Exome sequencing of high-grade pediatric gliomas, including supratentorial glioblastomas (GBM) and diffuse intrinsic pontine gliomas (DIPG) identified missense mutations Lys27Met (K27M) and Gly34Arg/Val (G34R/V) in genes encoding histone H3.3 (H3F3A) and H3.1 (HIST3H1B).1-3 The high occurrence and unique age distribution of histone H3 mutant tumors suggest that these mutations may block glial cell differentiation, perhaps in a mechanism akin to leukemic fusion proteins that “drive” cellular transformation via blockage of hematopoiesis.
H3K27 is methylated by the Polycomb Repressive Complex 2 (PRC2) for X-chromosome inactivation and gene silencing. Mutation or aberrant regulation of genes encoding chromatin regulators are highly tumor-type specific, as exemplified by the PRC2 catalytic subunit EZH2, which is overexpressed in a number of epithelial malignancies and lost or mutated in subsets of leukemia and lymphomas. These observations suggest that misregulation of H3K27 modification is common in tumorigenesis, but occurs only in very specific cell lineage and developmental contexts.
Recently, we and others observed nearly undetectable levels of H3K27me2/3 levels in gliomas encoding the K27M histone H3.4-6 We found that H3 K27M histone protein contributed to a minority of total H3 protein (3–17%) in these tumors. The invariant and heterozygous nature of the mutation (only 1 mutant H3 allele out of 32 encoding H3 per diploid cell) suggests that the K27M H3 histone promotes gliomagenesis through a powerful dominant-negative mechanism. In exploring the biochemical process, we found that H3 K27M peptides are potent inhibitors of PRC2 activity.4 Furthermore, H3 K27M peptides containing a photo-reactive methionine analog specifically crosslinked the SET-domain active site of EZH2, indicating that K27M likely competes with substrate binding and turnover.
We demonstrated that H3 K27M transgenes that contribute a few percent of total H3 protein were sufficient to severely reduce H3K27me3 amounts in cell culture. After surveying all possible amino acid substitutions, only K27I transgenes exhibited similar, albeit slightly less, inhibitory activity on PRC2 in vitro and on H3K27me3 levels in vivo. This biochemical observation may prove prescient; a single adenine-thymine transversion in the K27 “AAA” codon found in several H3 genes will produce the “ATA” codon encoding Isoleucine. While not yet reported, the existence of an H3 K27I mutation in pediatric gliomas or other cancers is conceivable and should be investigated. Norleucine, a leucine isomer, was the most potent inhibitor of PRC2 activity in vitro. The extended, unbranched aliphatic side chain of norleucine likely makes superior interactions with the aromatic and hydrophobic residues that line the SET domain active site.
The catalytic core of SET domains is highly conserved, and we explored the possibility that other histone methyltransferases (HMTs) might be similarly sensitive to K-to-M substitutions at their cognate peptidyl–lysine substrates. We found that K9M and K36M H3 transgenes in cultured cells decreased overall amounts of H3K9me3 and H3K36me3, respectively. Interestingly, H3 K4M transgenes reduced overall H3K4me2/3 amounts only slightly. This poor inhibitor activity may be a consequence of atypical SET domain proteins, or possibly the reported inhibition of FAD-dependent amine oxidases LSD1/2 by N-terminal H3 peptides containing K4M in vitro.7 Notably, Jumonji domain-containing histone demethylases, such as UTX, that target K9, K27, and K36 methylation lack an active site configuration of hydrophobic and aromatic co-enzymes and residues, and are therefore unlikely inhibited by K-to-M substitutions. Similarly, HMTs that lack SET domains, such as DOT1L, are not sensitive to K-to-M substitutions at their substrate site (i.e., H3 K79M).
How might K-to-M transgene-mediated inhibition occur in vivo? The estimated nuclear concentration of a unique histone H3 protein that makes up 3–5% of total H3 approaches 10µM. This histone concentration is similar to the IC50 we measured for K-to-M inhibition of SET domain proteins in vitro (1–6 µM). Additionally, specific deposition pathways exist to enrich histones H3.1 and H3.3 at distinct genomic locations.8 Yet, DIPG samples containing the K27M mutation in either H3.1 or H3.3 exhibit the same decrease in H3K27me3, indicating that nuclear concentration, rather than genomic location, is important for PRC2 inhibition.
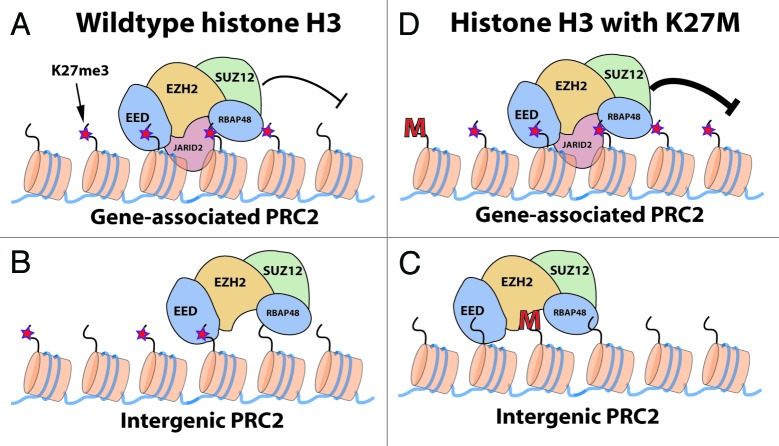
The notable lack of PRC2 subunit gene mutations detected in pediatric GBM or DIPG suggests that the H3 K27M mutation may promote a nuanced adjustment, rather than a en masse de-repression of PRC2 silenced genes (Fig. 1). Indeed, Chan et al. recently observed a marked decrease in the overall number of H3K27me3 peaks by ChIP sequencing in glioma cells containing H3 K27M.5 Surprisingly, the remaining H3K27me3 peaks in these cells spanned larger chromatin domains, had increased H3K27me3 density, and also correlated with enhanced gene silencing. How inhibition of some PRC2 complexes leads to enhanced PRC2-mediated gene repression remains a key mechanistic question.
Figure 1. At genes PRC2 molecules are recruited via Jarid2 and sequence-specific factors for H3K27me3-mediated silencing (A). Intergenic PRC2 may rely on positive feedback loops established through interaction between the H3K27me3 and the EED subunit (B). The intergenic PRC2-K27me3 positive feedback loop is possibly disrupted in glioma cells containing the H3 K27M protein (C). Contrastingly, genic H3K27me3 levels increase in K27M-containing cells, leading to enhanced gene silencing (D).
Additional questions remain, including the cell-of-origin of tumors containing the mutant histones, as well as how these mutations affect pathways that contribute to tumor initiation or progression. The high incidence and exclusivity of histone H3 mutations to pediatric gliomas highlights the importance of a specific cell-of-origin that is exquisitely susceptible to these mutations, as well as potential cooperating mutations that could collectively drive gliomagenesis. Indeed, p53-null postnatal mice transduced with H3 K27M failed to develop gliomas, indicating a requirement for developmental context and/or additional mutations.4 Future studies that address these questions will ultimately lead to more effective treatments for these tumors.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26356
References
- 1.Schwartzentruber J, et al. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 2.Wu G, et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Nat Genet. 2012;44:251–3. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khuong-Quang DA, et al. Acta Neuropathol. 2012;124:439–47. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis PW, et al. Science. 2013;340:857–61. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan KM, et al. Genes Dev. 2013;27:985–90. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venneti S, et al. Brain Pathol. 2013 doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karytinos A, et al. J Biol Chem. 2009;284:17775–82. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg AD, et al. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]