SUMMARY
Tumor cells commonly have increased glucose uptake and lactate accumulation. Lactate is produced from pyruvate by lactate dehydrogenase A (LDH-A), which is frequently overexpressed in tumor cells and is important for cell growth. Elevated transcription by c-Myc or HIF1α may contribute to increased LDH-A in some cancer types. Here, we show that LDH-A is acetylated at lysine 5 (K5) and that this acetylation inhibits LDH-A activity. Furthermore, the K5-acetylated LDH-A is recognized by the HSC70 chaperone and delivered to lysosomes for degradation. Replacement of endogenous LDH-A with an acetylation mimetic mutant decreases cell proliferation and migration. Importantly, K5 acetylation of LDH-A is reduced in human pancreatic cancers. Our study reveals a mechanism of LDH-A upregulation in pancreatic cancers.
INTRODUCTION
Alteration in cell metabolism is a common event in tumorigenesis, as indicated by the dramatic increase of glucose utilization. However, the increased glucose uptake in tumor cells often does not lead to a corresponding increase in oxidative phosphorylation even in the presence of sufficient oxygen supply. Instead, glycolysis is highly elevated in most cancer cells. This metabolic alteration, known as the Warburg effect (Warburg, 1956), is believed to benefit tumor cells not only by conditioning the microenvironment, but also by increasing the levels of glycolytic intermediates, many of which also serve as precursors for anabolic biosynthesis, to support increased cell growth (Koppenol et al., 2011; Vander Heiden et al., 2009). The fact that tumor cells have a dramatically increased glucose uptake has provided the basis for 18F-fluorodeoxyglucose-positron emission tomography technology, which is widely used for detecting tumors.
The last step of glycolysis is catalyzed by pyruvate kinase (PK), which converts phosphoenopyruvate to pyruvate. In normal non-proliferating cells, most, if not all, of pyruvate enters mitochondria, where it is converted to acetyl-CoA by the pyruvate dehydrogenase complex to fuel the tricarboxylic acid (TCA) cycle and oxidative phosphorylation for efficient energy production. In contrast, in cancer cells, and probably other highly proliferating cells, the influx of pyruvate into mitochondria and the TCA is not proportional to the increased glucose uptake; instead, more pyruvate is converted to lactate by lactate dehydrogenase (LDH). Therefore, a high conversion rate of pyruvate to lactate, hence high LDH, is commonly observed in cancer cells.
LDH is ahomo- or hetero-tetrameric enzyme composed of two subunits, M and H, encoded by two highly related genes, LDH-A (also known as LDHM, LDH1, GSD11, and PIG19) and LDH-B (also known as LDH-H, H-LDH, and LDH2), resulting in five different isozymes depending on the ratio of the M and H subunits (M4, M3H1, M2H2, M1H3, and H4). LDH enzyme catalyzes the reversible conversion of pyruvate to lactate using NAD+ as a cofactor. Although the physiologic significance of lactate accumulation in tumor cells, a dead-end product in cellular metabolism, is currently a topic of debate, it has long been known that many tumor cells express a high level of LDH-A (Goldman et al., 1964), including non-small cell lung cancer (Koukourakis et al., 2003), colorectal cancer (Koukourakis et al., 2006), and breast and gynecologic cancers (Koukourakis et al., 2009). In many tumors, elevated LDH-A levels have been correlated with poor prognosis and resistance to chemotherapy and radiation therapy. Further evidence linking an LDH-A increase to tumorigenesis comes from the findings that the LDH-A gene is a direct target of both Myc and HIF transcription factors (Lewis et al., 1997; Semenza et al., 1996; Shim et al., 1997). Inhibition of LDH-A by either RNA interference or pharmacologic agents blocks tumor progression in vivo (Fantin et al., 2006; Le et al., 2010; Xie et al., 2009), supporting an important role of elevated LDH-A in tumorigenesis and LDH-A as a potential therapeutic target.
We and others have recently discovered that a large number of non-nuclear proteins, especially those involved in intermediate metabolism, are acetylated (Choudhary et al., 2009; Kim et al., 2006; Wang et al., 2010; Zhao et al., 2010). In this report, we investigated LDH-A acetylation and its functional significance in tumorigenesis.
RESULTS
LDH-A Is Acetylated at Lysine 5
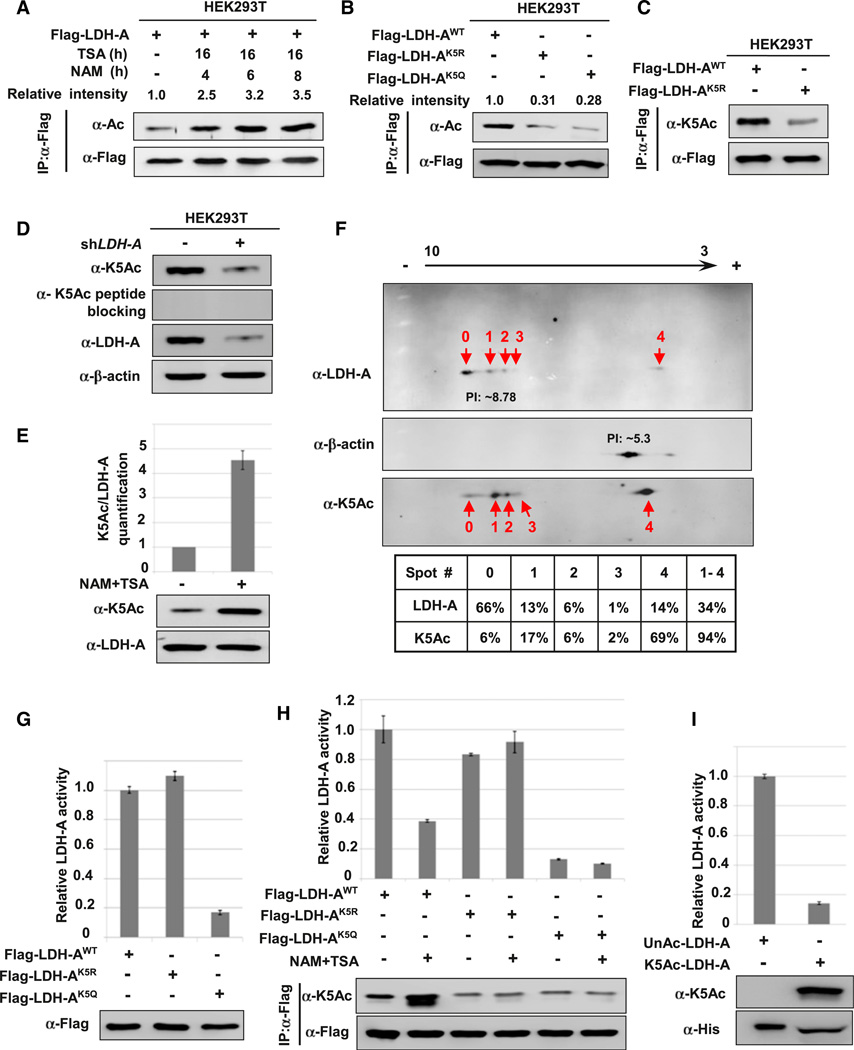
Eight putative acetylation sites were identified in LDH-A by mass spectrometry (Figure S1A available online; Choudhary et al., 2009). Western blotting with anti-acetyllysine antibody showed that LDH-A was indeed acetylated and its acetylation was enhanced approximately 3.5-fold after treatment with trichostatin A (TSA), an inhibitor of histone deacetylase HDAC I and II (Ekwall et al., 1997; Furumai et al., 2001), and nicotinamide (NAM), an inhibitor of the SIRT family of deacetylases (Avalos et al., 2005) (Figure 1A).
Figure 1. Acetylation at Lys-5 Decreases LDH-A Enzyme Activity.
(A) LDH-A is acetylated. Flag-LDH-A was transfected into 293T cells followed by treatment with deacetylase inhibitors TSA and NAM for indicated time. LDH-A acetylation and protein levels were analyzed by western blot with indicated antibody. Relative ratios of acetylation were calculated from normalizing against Flag-LDH-A.
(B) Mutation of K5 decreases LDH-A acetylation. The indicated plasmids were transfected into 293T cells and proteins were immunoprecipitated for western blotting.
(C) Characterization of acetyl-LDH-A(K5) antibody. The indicated plasmids were transfected into 293T cells, acetylation level of immunoprecipitated Flag-LDH-A was measured by direct western bloting using the acetyl-LDH-A (K5) antibody (α-K5Ac).
(D) Endogenous LDH-A is acetylated on lysine 5. Cell lysate from scramble or LDH-A shRNA knockdown stable cells were probed with indicated antibodies.
(E) Treatment with NAM and TSA increases endogenous LDH-A acetylation; 293T cells were treated with TSA and NAM. Endogenous LDH-A protein levels and acetylation of K5 were determined by western blotting with indicated antibodies (bottom panel). Relative K5-acetylated LDH-A over total LDH-A protein was quantified (top panel). Error bars represent ± SD for triplicate experiments.
(F) Quantitative analysis of endogenous LDH-A acetylation at K5 by isoelectric focusing (IEF) analysis; 293T cells lysate were separated by IEF, followed by western blotting using indicated antibodies. Relative LDH-A K5 acetylation and LDH-A protein levels for each spot were quantified by intensity, and the relative percentage of each spot is calculated and listed below the western blot panels.
(G) K5Q mutant decreases LDH-A enzyme activity. Flag-tagged wild-type and mutant LDH-A protein were expressed in 293T cells and purified by immuno-precipitation. The enzyme activity was measured and normalized against protein level. Relative enzyme activities of triplicate experiments with ± SD are presented.
(H) NAM and TSA treatment decreases the enzyme activity of wild-type, but not mutant LDH-A. Flag-tagged wild-type and mutant LDH-A protein were expressed in 293T cells and treated with or without NAM and TSA, then purified by immunoprecipitation. The LDH-A enzyme activity was measured and normalized against protein level. Relative enzyme activities of triplicate experiments ± SD are presented.
(I) Acetylated LDH-A has lower enzyme activity. Recombinant un-acetylated and K5-acetylated LDH-A protein were prepared by the system of genetically encoding Nε-acetyllysine in E. coli. The enzyme activity was measured and normalized against protein level. Relative enzyme activities of triplicate experiments ± SD are presented. See also Figure S1.
We then mutated each of eight putative acetylation sites individually to glutamine (Q), and examined their acetylation. Mutation of either K5 or K318, but not other lysine residues, to glutamine resulted in a significant reduction in LDH-A acetylation (Figure S1B). Arginine substitution of K5, but not K318, dramatically decreased the LDH-A acetylation by approximately 70% (Figure 1B; data not shown), indicating that K5, which is evolutionarily conserved from Caenorhabditis elegans to mammals (Figure S1C), is a major acetylation site in LDH-A.
We generated an antibody specifically recognizing the K5-acetylated LDH-A. The specificity of the anti-acetyl-LDH-A (K5) antibody was verified as it recognized the K5-acetylated peptide but not the unacetylated control peptide (Figure S1D). Western blotting using this antibody detected ectopically expressed wild-type, but only weakly recognized the K5R mutant LDH-A (Figure 1C). Moreover, this antibody detected the acetylated but not the unacetylated LDH-A that was expressed and purified from bacteria (Figure 1I). These characterizations demonstrate the specificity of our anti-acetyl-LDH-A(K5) antibody in recognizing the K5-acetylated LDH-A.
We used the anti-acetyl-LDH-A (K5) antibody to determine acetylation of endogenous LDH-A. Acetylation of LDH-A could readily be detected by the antibody. This signal was diminished by LDH-A knockdown and was completely blocked by the pre-incubation with the antigen peptide (Figure 1D), confirming the specificity of the anti-acetyl-LDH-A(K5) antibody. Treatment of cells with deacetylase inhibitors TSA and NAM strongly increased K5 acetylation of both endogenously (Figure 1E) and the ectopically expressed LDH-A (Figure S1E). To quantify LDH-A acetylation, we employed IEF (isoelectric focusing) to separate the acetylated protein based on the loss of positive charge due to lysine acetylation. The spot with highest pI, spot 0, showed the lowest relative acetylation, while the lowest pI spot 4 had the highest acetylation, indicating that the change of LDH-A pI is at least in part due to acetylation (Figure 1F). Assuming that spot 0 represented the unacetylated LDH-A while spot 4 represented the fully acetylated LDH-A, we estimated that approximately 20% of the LDH-A is acetylated on lysine 5. Therefore, a substantial fraction of endogenous LDH-A could be acetylated.
K5 Acetylation Inhibits LDH-A Enzyme Activity
To test the effect of K5 acetylation, the activity of LDH-AK5R and LDH-AK5Q mutants was compared with that of wild-type LDH-A. We found that LDH-AK5Q displayed only 18% of the wild-type activity, while the LDH-AK5R mutation had a minor effect on the LDH-A activity (Figure 1G). Consistent with an inhibitory effect of acetylation on LDH-A activity, inhibition of deacetylases by NAM and TSA treatment significantly decreased LDH-A enzyme activity by more than 60% (Figures 1H and S1F). Moreover, treatment of NAM and TSA had little effect on the activity of either LDH-AK5Q or LDH-AK5R mutants (Figure 1H).
To definitively demonstrate the effect of K5 acetylation on LDH-A activity, we employed the system of genetically encoding Nε-acetyllysine to prepare recombinant proteins in Escherichia coli (Neumann et al., 2008, 2009). This expression system produced LDH-A proteins with 100% acetylation at K5 due to the suppression of the K5-TAG stop codon by the Nε-acetyllysine-conjugated amber suppressor tRNA. We prepared both unacetylated and K5-acetylated LDH-A and compared their enzymatic activity. As shown in Figure 1I, K5-acetylated LDH-A showed significantly lower activity when compared with the un-acetylated LDH-A. Collectively, these results demonstrate that acetylation at lysine 5 inhibits LDH-A activity.
SIRT2 Decreases LDH-A Acetylation and Increases Its Enzyme Activity
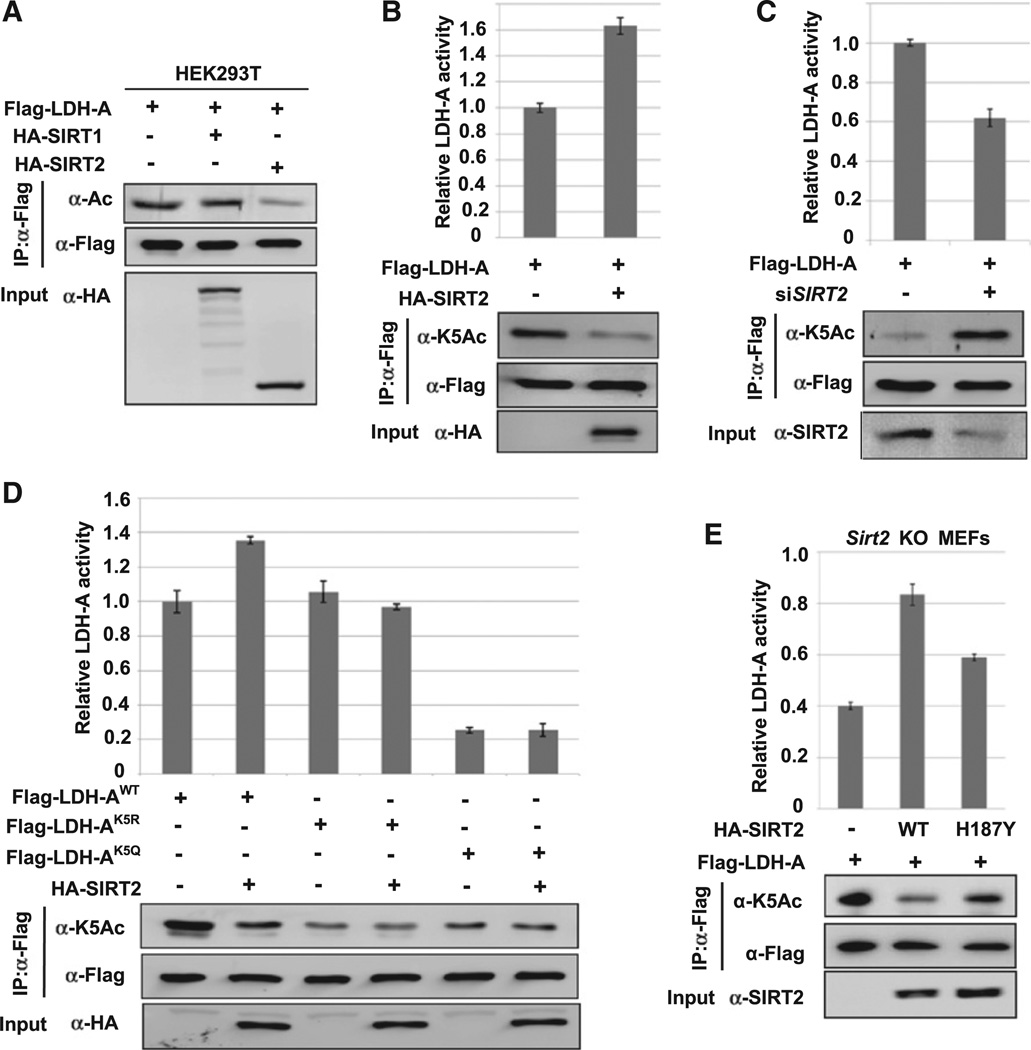
To identify the deacetylase responsible for LDH-A regulation, we first determined how inhibition of either SIRT or HDAC could affect LDH-A acetylation at lysine 5. Treatment of cells with SIRT inhibitor NAM, but not HDAC inhibitor TSA, increased acetylation at K5 (Figure S2), indicating that a SIRT deacetylase is probably involved in K5 deacetylation. To identify the specific SIRT, we co-expressed LDH-A with the two cytosolic SIRT deacetylases, SIRT1 and SIRT2, and found that SIRT2, but not SIRT1, decreased LDH-A acetylation (Figures 2A and 2B). Supporting this observation, knocking down SIRT2 significantly increased K5 acetylation (Figure 2C). Co-expression of SIRT2 increased the activity of the LDH-A by 63% along with the decreased lysine 5 acetylation (Figure 2B). Conversely, SIRT2 knockdown decreased LDH-A activity by 38% (Figure 2C). Together, these observations demonstrate a specific and prominent role of SIRT2 in the deacetylation and enzyme activation of LDH-A.
Figure 2. SIRT2 Deacetylates LDH-A at K5 and Increases LDH-A Activity.
(A) SIRT2 overexpression decreases LDH-A acetylation. 293T cells were transfected with indicated plasmids and LDH-A acetylation was determined by western blotting.
(B) SIRT2 decreases K5 acetylation and increases LDH-A activity. 293T cells were transfected with indicated plasmids, Flag-LDH-A was immuno-precipitated, and LDH-A activity was assayed. LDH-A activity was normalized against protein levels. Relative enzyme activities of triplicated experiments ± SD are presented.
(C) SIRT2 knockdown increases K5 acetylation and decreases LDH-A activity. 293T cells were transfected with indicated plasmids and SIRT2 siRNA oligonucleotides. LDH-A was immunoprecipitated and activity was assayed. LDH-A acetylation at K5 was determined by western blotting. Relative enzyme activities of triplicate experiments ± SD are presented.
(D) SIRT2 overexpression increases the activity of wild-type, but not acetylation-deficient K5R or K5Q mutant LDH-A. 293T cells were transfected with indicated plasmids, followed by immunoprecipitation and enzyme assay. Relative enzyme activities of triplicate experiments ± SD are presented.
(E) The deacetylase activity of SIRT2 is required to increase LDH-A activity. Sirt2 knockout MEFs were co-transfected with Flag-LDH-A and SIRT2 wild-type or the inactive mutant (H187Y). LDH-A was immunoprecipitated and enzyme activity was assayed. LDH-A activity was normalized against protein levels. Relative enzyme activities of triplicate experiments ± SD are presented.
See also Figure S2.
We also found that SIRT2 co-expression had no significant effect on the activity of LDH-AK5Q and LDH-AK5R mutants (Figure2D), indicating that SIRT2 stimulates LDH-A activity mostly via deacetylation of K5. Furthermore, re-expression of wild-type SIRT2, but not the inactive H187Y mutant, reduced LDH-A acetylation and increased LDH-A enzyme activity in Sirt2 knockout MEFs (Figure 2E). Collectively, these data support a critical role of SIRT2 enzyme activity in LDH-A regulation by deacetylating lysine 5.
Acetylation at K5 Decreases LDH-A Protein Level
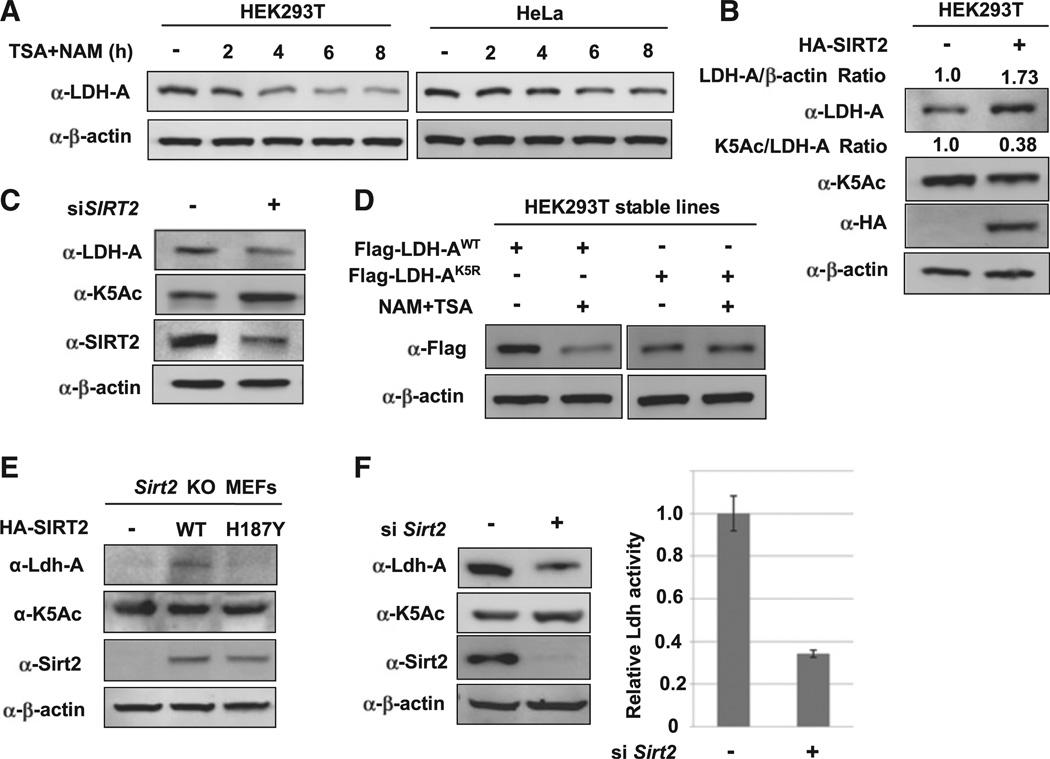
In addition to the effect on LDH-A enzyme activity, NAM and TSA treatment also led to a time-dependent reduction of LDH-A protein levels (Figures 3A and S3A). We then determined whether acetylation downregulating of LDH-A protein level occurs at or after transcription. Quantitative RT-PCR showed that NAM and TSA treatment had a minor effect on LDH-A mRNA levels (Figure S3B), indicating a posttranscriptional regulation of LDH-A protein by acetylation. To determine if acetylation could affect LDH-A protein level, we analyzed the effect of SIRT2 overexpression or knockdown on LDH-A protein. Overexpression of SIRT2 decreased LDH-A K5 acetylation and increased LDH-A protein in both 293T and pancreatic cancer cell line (Figures 3B and S3C). Conversely, SIRT2 knockdown increased LDH-A acetylation and concomitantly decreased the steady-state level of LDH-A protein (Figure 3C). These results indicate that acetylation may decrease LDH-A protein. Furthermore, we found that inhibition of deacetylases decreased the level of wild-type, but not the K5R mutant (Figure 3D). Based on these results, we propose that acetylation of K5 destabilizes LDH-A protein.
Figure 3. Acetylation at K5 Decreases LDH-A Protein Level.
(A) NAM and TSA treatment decreases endogenous LDH-A protein level. 293T and HeLa cells were either untreated or treated with NAM and TSA for different lengths of time, as indicated. The steady-state levels of LDH-A protein were determined by western blotting and normalized against β-actin.
(B) SIRT2 overexpression decreases endogenous LDH-A K5 acetylation and increases LDH-A protein level. Plasmid expressing SIRT2 was transfected into 293T cells, and endogenous K5 acetylation and LDH-A expression level were determined by western blotting.
(C) SIRT2 knockdown increases LDH-A K5 acetylation and decreases LDH-A protein level. siRNA oligo nucleotide targeting SIRT2 was transfected into 293T cells and the levels of endogenous LDH-A K5-acetylation, total LDH-A protein, and SIRT2 protein were determined by western blotting.
(D) NAM and TSA treatment decreases the level of wild-type, but not K5R mutant LDH-A. 293T cells stably expressing wild-type and K5R mutant LDH-A were either untreated or treated with NAM and TSA. The levels of K5-acetylated and total LDH-A protein were determined by western blotting.
(E) SIRT2 deacetylase activity is required to increase LDH-A protein level. Wild-type or H187Y mutant SIRT2 was expressed in Sirt2 knockout MEFs, and then endogenous Ldh-A protein level and acetylation at K5 were detected by WB.
(F) SIRT2 knockdown decreases LDH-A activity and protein level in mouse liver. siRNA oligo-nucleotides targeting mouse Sirt2 gene were injected into mouse tail vein and liver tissue was harvested to determine total LDH-A activity (left panel). SIRT2, LDH-A protein, and K5 acetylation were measured by western blotting (right panel). Error bars represent ± SD of triplicated experiments. See also Figure S3.
Next, we investigated the function of SIRT2 in regulation of LDH-A protein levels. We observed that re-expression of the wild-type, but not the H187Y mutant SIRT2, increased LDH-A protein level in Sirt2 knockout MEFs (Figure 3E). In addition, the relative K5 acetylation (the ratio of K5 acetylation over LDH-A protein level) was also reduced by expression of the wild-type, but not the H187Y mutant SIRT2. These data support the notion that the SIRT2 deacetylase activity plays a role in regulating LDH-A protein levels. To determine the function of SIRT2 in LDH-A regulation in vivo, we injected Sirt2 siRNA into mice via the tail vein, and Sirt2 was efficiently reduced in the mouse livers by western blot analysis (Figure 3F). We found that Ldh-A protein levels and activity were significantly decreased. As expected, the relative K5 acetylation was increased in Sirt2 knockdown livers (Figure 3F), indicating a critical function of SIRT2 in LDH-A regulation in vivo.
Acetylation Stimulates LDH-A Degradation by Chaperone-Mediated Autophagy
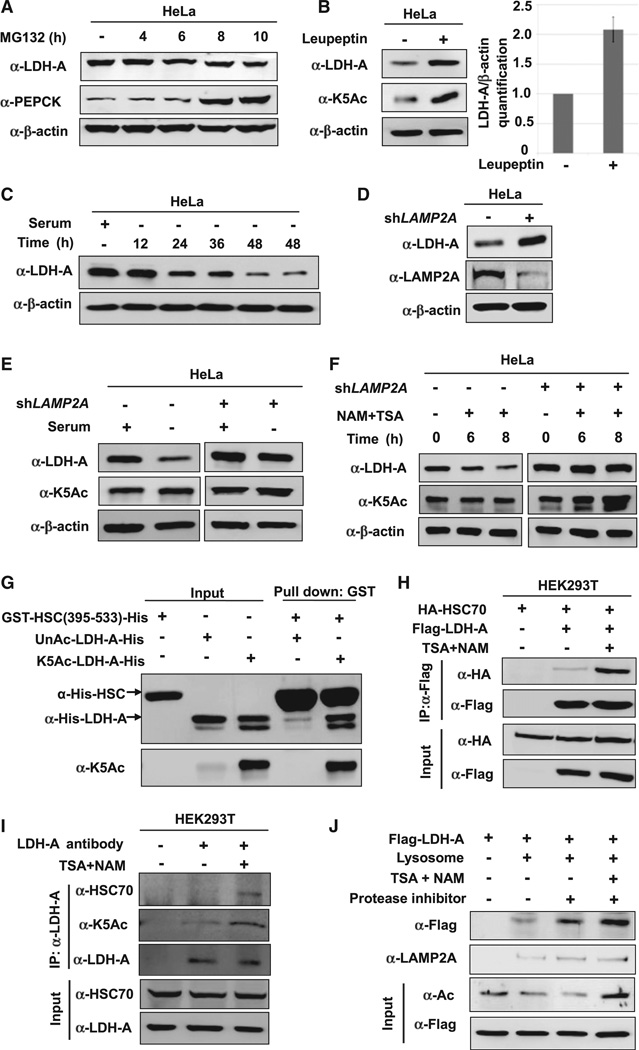
Inhibition of protein synthesis with cycloheximide (CHX) showed that LDH-A was a rather stable protein in HeLa cells with a half-life longer than 8 hr (Figure S4A). Treatment with the proteasome inhibitor MG132 did not increase LDH-A, but significantly increased the protein level of PEPCK (Figure 4A), a metabolic enzyme targeted by the proteasome for degradation (Jiang et al., 2011). These results indicate that the acetylation-induced decrease of LDH-A is mediated by a mechanism that is independent of proteasome.
Figure 4. Acetylation Promotes LDH-A Degradation via CMA.
(A) LDH-A is not degraded by the ubiquitin-proteasome system (UPS). HeLa cells were treated with a proteasome inhibitor MG132 and the LDH-A protein level was analyzed by western blotting. PEPCK, a known substrate of UPS, was included as a control.
(B) Leupeptin accumulates K5-acetylated and total LDH-A protein. HeLa cells were either untreated or treated with leupeptin for 48 hr. The levels of total and acetylated LDH-A were determined by western blotting. LDH-A level was normalized against β-actin. Error bars represent ± SD of triplicated experiments.
(C) Serum withdrawal decreases LDH-A protein. LDH-A level was determined by western blotting after serum withdrawal for different lengths of time, as indicated in HeLa cells.
(D) LAMP2A knockdown accumulates LDH-A. LAMP2A was stably knocked down in HeLa cells by shRNA. The knockdown efficiency and LDH-A protein level were determined by western blotting. (E and F) LAMP2A knockdown blocks the effect of serum deprivation or NAM and TSA treatment on LDH-A protein levels. HeLa cell pools stably expressing LAMP2A shRNA were cultured with or without serum (E) or NAM and TSA (F). The levels of K5-acetylated and total LDH-A protein were determined by western blotting.
(G) Acetylation at K5 increases LDH-A binding to the HSC70 C-terminal domain in vitro. Recombinant unacetylated and K5-acetylated LDH-A protein were prepared by the system of genetically encoding Nε-acetyllysine in E. coli. GST-HCS70 C-terminal domain (from 395 to 533 amino acids) was used in an in vitro binding assay to pulldown the purified LDH-A.
(H and I) Inhibition of deacetylases increases overexpressed or endogenous LDH-A-HSC70 binding. Indicated plasmids were co-transfected into 293T cells, followed by NAM and TSA treatment. LDH-A-HSC70 binding was determined by immunoprecipitation-western analysis (H). The 293T cells were untreated or treated with NAM and TSA (I). Endogenous LDH-A-HSC70 binding was determined by immunoprecipitation and western blot analysis.
(J) Inhibition of deacetylases promotes lysosomal uptake of LDH-A. Flag-tagged LDH-A was immunopurified from 293T cells untreated or treated with deacetylase inhibitors TSA and NAM. The immunoprecipitated LDH-A was incubated with the lysosomes isolated from rat liver. Lysosomes were re-isolated and the associated LDH-A (either inside or binding to the surface) were determined by western blotting.
See also Figure S4.
Autophagy is a major mechanism in intracellular degradation. Macro-autophagy is believed to be a nonselective bulk degradation of intracellular components, whereas chaperone-mediated autophagy (CMA) is a selective degradation for proteins, especially those with a long half-life (Mizushima et al., 2008). We treated cells with leupeptin, an inhibitor of lysosomal proteases that can block lysosome-dependent protein degradation (Jeong et al., 2009), and found that this treatment caused a significant accumulation of LDH-A protein and K5 acetylation (Figure 4B), confirming the involvement of lysosome in acetylation-induced LDH-A degradation. Two-dimensional PAGE analysis showed that leupeptin blocked LDH-A degradation in cells treated with deacetylase inhibitors (Figure S4B). Costaining of LDH-A and lysosomal marker also indicated that a fraction of LDH-A was colocalized with the lysosomal marker LAMP1 (Figure S4C), consistent with a role of lysosome in LDH-A degradation.
Prolonged serum starvation is known to activate CMA (Cuervo et al., 1995; Wing et al., 1991). We found that serum starvation caused a decrease of the steady-state level of LDH-A (Figure 4C), providing additional evidence for a CMA-dependent degradation of LDH-A. To rule out macro-autophagy in LDH-A degradation, we compared the subcellular localization of LDH-A with GFP-LC-3, which is a marker for autophagosome in the macro-autophagy pathway. As shown in Figure S4D, GFP-LC3 and LDH-A showed different subcellular localizations. Moreover, we determined LDH-A protein level in Atg5 knockout MEF cells, which is defective in macro-autophagy, and found that LDH-A protein levels were comparable in Atg5 wild-type and knockout MEF cells (Figure S4E).These data indicate that CMA, but not macro-autophagy, is responsible for LDH-A degradation.
During CMA, the HSC70 chaperone carries target proteins to the lysosomal receptor LAMP2A, which then translocates the target proteins into lysosome for degradation (Cuervo, 2010). To provide additional evidence for the role of CMA in LDH-A degradation, we found that LAMP2A knockdown significantly increased LDH-A protein (Figure 4D). Moreover, LAMP2A knockdown also blocked the LDH-A protein reduction caused by either serum starvation (Figure 4E) or inhibition of deacetylases (Figure 4F). These data support a model that acetylation promotes CMA-dependent degradation of LDH-A.
To explore the role of K5 acetylation in LDH-A degradation by CMA, we examined the interaction between LDH-A and HSC70. Co-immunoprecipitation showed that the acetylation mimetic K5Q mutant displayed a much stronger interaction with HSC70 than the wild-type LDH-A (Figure S4G). Fully acetylated or unacetylated recombinant LDH-A was prepared by the system of genetically encoded Nε-acetyllysine in E. coli, and their interaction with HSC70 was examined. The acetylated, but not the unacetylated, LDH-A could readily pull down endogenous HSC70 (Figure S4F). The C-terminal domain (amino acid residues 395–533) is the substrate binding domain of HSC70. We prepared recombinant HSC70 C-terminal domain and found it to preferentially pull down acetylated but not unacetylated LDH-A (Figure 4G). Consistently, treatment of cells with deacetylase inhibitors TSA and NAM significantly increased the binding between either ectopically expressed (Figure 4H) or endogenous LDH-A and HSC70 (Figure 4I). Collectively, these data demonstrate that LDH-A acetylation, in particular at lysine 5, promotes its interaction with HSC70.
To determine directly if LDH-A could be taken up by lysosomes, we incubated the immunopurified LDH-A with isolated lysosomes in vitro. The results showed LDH-A binding to isolated lysosomes (Figure 4J). When lysosomal protease was inhibited, more LDH-A was found with lysosome, presumably due to the accumulation of intralysosomal LDH-A. Notably, the LDH-A isolated from TSA- and NAM-treated cells showed more lysosomal binding/up-taken than LDH-A isolated from untreated cells. These data are consistent with a model that LDH-A acetylation increases its interaction with HSC70, binding to and being taken up by the lysosomes, and leading to its eventual degradation.
K5 Acetylation Impairs the Function of LDH-A in Supporting Cell Proliferation and Migration
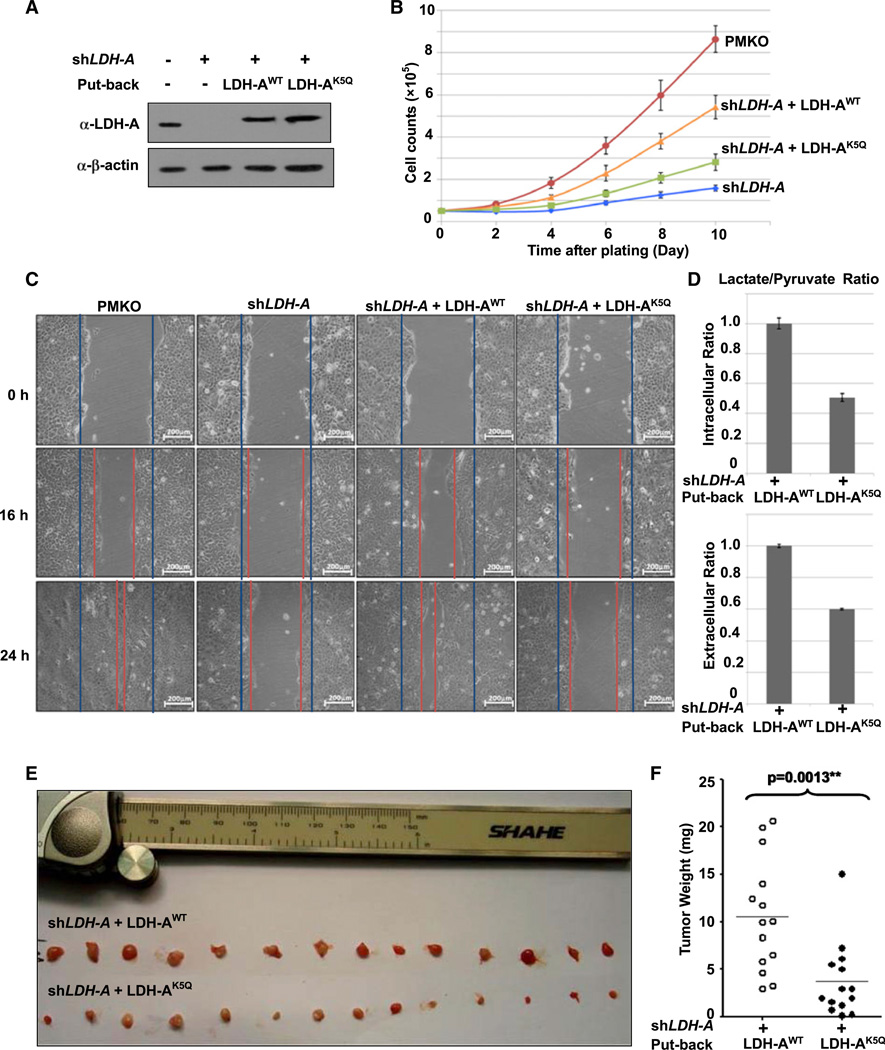
Elevated LDH-A protein levels are frequently seen in different types of tumors (Goldman et al., 1964). LDH-A is essential for cancer cell growth in vitro and in vivo (Fantin et al., 2006; Xie et al., 2009). We therefore investigated the effect of K5 acetylation of LDH-A on cell proliferation and migration. We knocked down endogenous LDH-A in the BxPC-3 pancreatic cancer cell line by shRNA and re-expressed shRNA-resistant wild-type and K5Q mutant LDH-A to a level similar to endogenous LDH-A (Figure 5A). Consistent with a previous report (Fantin et al., 2006), knocking down LDH-A caused a significant decrease of BxPC-3 cell proliferation that was substantially rescued by the re-expression of the wild-type LDH-A (Figure 5B). Notably, the LDH-AK5Q mutant was much less effective than the wild-type LDH-A in restoring LDH-A—knocking down cell proliferation. Similar effects were observed in 293 cells (Figure S5A). These results demonstrate that acetylation at Lys 5, which reduces the activity of LDH-A, impairs the ability of LDH-A in supporting BxPC-3 pancreatic cancer cell proliferation.
Figure 5. Acetylation Mimetic LDH-AK5Q Mutant Has Reduced Ability to Support Cell Proliferation and Cell Migration.
(A) Generation of LDH-A-expressing BxPC-3 stable cell lines. BxPC-3 cells stably knockdown LDH-A and re-express the shRNA-resistant wild-type or K5Q mutant were established. LDH-A knockdown efficiency and re-expression were determined by western blotting.
(B) LDH-AK5Q is compromised to support cell proliferation. LDH-AWT or LDH-AK5Q cells were seeded in each well. Cell numbers were counted every 48 hr. Error bars represent cell numbers ± SD for triplicate experiments.
(C) LDH-AK5Q mutant is compromised to support cell migration. BxPC-3 cells as described in panel A were analyzed for migration by a wound-healing assay. Scale bars are 200 mm.
(D) LDH-AK5Q decreases intracellular and extracellular lactate/pyruvate ratio. LDH-AWT or LDH-AK5Qcells were seeded in each well. Intracellular or extracellular pyruvate and lactate production were measured according to manufacturer’s protocol (BioVision). Error bars represent ± SD for triplicate experiments.
(E and F) LDH-AK5Q is defective in supporting tumor growth in vivo. Xenograft was performed using the BxPC-3 stable cell lines with LDH-A knockdown and re-expression of shRNA-resistant wild-type or K5Q mutant LDH-A as indicated. Seven weeks later, mice were sacrificed and tumor weight was measured. The p value was calculated by paired t test. See also Figure S5.
We then investigated the effect of LDH-AK5Q mutant on cell migration. Knockdown of LDH-A decreased cell migration in BxPC-3 (Figure 5C), 293, and 293T cells (Figures S5B and S5C), as determined by the wound-healing assay. Re-expression of wild-type, but not the K5Q mutant LDH-A restored cell migration, indicated that the acetylation at lysine-5 of LDH-A inhibits tumor cell migration.
LDH catalyzes the reversible conversion of pyruvate to lactate with LDH-A and LDH-B kinetically favoring the forward and the backward reactions, respectively (Ross et al., 2010). To confirm that the impaired ability of LDH-A K5Q mutant in supporting BxPC-3 cell proliferation and migration is due to its reduced catalytic activity, we measured pyruvate and lactate concentration in LDH-A knocking down cells that were re-introduced with either wild-type or K5Q mutant LDH-A. We found that the ratio of lactate to pyruvate was decreased by nearly one-half that of both intracellular (upper panel) and extracellular (low panel) levels in cells expressing K5Q mutant compared to cells expressing the wild-type LDH-A (Figure 5D). These results suggest LDH-A acetylation plays an important role in regulating the conversion of pyruvate to lactate.
It has been reported that lactate could drive cell migration (Bonuccelli et al., 2010; Végran et al., 2011). Therefore, we also determined the effect of lactate on migration in BxPC-3 cells. Consistently, we found that lactate promoted BxPC-3 cell migration (Figure S5D). These data indicate that K5 acetylation of LDH-A decreases lactate production, thereby restraining BxPC-3 pancreatic cancer cell migration.
To address the biologic significance of K5 acetylation in tumor growth, we performed xenograft experiments using the BxPC-3 stable cell lines with LDH-A knockdown and re-expression of shRNA-resistant wild-type or K5Q mutant LDH-A. As shown in Figures 5E and 5F, the K5Q mutant-expressing BxPC-3 cells displayed tumor growth significantly slower than the wild-type LDH-A-expressing cells. Taken together, these data indicate that LDH-A K5 acetylation impairs its function in catalyzing pyruvate to lactate conversion, and then inhibits cell proliferation and tumor growth.
K5 Acetylation of LDH-A Is Downregulated in Pancreatic Cancer
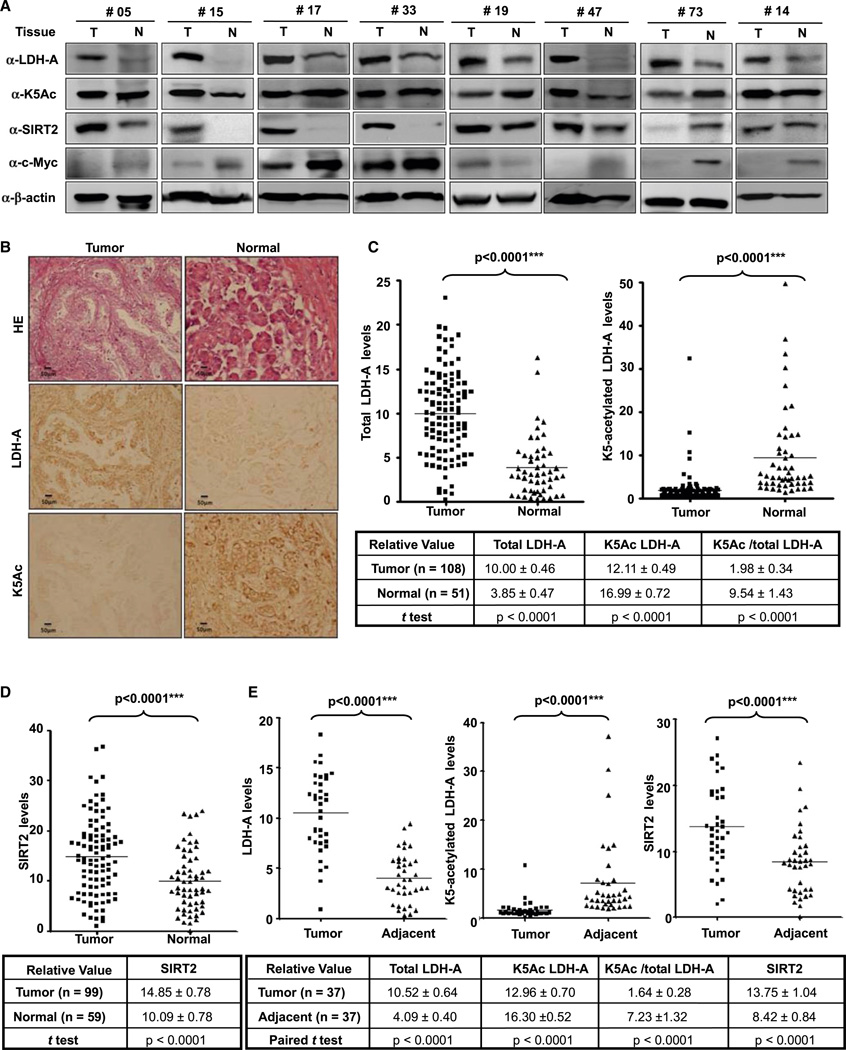
Pancreatic ductal adenocarcinoma cancer (PDAC) is the fourth leading cause of cancer death, with less than 5% 5 year survival after diagnosis. Pharmacologic inhibition of LDH-A has been reported to suppress the progression of pancreatic tumors in a xenograft model (Le et al., 2010). The finding that acetyl-mimetic substitution at lysine-5 impairs the ability of LDH-A to support BxPC-3 pancreatic cancer cell proliferation and tumor growth prompted us to examine both the K5 acetylation and total LDH-A protein in human cancers. We collected a total of 127 primary human pancreatic cancer samples, including 65 pairs that had surrounding normal pancreatic ducts tissues. We first carried out a direct immunoblotting analysis of a panel of 19 pairs of primary pancreatic tumors (T) and their adjacent normal tissues (N), for which we were able to obtain sufficient amounts of proteins. This analysis revealed that, when compared to normal pancreatic tissues, eight pairs showed a significant increase of the steady-state levels of total LDH-A protein without a corresponding increase of K5 acetylation (Figure 6A). Therefore, these eight pairs of tumor samples had a decreased ratio of K5-acetylated versus total LDH-A proteins. Quantification of six pairs (two pairs exhibiting levels of LDH-A in the normal tissues too low to be reliably quantified) confirmed that both the increase of total LDH-A (p < 0.0001) and the decrease in the ratio of K5-acetylated LDH-A versus total LDH-A proteins (p = 0.0031) in tumor cells are statistically significant (Figure S6A). Of the remaining 11 pairs, the total LDH-A protein was increased in four pairs, unchanged in four pairs, and decreased in three pairs in tumor tissues when compared to the adjacent normal tissues (Figure S6B). The ratio of K5-acetylated versus total LDH-A was not significantly decreased in these 11 pairs.
Figure 6. K5-Acetylation of LDH-A Is Downregulated in Pancreatic Cancer.
(A) Total LDH-A and SIRT2 protein are increased and K5-acetylated LDH-A decreased in pancreatic cancer tissues compared to adjacent tissues. The levels of LDH-A protein, K5 acetylation, SIRT2, and c-Myc in 19 pairs of pancreatic cancer and adjacent normal tissues were analyzed by western blotting. Eight pairs that exhibited clear inverse correlation between K5-acetylated and total LDH-A and positive correlation between SIRT2 and total LDH-A are shown. See also Figure S6A for the complete western blotting of the other 11 pairs.
(B and C) Immunohistochemical stainings of K5-acetylated and total LDH-A proteins in tumor and adjacent normal tissues. One example is shown in (B) and the statistical analysis of all samples is shown in (C). Scale bars are 50 µm. The intensities of the total (left panel) and K5-acetylated (right panel) LDH-A proteins were quantified using the Motic Images Advanced software, followed by statistical analysis. A total of 108 pancreatic cancer tissues and 51 adjacent normal pancreatic tissues were analyzed. The mean value of multiple samples and standard deviation are presented.
(D) Immunohistochemical staining of SIRT2 proteins in tumor and adjacent normal pancreatic cancer tissues. The statistical analysis of 99 tumor and 59 normal samples is shown. The intensities of SIRT2 proteins were quantified using the Motic Images Advanced software, followed by statistical analysis. The mean value of multiple samples and standard deviation is presented.
(E) LDH-A protein levels show negative correlation with K5 acetylation, and positive correlation with SIRT2 protein in pancreatic tumors. Among the 39 paired pancreatic cancer tissues that had been examined for all three signals (LDH-A, K5Ac, and SIRT2), 37 cases showed high LDH-A protein levels in tumors compared with adjacent tissues. These tumors also exhibited increased SIRT2 and decreased acetylation at K5. See also Figure S6.
C-Myc has been implicated in transcription regulation of many metabolic genes, including LDH-A (Shim et al., 1997). We also examined c-Myc protein levels in these 19 pairs of pancreatic tissues. However, we did not find an increase of c-Myc in pancreatic tumor tissues or a positive correlation between c-Myc and LDH-A protein levels (Figures 6A and S6B). Therefore, the reduced LDH-A K5 acetylation correlates with the increased LDH-A protein levels in the pancreatic tumors.
To substantiate the finding that K5-aetylated LDH-A is significantly decreased in some pancreatic tumors, we explored the feasibility of determining the level of both total and K5-acetylated LDH-A by immunohistochemistry in paraffin-embedded tissues to expand our study. The anti-acetyl-LDH-A(K5) antibody was characterized by its suitability for immunohistochemistry. We found that this antibody could detect strong signals that were specifically blocked by the acetyl-K5 antigen peptide in paraffin-embedded tissues (Figure S6C). Taking the advantage of this reagent, we then performed immunohistochemistry in 108 pancreatic cancer samples, including 46 samples that had the adjacent normal pancreatic ducts tissues. In most samples, we observed that the levels of total LDH-A were higher and the levels of relative K5-acetylated LDH-A were lower in the tumor tissues than in the adjacent normal tissues (Figure 6B). Statistical analyses of quantified images indicated that the differences between tumor and normal tissues in total LDH-A protein levels (p < 0.0001), in K5-acetylated LDH-A (p < 0.0001), and in the ratio of K5-acetylated LDH-A versus total LDH-A proteins (p < 0.0001) are all highly significant, comparing either the 108 tumor samples to the 51 normal pancreatic ducts samples (Figure 6C), or the 46 tumor samples with their adjacent normal tissues (Figure S6D). We also found that SIRT2 expression was increased in pancreatic tumor tissues compared to adjacent normal tissues (Figures 6A, 6D, and S6E).
Although more than 100 case tumors were collected, most pancreatic tumors are very small, and the number of paired paraffin sections with both tumor and adjacent on the same slide is hence limited. We determined the levels of LDH-A, K5-acetylated LDH-A, and SIRT2 in only 39 paired tissues. Among these pairs, high LDH-A protein level is found in 37 pairs of tumor compared with adjacent tissue. These tumors also exhibited increased SIRT2 and decreased acetylation at K5 as shown in Figure 6E. The tumor sample analyses demonstrate that LDH-A protein levels have a negative correlation with K5 acetylation and a positive correlation with SIRT2 levels in pancreatic tumors. These data also indicate that LDH-A and K5 acetylation may be potential biomarkers for pancreatic tumor.
The development of pancreatic cancer can be divided into five stages according to their location, size, and metastatic features: stage 0 (carcinoma in situ found in the lining of the pancreas), stage I (found only in pancreas with size smaller [IA] or larger [IB] than 2 cm), stage II (spread to nearby tissue, either including [IIB] or excluding [IIA] the lymph nodes), stage III (spread to major blood vessels near the pancreas), and stage IV (spread to distant organs). To determine whether LDH-A K5-acetylation level is related to the pancreatic tumor progression, we analyzed the levels of K5-acetylated as well as total LDH-A in the panel of 108 pancreatic tumors according to their stages. LDH-A protein level was significantly increased in all cancer stages when compared to normal tissues (Figure S6F, left panel), but no significant difference was detected between different stages (Figure S6G). The levels of K5-acetylated LDH-A were decreased significantly in all cancer stages when compared to normal tissues (Figure S6F, right panel), and there appeared to be a progressive decrease in the levels of K5-acetylated LDH-A from stage IA to stage IB (p = 0.009) and then to stage IIA (p = 0.0068 versus IA, Figure S6H). There was no significant difference in the levels of K5-acetylated LDH-A among stages IIA, IIB, III, and IV. Taken together, these data suggest a possible role of K5 acetylation contributing to pancreatic cancer initiation, but not progression to the advanced stages.
DISCUSSION
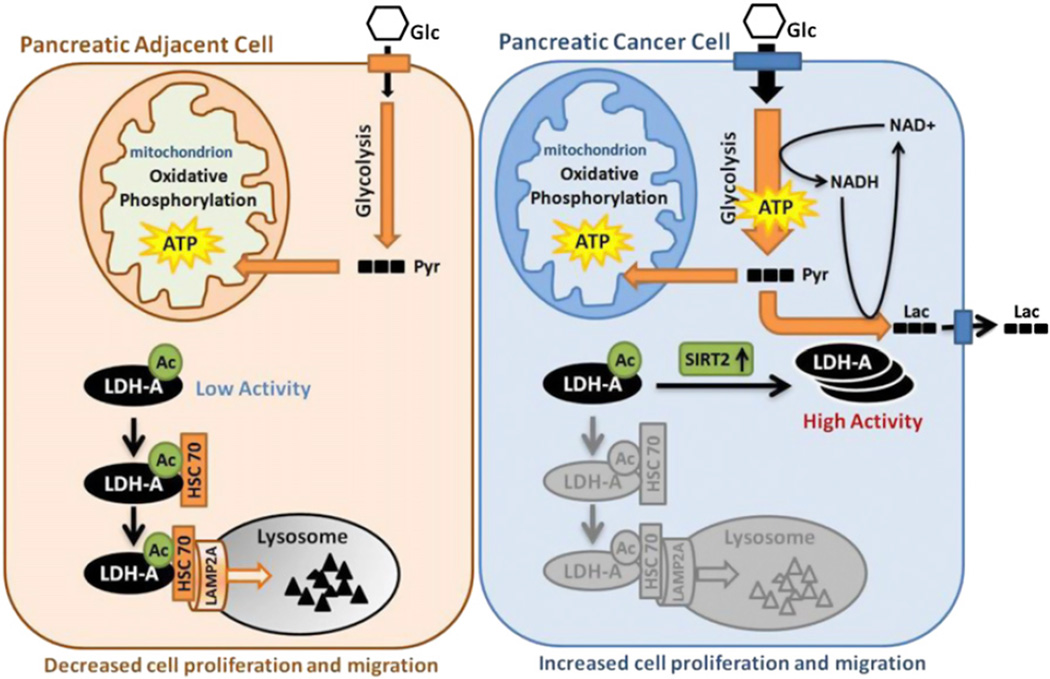
Reprogramming of energy metabolism, including elevated glycolysis, is a hallmark of cancer (Hanahan and Weinberg, 2011). To support rapid cell growth, glucose uptake and metabolic intermediates for macromolecule biosynthesis are dramatically increased in cancer cells. In particular, glycolysis is highly elevated. Among the glycolytic enzymes, LDH is unique because it is essential to maintain high glycolysis rate by regenerating NAD+ required in early steps in glycolysis (Bui and Thompson, 2006). Moreover, LDH channels pyruvate to lactate instead of converting it to acetyl-CoA for oxidative phosphorylation, a commonly observed phenomenon in many tumor cells. In this study, we uncovered a mechanism of LDH-A regulation that contributes to its increased protein level and activity to meet the elevated lactate production in tumor cells (Figure 7). We demonstrate that acetylation at K5 inhibits LDH-A enzyme activity and promotes its lysosomal degradation via CMA. In pancreatic cancer tissues, SIRT2 deacetylates LDH-A and increases its activity and protein level, thereby accelerating glycolysis and lactate production, leading to increased cell proliferation and migration.
Figure 7. Working Model.
Acetylation at K5 inhibits LDH-A enzyme activity and promotes its lysosomal degradation via CMA. In pancreatic cancer tissues, SIRT2 deacetylates LDH-A and increases its activity and protein level, thereby accelerating glycolysis and lactate production, leading to increased cell proliferation and migration. Glc, glucose; Pyr, pyruvate; Lac, lactate; Ac, acetylation.
LDH-A upregulation is commonly observed in cancers. This is in part due to transcriptional activation by the increased Myc and HIF in cancers. In this study, we report another mechanism in regulation of LDH-A protein levels. Acetylation plays an important role in posttranslational regulation of LDH-A by two mechanisms. First, acetylation directly inhibits LDH-A enzymatic activity. Second, acetylation stimulates CMA-mediated degradation of LDH-A. Notably, the relative acetylation of LDH-A is reduced in pancreatic cancer. We propose that the decreased LDH-A acetylation in cancer cells may contribute to the elevated LDH-A protein levels and activity as well as tumorigenesis (Figure 7).
A key step in CMA regulation is the interaction between chaperone HSC70 and target proteins. It has been reported that posttranslation modifications can regulate this process (Cuervo, 2010). For LDH-A, acetylation enhances the interaction between LDH-A and HSC70 (Figure 7). We show that HSC70 selectively interacts with acetylated proteins and thereby preferentially promotes lysosome-dependent degradation of the acetylated LDH-A. The three-dimensional structure of LDH indicates that lysine 5 is located in the N-terminal alpha-helix region of LDH-A, which is structurally separated from the catalytic domain (Read et al., 2001). Therefore, the K5-containing helix can be available for interaction with other proteins. Chaperone normally interacts with unfolded proteins that often have an exposed hydrophobic surface. It is conceivable that lysine acetylation increases surface hydrophobicity of the K5 helix in LDH-A and therefore promotes its interaction with the HSC70 chaperone. Further structural studies will be needed to obtain a precise understanding of how HSC70 recognizes acetylated target proteins.
Fantin and colleagues reported that LDH-A knockdown could inhibit tumor cell proliferation, especially under hypoxia (Fantin et al., 2006). A unique feature of LDH-A is that it acts at the end of the glycolytic pathway and catalyzes pyruvate to produce lactate, which is often accumulated in cancer cells (Figure 7). Many studies have shown that lactate can condition the microenvironment, which promotes interaction between cancer cells and stromal cells, eventually resulting in cancer cell invasion. Indeed, the ratio of lactate to pyruvate is significantly decreased in the acetylation mimetic K5Q mutant-expressing cells. Moreover, K5Q mutant is compromised in its ability to support proliferation and migration of BxPC-3 cells, most likely due to the decreased LDH-A activity. This may potentially explain why cancer cells have reduced LDH-A acetylation and increased LDH-A protein levels.
We observed that LDH-A expression positively correlates with SIRT2 expression in pancreatic cancer tissues, suggesting that SIRT2 may have oncogenic function in pancreatic cancer. However, SIRT2 has been reported as a tumor suppressor gene in a knockout mouse model (Kim et al., 2011). Notably, SIRT1 has been also suggested to act as both tumor promoter and suppressor in a context-dependent manner. Therefore, it is possible that SIRT2 may promote tumor growth under one circumstance, such as in human pancreatic cancer, and suppress tumor growth under another circumstance, such as hepatocellular carcinoma in Sirt2 knockout mice. A noticeable difference in these two systems is that SIRT2 expression is increased at the initial stage of pancreatic cancer while the mouse model has a complete deletion even before tumor development. Therefore, the functions of both SIRT1 and SIRT2 in cancer development may be context-dependent.
Previous studies have indicated an important role of LDH-A in tumor initiation and progression (Koukourakis et al., 2006; Le et al., 2010). LDH-A overexpression in pancreatic β cells led to increased mitochondrial membrane potential in many carcinomas (Ainscow et al., 2000; Chen, 1988). We showed that LDH-A is significantly increased in pancreatic cancer tissues compared to adjacent normal tissues. Consistently, LDH-A K5 acetylation was significantly decreased in pancreatic cancer tissues but not further increased during late stage tumor progression, indicating that LDH-A acetylation at K5 may play a role in pancreatic cancer initiation. Our study indicates an important mechanism of LDH-A regulation by acetylation and LDH-A K5 acetylation as a potential pancreatic cancer initiation marker.
EXPERIMENTAL PROCEDURES
LDH-A Enzyme Assay
Flag-LDH-A was ectopically expressed, immunoprecipitated, and eluted using 250 µg/ml of Flag peptide. The eluent was added to a reaction buffer containing 0.2M Tris-HCl (pH 7.3), 30 mM pyruvate, and 6.6 mM NADH. The change in absorbance (340 nm) resulting from NADH oxidation was measured using a F-4600 fluorescence spectrophotometer (HITACHI).
Genetically Encoding Nε-Acetyllysine in Recombinant Proteins
To generate a homogenously K5-acetylated LDH-A construct, we used a three-plasmid system as described (Neumann et al., 2008, 2009). This system allows for the site-specific incorporation of N-acetyllysine by way of a Methanosarcina barkeri acetyl-lysyl-tRNA synthetase/tRNACUA pair that responds to the amber codon. We cloned wild-type LDH-A into pTEV-8 (pET-21b as backboned with TEV cleavage site) producing a C-terminal His6-tagged construct, and incorporated an amber codon at lysine 5 (AAG to TAG by site-directed mutagenesis). Cells were induced at an OD600 of 0.6 with 0.5 mM IPTG. The amber construct was overexpressed in LB with spectinomycin (50 µg/ml), kanamycin (50 µg/ml), and ampicillin (150 µg/ml), in addition to 2 mM N-acetyllysine (Sigma-Aldrich) and 20 mM nicotinamide at the time of induction. Both LDH-A and K5-acetylated LDH-A protein are purified for enzyme activity analysis.
Pancreatic Cancer Model by Xenograft
BxPC-3 stable cell lines with LDH-A knockdown and re-expressed shRNA resistant wild-type or K5Q mutant LDH-A were prepared; 7.5 × 106 cells in PBS were subcutaneously injected into each of 14 nude mice, purchased from SLAC. Shanghai. Every mouse was injected LDH-AWT cells on left side and LDH-AK5Q on right side. Seven weeks later, all mice were sacrificed and tumors were harvested, followed by photography and weighing. The animal protocols were approved by the Animal Welfare Committee of Shanghai Medical College, Fudan University.
Pancreatic Tumor Samples and Immunohistochemistry
Pancreatic tumor samples were acquired from Affiliated Shanghai Tenth People’s Hospital of Tongji University. A physician obtained informed consent from the patients. The procedures related to human subjects were approved by Ethic Committee of the Institutes of Biomedical Sciences (IBS), Fudan University. Immunohistochemistry (IHC) was performed as previously described (Lei et al., 2006). To quantify the IHC result of positive staining, the tissue areas of five ducts (173 µm2) in each sample were microscopically examined and analyzed by an experienced pathologist. Images were captured using a charge-coupled device camera and analyzed using Motic Images Advanced software (version 3.2, Motic China Group). Average of staining score was calculated by dividing the positive areas with total areas. Data obtained were expressed as mean values ± SD. Differences were considered significant if the p value was less than 0.05.
Supplementary Material
Significance.
This study uncovers a critical role and the mechanism of acetylation in the regulation of lactate dehydrogenase A (LDH-A), which is elevated in cancer cells. Lysine-5 acetylation inhibits LDH-A by two mechanisms: decreasing enzymatic activity and increasing degradation by a chaperone-mediated autophagy. Moreover, LDH-A lysine-5 acetylation inversely correlates with pancreatic cancer initiation. Therefore, acetylation plays an important role in the regulation of cell growth and cancer metabolism.
ACKNOWLEDGMENTS
We thank the members of the Fudan MCB laboratory for discussions throughout this study. We also thank Dr. Liming Wei for IEF assay. This work was supported by the Chinese Ministry of Sciences and Technology 973 (grant nos. 2009CB918401, 2011CB910600, and NCET-09-0315), the NSFC (grant nos. 31271454 and 81225016), NSFC-NIH (grant no. 81110313), the 100 Talents Program of Shanghai Health, the Scholar of ‘‘Dawn’’ Program of Shanghai Education Commission, Shanghai Outstanding Academic Leader, and the Shanghai Key basic research program (12JC1401100) to Q.Y.L.; NIH grants (to Y.X. and K.L.G.); and Fudan University Medical School Graduate Student Ming Dao Project funds (to D.Z.). This work was also supported by the Chinese Ministry of Education 985 Program. This work is dedicated to the memory of Zhen Yu, who prepared the K5 acetylation antibody. Y.-H.X. and Q.-Y.L. are members of the Chinese Hippo Consortium.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.ccr.2013.02.005.
REFERENCES
- Ainscow EK, Zhao C, Rutter GA. Acute overexpression of lactate dehydrogenase-A perturbs beta-cell mitochondrial metabolism and insulin secretion. Diabetes. 2000;49:1149–1155. doi: 10.2337/diabetes.49.7.1149. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Bonuccelli G, Tsirigos A, Whitaker-Menezes D, Pavlides S, Pestell RG, Chiavarina B, Frank PG, Flomenberg N, Howell A, Martinez-Outschoorn UE, et al. Ketones and lactate ‘‘fuel’’ tumor growth and metastasis: Evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle. 2010;9:3506–3514. doi: 10.4161/cc.9.17.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T, Thompson CB. Cancer’s sweet tooth. Cancer Cell. 2006;9:419–420. doi: 10.1016/j.ccr.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Chen LB. Mitochondrial membrane potential in living cells. Annu. Rev. Cell Biol. 1988;4:155–181. doi: 10.1146/annurev.cb.04.110188.001103. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol. Metab. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Furumai R, Komatsu Y, Nishino N, Khochbin S, Yoshida M, Horinouchi S. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc. Natl. Acad. Sci. USA. 2001;98:87–92. doi: 10.1073/pnas.011405598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Kaplan NO, Hall TC. Lactic Dehydrogenase in Human Neoplastic Tissues. Cancer Res. 1964;24:389–399. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20:487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Sivridis E, Bougioukas G, Didilis V, Gatter KC, Harris AL Tumour and Angiogenesis Research Group. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer. 2003;89:877–885. doi: 10.1038/sj.bjc.6601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL Tumour Angiogenesis Research Group. Lactate dehydrogenase 5 expression in operable colorectal cancer: strong association with survival and activated vascular endothelial growth factor pathway—a report of the Tumour Angiogenesis Research Group. J. Clin. Oncol. 2006;24:4301–4308. doi: 10.1200/JCO.2006.05.9501. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Kontomanolis E, Giatromanolaki A, Sivridis E, Liberis V. Serum and tissue LDH levels in patients with breast/gynaecological cancer and benign diseases. Gynecol. Obstet. Invest. 2009;67:162–168. doi: 10.1159/000183250. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Lewis BC, Shim H, Li Q, Wu CS, Lee LA, Maity A, Dang CV. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol. Cell. Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read JA, Winter VJ, Eszes CM, Sessions RB, Brady RL. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins. 2001;43:175–185. doi: 10.1002/1097-0134(20010501)43:2<175::aid-prot1029>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Ross JM, Oberg J, Brené S, Coppotelli G, Terzioglu M, Pernold K, Goiny M, Sitnikov R, Kehr J, Trifunovic A, et al. High brain lactate is a hallmark of aging and caused by a shift in the lactate dehydrogenase A/B ratio. Proc. Natl. Acad. Sci. USA. 2010;107:20087–20092. doi: 10.1073/pnas.1008189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Végran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011;71:2550–2560. doi: 10.1158/0008-5472.CAN-10-2828. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wing SS, Chiang HL, Goldberg AL, Dice JF. Proteins containing peptide sequences related to Lys-Phe-Glu-Arg-Gln are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem. J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Valera VA, Merino MJ, Amato AM, Signoretti S, Linehan WM, Sukhatme VP, Seth P. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol. Cancer Ther. 2009;8:626–635. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.