Scholar]
- 8. Tan Y, Ruan H, Demeter MR, Comb MJ (1999) p90(RSK) blocks bad-mediated cell death via a protein kinase C-dependent pathway. J Biol Chem 274: 34859–34867. [DOI] [PubMed] [Google Scholar]
- 9. Hauge C, Frodin M (2006) RSK and MSK in MAP kinase signalling. J Cell Sci 119: 3021–3023. [DOI] [PubMed] [Google Scholar]
- 10. Poirier R, Jacquot S, Vaillend C, Soutthiphong AA, Libbey M, et al. (2007) Deletion of the Coffin-Lowry Syndrome Gene Rsk2 in Mice is Associated With Impaired Spatial Learning and Reduced Control of Exploratory Behavior. Behav Genet 37: 31–50. [DOI] [PubMed] [Google Scholar]
- 11. Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, et al. (2004) ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 117: 387–398. [DOI] [PubMed] [Google Scholar]
- 12. Kohn M, Hameister H, Vogel M, Kehrer-Sawatzki H (2003) Expression pattern of the Rsk2, Rsk4 and Pdk1 genes during murine embryogenesis. Gene Expr Patterns 3: 173–177. [DOI] [PubMed] [Google Scholar]
- 13. Viriot L, Peterkova R, Peterka M, Lesot H (2002) Evolutionary implications of the occurrence of two vestigial tooth germs during early odontogenesis in the mouse lower jaw. Connect Tissue Res 43: 129–133. [DOI] [PubMed] [Google Scholar]
- 14. Fleischmannova J, Matalova E, Tucker AS, Sharpe PT (2008) Mouse models of tooth abnormalities. Eur J Oral Sci 116: 1–10. [DOI] [PubMed] [Google Scholar]
- 15. Aberg T, Cavender A, Gaikwad JS, Bronckers AL, Wang X, et al. (2004) Phenotypic Changes in Dentition of Runx2 Homozygote-null Mutant Mice. J Histochem Cytochem 52: 131–140. [DOI] [PubMed] [Google Scholar]
- 16. Zhang M, Chen W, Smith SM, Napoli JL (2001) Molecular characterization of a mouse short chain dehydrogenase/reductase active with all-trans-retinol in intact cells, mRDH1. J Biol Chem 276: 44083–44090. [DOI] [PubMed] [Google Scholar]
- 17. Bloch-Zupan A, Decimo D, Loriot M, Mark MP, Ruch JV (1994) Expression of nuclear retinoic acid receptors during mouse odontogenesis. Differentiation 57: 195–203. [DOI] [PubMed] [Google Scholar]
- 18. Bloch-Zupan A, Mark MP, Weber B, Ruch JV (1994) In vitro effects of retinoic acid on mouse incisor development. Arch Oral Biol 39: 891–900. [DOI] [PubMed] [Google Scholar]
- 19. Mark MP, Bloch-Zupan A, Ruch JV (1992) Effects of retinoids on tooth morphogenesis and cytodifferentiations, in vitro. Int J Dev Biol 36: 517–526. [PubMed] [Google Scholar]
- 20. Mark MP, Bloch-Zupan A, Wolf C, Ruberte E, Ruch JV (1991) Involvement of cellular retinoic acid-binding proteins I and II (CRABPI and CRABPII) and of the cellular retinol-binding protein I (CRBPI) in odontogenesis in the mouse. Differentiation 48: 89–98. [DOI] [PubMed] [Google Scholar]
- 21. Wan X, Ji W, Mei X, Zhou J, Liu JX, et al. (2010) Negative feedback regulation of Wnt4 signaling by EAF1 and EAF2/U19. PLoS One 5: e9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao W, Zhang Q, Habermacher G, Yang X, Zhang AY, et al. (2008) U19/Eaf2 knockout causes lung adenocarcinoma, B-cell lymphoma, hepatocellular carcinoma and prostatic intraepithelial neoplasia. Oncogene 27: 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wise GE, He H, Gutierrez DL, Ring S, Yao S (2011) Requirement of alveolar bone formation for eruption of rat molars. Eur J Oral Sci 119: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wright TJ, Ladher R, McWhirter J, Murre C, Schoenwolf GC, et al. (2004) Mouse FGF15 is the ortholog of human and chick FGF19, but is not uniquely required for otic induction. Dev Biol 269: 264–275. [DOI] [PubMed] [Google Scholar]
- 25. Ji Q, Luo ZX, Yuan CX, Wible JR, Zhang JP, et al. (2002) The earliest known eutherian mammal. Nature 416: 816–822. [DOI] [PubMed] [Google Scholar]
- 26. Peterka M, Vonesch JL, Ruch JV, Cam Y, Peterkova R, et al. (2000) Position and growth of upper and lower tooth primordia in prenatal mouse—3D study. J Craniofac Genet Dev Biol 20: 35–43. [PubMed] [Google Scholar]
- 27. Viriot L, Lesot H, Vonesch JL, Ruch JV, Peterka M, et al. (2000) The presence of rudimentary odontogenic structures in the mouse embryonic mandible requires reinterpretation of developmental control of first lower molar histomorphogenesis. Int J Dev Biol 44: 233–240. [PubMed] [Google Scholar]
- 28. Peterkova R, Peterka M, Lesot H (2003) The developing mouse dentition: a new tool for apoptosis study. Ann N Y Acad Sci 1010: 453–466. [DOI] [PubMed] [Google Scholar]
- 29.Porntaveetus T, Ohazama A, Choi HY, Herz J, Sharpe PT (2011) Wnt signaling in the murine diastema. Eur J Orthod. [DOI] [PMC free article] [PubMed]
- 30. Ohazama A, Blackburn J, Porntaveetus T, Ota MS, Choi HY, et al. (2010) A role for suppressed incisor cuspal morphogenesis in the evolution of mammalian heterodont dentition. Proc Natl Acad Sci U S A 107: 92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, et al. (2006) Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell 11: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Charles C, Hovorakova M, Ahn Y, Lyons DB, Marangoni P, et al. (2011) Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development 138: 4063–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohazama A, Haycraft CJ, Seppala M, Blackburn J, Ghafoor S, et al. (2009) Primary cilia regulate Shh activity in the control of molar tooth number. Development 136: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kassai Y, Munne P, Hotta Y, Penttila E, Kavanagh K, et al. (2005) Regulation of mammalian tooth cusp patterning by ectodin. Science 309: 2067–2070. [DOI] [PubMed] [Google Scholar]
- 35. Murashima-Suginami A, Takahashi K, Sakata T, Tsukamoto H, Sugai M, et al. (2008) Enhanced BMP signaling results in supernumerary tooth formation in USAG-1 deficient mouse. Biochem Biophys Res Commun 369: 1012–1016. [DOI] [PubMed] [Google Scholar]
- 36. Ohazama A, Johnson EB, Ota MS, Choi HY, Porntaveetus T, et al. (2008) Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One 3: e4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA (1998) sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92: 253–263. [DOI] [PubMed] [Google Scholar]
- 38. Kim HJ, Bar-Sagi D (2004) Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol 5: 441–450. [DOI] [PubMed] [Google Scholar]
- 39. Tucker AS, Headon DJ, Courtney JM, Overbeek P, Sharpe PT (2004) The activation level of the TNF family receptor, Edar, determines cusp number and tooth number during tooth development. Dev Biol 268: 185–194. [DOI] [PubMed] [Google Scholar]
- 40. Pispa J, Mikkola ML, Mustonen T, Thesleff I (2003) Ectodysplasin, Edar and TNFRSF19 are expressed in complementary and overlapping patterns during mouse embryogenesis. Gene Expr Patterns 3: 675–679. [DOI] [PubMed] [Google Scholar]
- 41. Mustonen T, Pispa J, Mikkola ML, Pummila M, Kangas AT, et al. (2003) Stimulation of ectodermal organ development by Ectodysplasin-A1. Dev Biol 259: 123–136. [DOI] [PubMed] [Google Scholar]
- 42. Peterkova R, Lesot H, Viriot L, Peterka M (2005) The supernumerary cheek tooth in tabby/EDA mice-a reminiscence of the premolar in mouse ancestors. Arch Oral Biol 50: 219–225. [DOI] [PubMed] [Google Scholar]
- 43. Peterkova R, Kristenova P, Lesot H, Lisi S, Vonesch JL, et al. (2002) Different morphotypes of the tabby (EDA) dentition in the mouse mandible result from a defect in the mesio-distal segmentation of dental epithelium. Orthod Craniofac Res 5: 215–226. [DOI] [PubMed] [Google Scholar]
- 44. Lesot H, Peterkova R, Kristenova P, Lisi S, Peterka M (2003) [Effect of the Tabby mutation on the dentition of mice]. Bull Group Int Rech Sci Stomatol Odontol 45: 1–11. [PubMed] [Google Scholar]
- 45. Charles C, Pantalacci S, Peterkova R, Tafforeau P, Laudet V, et al. (2009) Effect of eda loss of function on upper jugal tooth morphology. Anat Rec (Hoboken) 292: 299–308. [DOI] [PubMed] [Google Scholar]
- 46. Kristenova P, Peterka M, Lisi S, Gendrault JL, Lesot H, et al. (2002) Different morphotypes of functional dentition in the lower molar region of tabby (EDA) mice. Orthod Craniofac Res 5: 205–214. [DOI] [PubMed] [Google Scholar]
- 47. Ghoda L, Lin X, Greene WC (1997) The 90-kDa ribosomal S6 kinase (pp90rsk) phosphorylates the N-terminal regulatory domain of IkappaBalpha and stimulates its degradation in vitro. J Biol Chem 272: 21281–21288. [DOI] [PubMed] [Google Scholar]
- 48. Schouten GJ, Vertegaal AC, Whiteside ST, Israel A, Toebes M, et al. (1997) IkappaB alpha is a target for the mitogen-activated 90 kDa ribosomal S6 kinase. Embo J 16: 3133–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Courtois G, Smahi A, Reichenbach J, Doffinger R, Cancrini C, et al. (2003) A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest 112: 1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boran T, Lesot H, Peterka M, Peterkova R (2005) Increased apoptosis during morphogenesis of the lower cheek teeth in tabby/EDA mice. J Dent Res 84: 228–233. [DOI] [PubMed] [Google Scholar]
- 51. Coin R, Lesot H, Vonesch JL, Haikel Y, Ruch JV (1999) Aspects of cell proliferation kinetics of the inner dental epithelium during mouse molar and incisor morphogenesis: a reappraisal of the role of the enamel knot area. Int J Dev Biol 43: 261–267. [PubMed] [Google Scholar]
- 52. Lesot H, Peterkova R, Viriot L, Vonesch JL, Tureckova J, et al. (1998) Early stages of tooth morphogenesis in mouse analyzed by 3D reconstructions. Eur J Oral Sci 106 Suppl 164–70. [DOI] [PubMed] [Google Scholar]
- 53. Viriot L, Peterkova R, Vonesch JL, Peterka M, Ruch JV, et al. (1997) Mouse molar morphogenesis revisited by three-dimensional reconstruction. III. Spatial distribution of mitoses and apoptoses up to bell-staged first lower molar teeth. Int J Dev Biol 41: 679–690. [PubMed] [Google Scholar]
- 54. Guimiot F, Delezoide AL, Hanauer A, Simonneau M (2004) Expression of the RSK2 gene during early human development. Gene Expr Patterns 4: 111–114. [DOI] [PubMed] [Google Scholar]
- 55. Zeniou M, Ding T, Trivier E, Hanauer A (2002) Expression analysis of RSK gene family members: the RSK2 gene, mutated in Coffin-Lowry syndrome, is prominently expressed in brain structures essential for cognitive function and learning. Hum Mol Genet 11: 2929–2940. [DOI] [PubMed] [Google Scholar]
- 56. Cho YY, He Z, Zhang Y, Choi HS, Zhu F, et al. (2005) The p53 protein is a novel substrate of ribosomal S6 kinase 2 and a critical intermediary for ribosomal S6 kinase 2 and histone H3 interaction. Cancer Res 65: 3596–3603. [DOI] [PubMed] [Google Scholar]
- 57. Lagronova-Churava S, Spoutil F, Vojtechova S, Lesot H, Peterka M, et al. (2013) The dynamics of supernumerary tooth development are differentially regulated by Sprouty genes. J Exp Zool B Mol Dev Evol 320: 307–320. [DOI] [PubMed] [Google Scholar]
- 58.Liu Z, Li T, Reinhold MI, Naski MC (2012) MEK1-RSK2 contributes to hedgehog signaling by stabilizing GLI2 transcription factor and inhibiting ubiquitination. Oncogene. [DOI] [PubMed]
- 59. Schneider A, Mehmood T, Pannetier S, Hanauer A (2011) Altered ERK/MAPK signaling in the hippocampus of the mrsk2_KO mouse model of Coffin-Lowry syndrome. J Neurochem 119: 447–459. [DOI] [PubMed] [Google Scholar]
- 60. Mitsiadis TA, Luukko K (1995) Neurotrophins in odontogenesis. Int J Dev Biol 39: 195–202. [PubMed] [Google Scholar]
- 61. Tummers M, Thesleff I (2009) The importance of signal pathway modulation in all aspects of tooth development. J Exp Zoolog B Mol Dev Evol 312B: 309–319. [DOI] [PubMed] [Google Scholar]
- 62.Bloch-Zupan A, Sedano H, Scully C (2012) Dento/Oro/Craniofacial Anomalies and Genetics. London: Elsevier Inc.
- 63. Richtsmeier JT, Paik CH, Elfert PC, Cole TM 3rd, Dahlman HR (1995) Precision, repeatability, and validation of the localization of cranial landmarks using computed tomography scans. Cleft Palate Craniofac J 32: 217–227. [DOI] [PubMed] [Google Scholar]
- 64. Richtsmeier JT, Baxter LL, Reeves RH (2000) Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev Dyn 217: 137–145. [DOI] [PubMed] [Google Scholar]
- 65. Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, et al. (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol 9: e1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chotteau-Lelievre A, Dolle P, Gofflot F (2006) Expression analysis of murine genes using in situ hybridization with radioactive and nonradioactively labeled RNA probes. Methods Mol Biol 326: 61–87. [DOI] [PubMed] [Google Scholar]
- 67. Laugel-Haushalter V, Paschaki M, Thibault-Carpentier C, Dembele D, Dolle P, et al. (2013) Molars and incisors: show your microarray IDs. BMC Res Notes 6: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of the craniofacial anatomical landmarks used for X-Ray microtomographic analysis of Rsk mutant mice.
(TIF)
Molar landmarks used for the analysis of X-Ray tomography images.
(TIF)
Three-dimensional isosurface rendering of a right mandibular incisor from a wild-type mouse, in lateral (A) and dorsal (B) views, depicting the distances measured for the morphometric analysis.
(TIF)
Supporting Information file containing Tables S1–S3. Table S1 in file S1. Molar root numbers in WT and Rsk2-/Y mice. UR: upper right. UL: upper left. LR: lower right. LL: lower left. M1: first molar. M2: second molar. M3: third molar. ST: supernumerary tooth. Table S2 in file S1. Template plasmids used for in situ hybridization. Table S3 in file S1. Sequences of primers used for quantitative RT-PCR experiments.
(DOCX)