Abstract
Our recently published work suggests that DNA helicases such as the Werner syndrome helicase (WRN) represent a novel class of proteins to target for anticancer therapy. Specifically, pharmacological inhibition of WRN helicase activity in human cells defective in the Fanconi anemia (FA) pathway of interstrand cross-link (ICL) repair are sensitized to the DNA cross-linking agent and chemotherapy drug mitomycin C (MMC) by the WRN helicase inhibitor NSC 617145.1 The mechanistic basis for the synergistic interaction between NSC 617145 and MMC is discussed in this paper and extrapolated to potential implications for genetic or chemically induced synthetic lethality provoked by cellular exposure to the WRN helicase inhibitor under the context of relevant DNA repair deficiencies associated with cancers or induced by small-molecule inhibitors. Experimental data are presented showing that small-molecule inhibition of WRN helicase elevates sensitivity to MMC-induced stress in human cells that are deficient in both FANCD2 and DNA protein kinase catalytic subunit (DNA-PKcs). These findings suggest a model in which drug-mediated inhibition of WRN helicase activity exacerbates the deleterious effects of MMC-induced DNA damage when both the FA and NHEJ pathways are defective. We conclude with a perspective for the FA pathway and synthetic lethality and implications for DNA repair helicase inhibitors that can be developed for anticancer strategies.
Keywords: DNA repair, Fanconi anemia, Werner syndrome, anticancer therapy, helicase, small molecule, synthetic lethality
Introduction
The emergence of DNA repair inhibitors as tools to understand DNA damage response pathways and improve anticancer therapy
Synthetic lethality was classically used to describe a genetic interaction in which deficiency in a pair of genes resulted in cell death. In recent years, chemically induced synthetic lethality has become mainstream with the discovery of poly(ADP ribose) polymerase (PARP) inhibitors that exploit the existing vulnerability of BRCA1- or BRCA2-deficient tumors that are defective in homologous recombination (HR) repair.2,3 Since then, the field has blossomed with a potentially broader scope for PARP inhibitors4 as well as newly discovered DNA repair inhibitors that exert synthetic lethal effects in distinct genetic backgrounds.5 Targeting tumors that have defects in DNA repair pathways with pharmacological inhibitors has attracted considerable interest.6 The effort to develop chemically induced synthetic lethal strategies for killing cancer cells has been championed with the increased awareness that tumors can become resistant to conventional DNA damaging therapies or radiation. Here, we provide a perspective that chemical DNA helicase inhibitors may be a useful class of drugs for not only understanding DNA damage response pathways but also for developing improved anticancer treatment strategies. We place particular emphasis on a newly described chemical inhibitor of the DNA unwinding (helicase) activity catalyzed by the Werner syndrome protein (WRN), because the RecQ helicase plays a prominent role in replication fork progression after DNA damage or fork arrest that might allow rapidly dividing cancerous cells to deal with replicative lesions.7 Moreover, WRN is considered a replication caretaker, enabling cells to elicit an appropriate checkpoint response,8 and plays an instrumental role in molecular mechanisms that suppress premature replicative senescence.9 WRN and other DNA helicases may represent good biomarkers or targets for chemotherapy.10 Tumors with a pre-existing DNA repair deficiency may be hypersensitive to a WRN helicase inhibitor, enabling scientists to study cross-talk between DNA damage response/repair pathways and target an Achilles’ heel of cancer by a novel approach.
Interference with cross-link repair by WRN helicase inhibitor
A small molecule (NSC 19630) from the NCI Diversity Set was discovered from an in vitro ATP-dependent DNA unwinding assay screen that inhibited WRN helicase activity in a potent and specific manner.11 NSC 19630 was demonstrated to impair proliferation of human endocervical carcinoma cells grown in culture and induce DNA damage and apoptosis in a WRN-dependent manner.11 Recently, we characterized a small molecule structurally related to the original WRN helicase inhibitor, designated NSC 617145, and demonstrated that pharmacological inhibition of WRN helicase activity by NSC 617145 sensitized cancer cells to a low (9.6 nM) concentration of the DNA cross-linking agent and chemotherapy drug mitomycin C (MMC), resulting in decreased cell proliferation and the accumulation of DNA damage and chromosomal abnormalities.1 This led us to ask if mutant cells from Fanconi anemia (FA) patients, which are defective in a major pathway of cross-link resistance,12 would be further sensitized to MMC when the cells were co-treated with the WRN helicase inhibitor. Indeed, we observed that human cells harboring autosomal recessive mutations in either the Fanconi anemia (FA) group A (FANCA) or group D (FANCD2) genes were hypersensitive to low concentrations of MMC when they were co-treated with the WRN helicase inhibitor NSC 617145. Under these conditions, NSC 617145-treated FA-D2 mutant cells displayed aberrant processing of DNA cross-links, resulting in ATM activation and accumulation of DNA–protein kinase catalytic subunit (PKcs) pS2056 foci, suggesting an increased number of double-strand break (DSB)s processed by error-prone nonhomologous end-joining (NHEJ). The effect of WRN helicase inhibitor NSC 617145 was WRN-dependent, because depletion of WRN protein by RNA interference (RNAi) negated the anti-proliferative and DNA damage-inducing effect of the compound, similar to our previous observation with the structurally related compound NSC 19630.11 In the absence of the WRN helicase inhibitor, depletion of WRN by RNAi did not exert a negative effect on MMC resistance in FA mutant cells, suggesting that WRN-independent pathway(s) confer residual resistance to the DNA cross-linker when the FA pathway is defective. Based on the collective experimental evidence, we concluded that small-molecule inhibition of WRN helicase activity rather than the absence of WRN protein altogether exacerbated MMC-induced DNA damage that accumulated in the absence of a functionally intact FA pathway.
Understanding the basis for the increased MMC-induced cytotoxicity that occurs when WRN helicase is inhibited by a small molecule will require further study. MMC forms interstrand cross-links (ICLs) in the minor groove, which causes minor widening in addition to intrastrand cross-links and mono-adducts.13 Given that MMC is considered to induce a relatively non-distorting cross-link, it is not likely to create a DNA substrate that is directly acted upon by nucleotide excision repair (NER). Rather, the cross-link is expected to be discovered by the cellular DNA repair machinery via a mechanism involving unwinding of duplex DNA that occurs during replication or possibly transcription. Because WRN, like several other RecQ helicases (BLM, RECQ1, RECQ4), is involved in the cellular DNA replication stress response,10 we will focus our discussion on the replication fork scenario for ICL repair (Fig. 1). Presumably, the processing of a DNA cross-link leads to a cross-link remnant attached to one strand, resulting in a DSB due to a broken replication fork (for a recent review of ICL repair, see ref. 14). The lagging strand then serves as a template for translesion synthesis (TLS) to create a duplex molecule that can be subsequently repaired by HR. Restoration of fork integrity and resumption of DNA synthesis is paramount to the preservation of genomic stability and maintenance of cellular homeostasis.
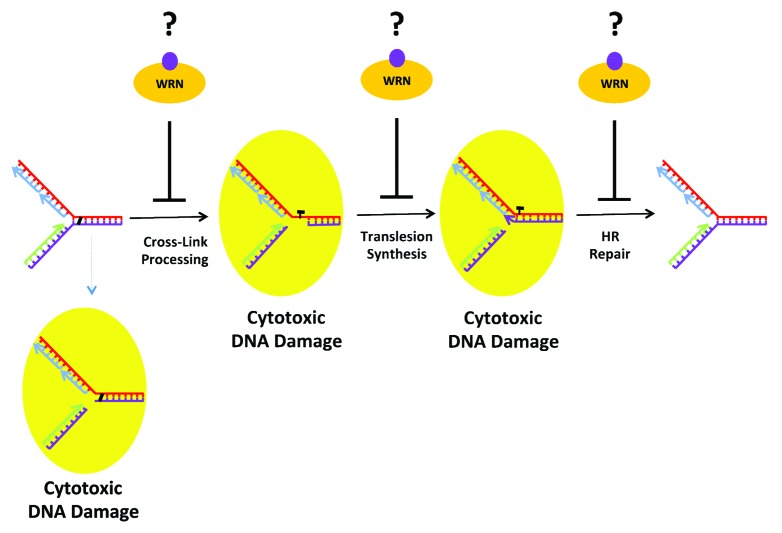
Figure 1. Potential sites of action where the WRN helicase inhibitor interferes with DNA cross-link repair during S phase, resulting in the accumulation of double-strand breaks and chromosomal instability. See text for details.
As depicted in Figure 1, there are several nexus where interference of a key step by a WRN helicase inhibitor can derail ICL repair during S phase and lead to the genesis of broken replication forks. One possibility is that the WRN helicase inhibitor interferes with DNA incisions on each side of the 2 covalently linked nucleotides, the so-called unhooking step, by blocking access or function of structure-specific incision endonucleases (XPF-ERCC1, MUS81-EME1, or SLX1) at the site of DNA damage. This may involve masking the scaffolding provided by SLX4, or preventing further processing of the incised cross-link by the nucleases FAN1 or SNM1A. Experimental studies using a reconstituted in vitro system provided evidence for a role of WRN helicase in ICL processing of a DNA substrate harboring a psoralen-ICL, a form of damage that locally constrains and distorts the DNA double helix.15 Moreover, results from cellular genetic rescue experiments suggested that WRN helps to process psoralen-ICL by its helicase activity.16 NSC 617145 may sensitize cancer cells to ICLs induced by MMC by a mechanism of action whereby WRN helicase inhibitor blocks initial cross-link processing; however, an effect of the small molecule on ICL unhooking remains to be shown.
WRN was previously shown to interact with several translesion DNA polymerases,17 suggesting a second site of action for the WRN helicase inhibitor. Interference with TLS mediated by an interaction of the WRN helicase-inhibitor drug complex with the damaged DNA molecule and associated histones or DNA metabolic proteins would be consistent with the demonstrated enriched association of WRN protein with chromatin when cells are treated with the WRN helicase inhibitor NSC 617145.1 Therefore, it would be of interest to determine what chromatin-associated factors are bound by the WRN-helicase inhibitor complex, and if WRN is sequestered at the site of a defectively processed DNA cross-link. It is generally believed that recruitment of TLS polymerases to sites of action is governed by protein interactions. We would predict that disruption of timely recruitment of DNA repair proteins to sites of ICLs occurs in a WRN-dependent manner, and that the trapped WRN helicase inhibitor-protein complex elicits a unique situation predisposing cells to elevated genomic instability.
Third, NSC 617145 bound to WRN may interfere with HR repair of the replication fork-associated DSBs. This possibility would be consistent with the elevated RAD51 foci in FANCD2 mutant cells exposed to low doses of MMC and NSC 617145, suggesting WRN helicase inhibition interferes with later steps of HR at ICL-induced DSBs. Indeed, the elevated RAD51 foci observed in FA-D2 cells co-treated with sub-toxic doses of MMC and NSC 617145 may reflect hyper-recombination triggered by a WRN helicase-inhibitor drug complex. As discussed nicely in a recent review by Kottemann and Smogorewska,14 competition for DSBs between protein factors implicated in HR vs. NHEJ may determine the fate of DNA repair and extent of chromosomal instability incurred by the lesion. Normally, the FA pathway plays a pivotal role in pathway choice; however, in the absence of an intact FA pathway, exposure to the WRN helicase inhibitor may conceal the DNA end from HR machinery, leading to illegitimate repair by NHEJ. By analogy, it has been suggested that the DNA end-binding factor 53BP1 exclude HR factors from their role in HR repair in BRCA1 mutant mammalian cells by preventing their access to the DNA break sites.18,19 Thus, drug-mediated inhibition of WRN helicase activity may serve to activate the toxic NHEJ pathway in the cellular context of FA pathway deficiency and DSBs incurred by MMC-induced DNA damage. If so, the paradigm would follow that of reported genetic interactions between the FA pathway and NHEJ in mammalian cells.20,21 Alternatively, defective resolution of HR intermediates by pharmacological inhibition of WRN helicase activity would lead to NHEJ activation and genomic instability, as observed experimentally by the accumulation of DNA-PKcs pS2056 foci and chromosomal aberrations in FA-D2 cells exposed to the synergistic concentrations of MMC and NSC 617145.
Previous evidence has implicated a role of WRN in TLS22 and HR.23,24 This coupled with our observations that NSC 617145 and MMC synergistically induce DNA damage and chromosomal instability, activate ATM, and cause accumulation of DNA-PKcs pS2056 foci suggest it is likely that inhibition of WRN helicase activity by NSC 617145 interferes with TLS or HR. NSC 617145-mediated WRN helicase inhibition may hamper HR by blocking the functions of FA proteins (PALB2, BRCA2, RAD51C) or other HR proteins. Understanding the precise molecular steps whereby the WRN helicase inhibitor induces cellular DNA damage will not only yield insight to the compensatory mechanisms involving WRN that cells use to cope with cross-links and other forms of DNA adducts, but also to the potential improvement of anticancer therapies, since MMC and other DNA cross-linking drugs are prominently used in the clinic to treat leukemia and other types of cancer.13
Results
Inhibition of WRN helicase elevates sensitivity to endogenous or MMC-induced stress in cells that are deficient in both the FA and NHEJ pathways
Interestingly, previous work demonstrated that depletion of NHEJ factors (Ku80 or DNA-PKcs) or inhibition of Ligase IV suppressed the toxic effects of MMC observed in mammalian cells that are defective in the FA pathway.20,21 It was reasoned that NHEJ is toxic in FA-deficient cells, and that the FA pathway serves to channel MMC-induced DNA damage into the HR pathway, which is characterized by far greater fidelity than error-prone NHEJ. The interaction of WRN with KU protein25 and results from cell-based plasmid end-joining assays23 had suggested a possible role of WRN in NHEJ. This might lead to the hypothesis that co-treatment of FA-deficient cells with MMC and NSC 617145 would suppress DNA damage accumulation and chromosomal instability if the WRN helicase inhibitor acts by blocking WRN’s function in NHEJ. However, this hypothesis may be oversimplified because of the complex relationships between DNA repair pathways and how they are regulated.26 For example, as mentioned above, a mouse study of DNA cross-link repair demonstrated that deletion of the DNA double-strand break responders 53BP1 or KU exacerbates genomic instability in cells lacking FANCD2.19 Another possible outcome of WRN helicase inhibition hinges on the possibility that WRN helicase is engaged in a pathway (e.g., alternative NHEJ27) besides error-prone NHEJ in certain cancers when cross-linked DNA damage builds up in FA-deficient cells. In that scenario, WRN helicase inhibition may lead to enhanced processing of MMC-induced DNA damage (DSBs) by NHEJ, resulting in genomic instability.
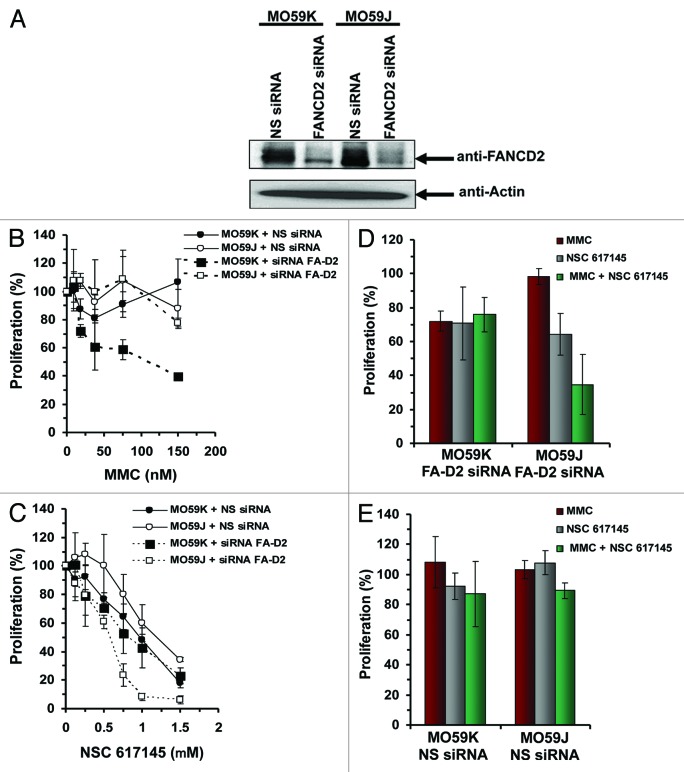
To explore these possibilities, we examined the effect of NSC 617145 and MMC co-treatment on FA-D2−/− cells when NHEJ was defective. For this, we utilized human glioblastoma MO59K (DNA-PKcs-proficient) and MO59J (DNA-PKcs-deficient) cells. Initially, we examined the effect of FANCD2 depletion on MMC sensitivity of DNA-PKcs-deficient and -proficient cells. FANCD2 expression in MO59K and MO59J cells was reduced by ≥90% compared with siRNA control (Fig. 2A). FANCD2 depletion in MO59K cells resulted in reduced proliferation upon MMC exposure, whereas FANCD2-depleted MO59J cells were MMC-resistant (Fig. 2B), consistent with previous results.20 There was no significant difference in MMC sensitivity of MO59K and MO59J cells transfected with NS-siRNA (Fig. 2B).
Figure 2. Effect of NSC 617145 exposure on FA-D2 mutant and NHEJ-deficient cells upon co-treatment with MMC. (A) Western blot analysis of FANCD2 knockdown in DNA-PKcs-proficient and -deficient cell lines. DNA-PKcs-proficient (MO59K) and DNA-PKcs-deficient (MO59J) cells were transfected with siRNA targeted specifically against FANCD2 (FANCD2 siRNA) or non-specific siRNA (NS siRNA) for 4 h. Cell lysates were then prepared and analyzed by immunoblotting with anti-FANCD2 antibody. As a control blot was reprobed with anti-actin antibody. The arrow indicates the position of the FANCD2 and actin bands. (B and C) DNA-PKcs-proficient (MO59K) and -deficient (MO59J) cells lines were transfected with FANCD2-specific siRNA (siRNA FA-D2) or non-specific siRNA (NS siRNA). Sensitivity of these cell lines to MMC (B) or NSC 617145 (C) was determined with WST-1 reagent. Percent proliferation was calculated.(D and E) DNA-PKcs-proficient (MO59K) and -deficient (MO59J) cells were transfected with FANCD2-specific siRNA (siRNA FA-D2) (D) or NS siRNA (E). Cells were treated with MMC (19 nM), NSC 617145 (0.5 μM), or both for 3 d. Cell proliferation was determined with WST-1 reagent. Cell proliferation data are mean of at least 2 independent experiments. Error bars represents standard deviations.
We then assessed the effect of pharmacological WRN helicase inhibition on proliferation of cells deficient in both DNA-PKcs and FANCD2. DNA-PKcs proficient (MO59K) cells transfected with NS-siRNA showed moderately greater sensitivity to NSC 617145 as compared with DNA-PKcs deficient (MO59J) cells transfected with NS-siRNA (Fig. 2C). This result suggested that when NHEJ is operational, WRN helicase inhibition causes greater genomic instability. Exposure of FANCD2-depleted, DNA-PKcs-deficient cells to NSC 617145 resulted in a dose-dependent decrease in proliferation, culminating in approximately 95% inhibition at 1 nM WRN inhibitor (Fig. 2C). This reduction in proliferation was significantly greater than that for FANCD2-depleted, DNA-PKcs-proficient cells exposed to 1 μM NSC 617145 (Fig. 2C), suggesting WRN helicase inhibition in cells deficient in both FA and NHEJ pathways further compromises their ability to proliferate. In the absence of exogenous stress, WRN helicase activity may help to protect FANCD2-deficient cells from error-prone NHEJ that is deleterious to genomic stability.
Cells deficient for DNA-PKcs and FANCD2 exposed to 19 nM MMC and NSC 617145 (0.5 μM) displayed significantly reduced proliferation compared with similarly treated DNA-PKcs-proficient cells depleted of FANCD2 (Fig. 2D). In NS-siRNA experiments in which FANCD2 status was unaffected, DNA-PKcs-proficient and -deficient cells exposed to MMC and NSC 617145 displayed a very similar level of proliferation (Fig. 2E). These results suggest that the combined deficiency in the FA pathway and NHEJ renders human cells even further sensitive to MMC-induced DNA damage when the WRN helicase is pharmacologically inhibited by a small molecule. We would suggest that under these conditions, when the FA pathway and NHEJ fail to operate efficiently, then the targeted inhibition of WRN helicase activity leaves cells further compromised in their ability to tolerate cytotoxic MMC-induced DNA damage.
Discussion
Implications for anticancer therapy targeting DNA repair with helicase inhibitors
Experimental data from our lab and others would suggest that WRN (1) as well as other DNA damage response factors may be candidates for chemically induced synthetic lethality in FA-deficient tumors28 and other cancers.5 However, depending on the molecular target for pharmacological inhibition, different results may be observed. For example, treatment of FA-deficient cells with the WRN helicase inhibitor sensitized them to low doses of MMC. In contrast, treatment of FA-deficient cells with a DNA-PKcs inhibitor rendered them resistant to MMC.20 These findings demonstrate that in the setting of a FA pathway deficiency, pharmacological inhibition of 2 DNA repair enzymes (a helicase implicated in recombinational repair and a normal replication stress response, or a kinase implicated in NHEJ) operates by distinctive mechanisms. In this study, we found that treatment of human cells deficient in both FANCD2 and DNA-PKcs with the WRN helicase inhibitor rendered them sensitive to MMC. This may be akin to the recent observation that cells from FANCD2−/− 53BP1−/−mice showed hypersensitivity to cisplatin and MMC.19 Thus, in certain contexts, the FA pathway and NHEJ collectively help to relieve stress in cells that become overburdened with accumulated ICLs. It was also reported that mice double null for FANCD2 and Ku80 were inviable, leading the authors to suggest that a combined deficiency in the FA pathway and NHEJ posed extreme developmental defects.19 Our work would suggest that inhibition of WRN helicase activity by a small molecule further prevents compensatory mechanism(s) to deal with ICL-induced DNA damage. This may be relevant for personalized medicine to combat cancer in which targeted therapy exploits existing genetic deficiencies of the tumor, such as those in DSB repair.29
The outcome for cell survival from treatment with chemotherapy agents may also depend on the type of drug used to impose replication stress. From our recent work, we observed that the WRN helicase inhibitor did not sensitize FA-deficient cells to the replication inhibitor hydroxyurea,1 suggesting that impairment of WRN unwinding during processing of cross-linked DNA molecules that accumulate when the FA pathway is defective is responsible for cytotoxicity rather than an effect that is simply related to fork stalling. Interference of WRN helicase function at sites of replication fork-associated DNA damage is also likely to be the case for the synergism between the WRN helicase inhibitor NSC 19630 and the topoisomerase inhibitor/chemotherapy drug topotecan, resulting in the inhibition of cell proliferation and induction of DNA damage.11 Exposure to NSC 19630 also sensitized cancer cells to the G-quadruplex-binding compound telomestatin or a PARP inhibitor,11 providing further proof-of-principle for chemically induced synthetic lethality that involves small-molecule modulation of WRN helicase function. A WRN-specific helicase inhibitor may interfere with the role of WRN to accurately replicate human G-rich telomeric sequences, which may ultimately lead to the loss of telomeres and cellular senescence.30 Chemically induced synthetic lethality by means of G4 targeting ligands that act synergistically with a mutation or drug inhibition of a DNA repair protein has attracted interest and may be a useful strategy for compounds that target G4 resolving enzymes such as WRN, BLM, FANCJ, and PIF1.31 In addition, proteins such as WRN helicase that regulate chromatin structure may be a bull’s eye for anticancer drugs. For example, the introduction of a WRN helicase-inactivating allele was found to inhibit immortalization of mouse cells lacking Scaffold attachment factor 1, a protein important for higher-order chromatin structure.32 Future studies will hopefully yield new insights to improving cancer therapy strategies by targeting DNA helicases.10
Perspective for the FA pathway and synthetic lethality
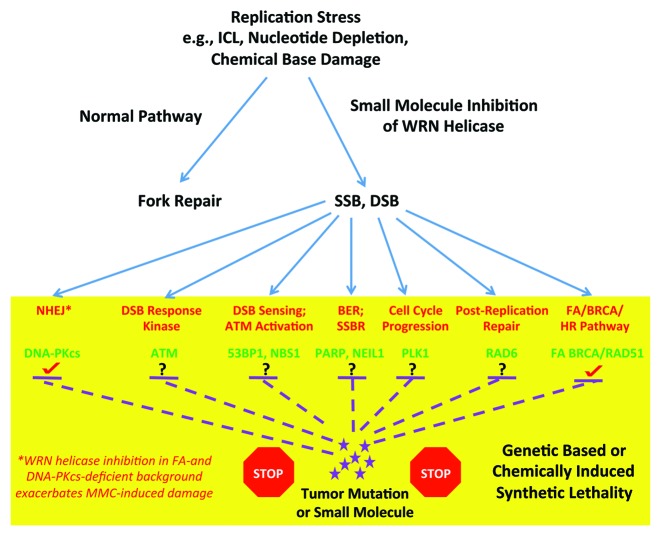
It has been postulated that the FA pathway may serve as a good target for the development of anticancer therapeutic molecules. First, inhibition of the FA pathway itself by small-molecule inhibitors or siRNA depletion of key FA proteins may sensitize cancer cells to DNA damaging agents, particularly those that impose replication stress. Second, inhibition of the FA pathway by the aforementioned small molecules or RNAi in cancers that are already defective in key DNA damage response factors may result in a synthetic lethal situation. Third, tumors that are already deficient in the FA pathway may be sensitized to inhibitors or depletion of certain DNA damage response or DNA repair factors. This aspect may be highly relevant, because it is estimated that 15% of all cancers harbor defects in the FA pathway.33 Moreover, a diverse spectrum of solid tumor (e.g., head and neck, lung, ovarian, cervical) and hematological cancers are epigenetically silenced in expression of wild-type FA genes.34 Therefore, exploitation of FA pathway defects may be one way to target cancers. Examples of synthetic lethal interactions with the FA pathway include proteins such as a DSB response kinase (e.g., ATM34 or ATR35), a factor responsible for DSB sensing or ATM activation (e.g., NBS1), PARP1 implicated in base excision repair (BER) and single-strand break repair (SSBR), a protein such as PLK1 involved in cell cycle progression, as well as other proteins (e.g., NEIL1, 53BP1, RAD6, TREX1). Based on our finding that FANCA- or FANCD2-deficient cells are rendered even more sensitive to the DNA cross-linking agent MMC by WRN helicase inhibition, we would suggest that compounds which inhibit helicase activity of WRN or perhaps other DNA repair helicases represent a new class of small molecules that might be exploited to tailor anticancer drugs that act through a mechanism that behaves synergistically when the FA pathway is defective. It would be of interest to test if cancer cells that are deficient in other factors required for a robust DNA damage response or DNA repair, such as those depicted in Figure 3, are hypersensitive to a WRN helicase inhibitor or helicase inhibitor combined with a DNA damaging agent. These studies may lead to new insights for therapeutic approaches to circumvent the resistance of tumors to DNA damaging treatments (e.g., cross-linking agents) commonly observed.
Figure 3. Potential pathways whereby pharmacological inhibition of WRN helicase activity may impose genetic-based or chemically induced synthetic lethality. See text for details.
Materials and Methods
Cell culture
DNA-PKcs-deficient (MO59J) and corrected (MO59K) cells were grown in Dulbecco modified Eagle medium: nutrient mixture F-12 (DMEM/F12) with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 1% l-glutamine, and 0.5% MEM non-Essential amino acids at 37 °C in 5% CO2.
Cell proliferation assays
Proliferation was measured using WST-1 assay (Roche) as described.11
siRNA transfection and western blot analysis
FANCD2 siRNA (5′-AACAGCCAUG GAUACACUUG AUU-3′) and control siRNA (ThermoFisher Scientific Biosciences) was transfected in DNA-PKcs-deficient and corrected cells using Lipofectamine 2000 as per the manufacturer’s protocol (Invitrogen). Cells were plated to 50–60% confluence in 10 cm dishes 24 h prior to transfection. siRNA (0.6 nmol) was mixed with 30 μl of Lipofectamine 2000 in 3 ml of Opti-MEM (Invitrogen). The mixture was added to cells which were subsequently incubated for 4 h. Forty-eight hours after the transfection, cells were harvested for preparing lysate or treated with small-molecule compound or DMSO and/ or mitomycin C (MMC) at the indicated concentrations, and cell proliferation was measured using WST-1 reagent (Roche) as described above.
For lysate preparation, cells were washed twice with 1× PBS. RIPA buffer (10 mM sodium phosphate [pH 7.2], 300 mM NaCl, 0.1% SDS, 1% NP-40, 1% sodium deoxycholate, 2 mM EDTA) was added to the cells, and the cells were incubated at 4°C for 30 min. Cells were scrapped, and the suspension was further incubated on ice for 30 min. Cell suspension was centrifuged at 18 500 × g for 10 min at 4 °C, and supernatant was collected. Twenty μg of the lysate was loaded on 8–16% SDS-PAGE. Protein was transferred onto a PVDF membrane, and blot was probed with anti-FANCD2 antibody (1:500, Abcam). For secondary antibody, peroxidase-labeled anti-rabbit IgG (GE healthcare) was used. Blot was developed using ECL Plus Western Blot Detection Kit as per manufacturer’s protocol (Amersham). As a loading control, blot was stripped and then re-probed with anti-actin antibody (1:5000, Sigma).
Acknowledgments
This work was supported by the Intramural Research program of the National Institutes of Health, National Institute on Aging, and the Fanconi Anemia Research Fund (RMB).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26320
References
- 1.Aggarwal M, Banerjee T, Sommers JA, Iannascoli C, Pichierri P, Shoemaker RH, Brosh RM., Jr. Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer Res. 2013;73:5497–507. doi: 10.1158/0008-5472.CAN-12-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 3.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 4.Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10:1192–9. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 6.Reinhardt HC, Jiang H, Hemann MT, Yaffe MB. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 2009;8:3112–9. doi: 10.4161/cc.8.19.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidorova JM, Li N, Folch A, Monnat RJ., Jr. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franchitto A, Pichierri P. Understanding the molecular basis of common fragile sites instability: role of the proteins involved in the recovery of stalled replication forks. Cell Cycle. 2011;10:4039–46. doi: 10.4161/cc.10.23.18409. [DOI] [PubMed] [Google Scholar]
- 9.Pichierri P, Ammazzalorso F, Bignami M, Franchitto A. The Werner syndrome protein: linking the replication checkpoint response to genome stability. Aging (Albany NY) 2011;3:311–8. doi: 10.18632/aging.100293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosh RM., Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13:542–58. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci U S A. 2011;108:1525–30. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–80. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–63. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski RJ. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J Biol Chem. 2005;280:40559–67. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- 16.Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML, Paull TT, Campisi J, Seidman M, Bohr VA. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34:2751–60. doi: 10.1093/nar/gkl362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci U S A. 2007;104:10394–9. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunting SF, Nussenzweig A. Dangerous liaisons: Fanconi anemia and toxic nonhomologous end joining in DNA crosslink repair. Mol Cell. 2010;39:164–6. doi: 10.1016/j.molcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunting SF, Callén E, Kozak ML, Kim JM, Wong N, López-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46:125–35. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 21.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–23. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi J, Okui M, Asaithamby A, Burma S, Chen BP, Tanimoto K, Matsuura S, Komatsu K, Chen DJ. WRN participates in translesion synthesis pathway through interaction with NBS1. Mech Ageing Dev. 2010;131:436–44. doi: 10.1016/j.mad.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–9. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- 24.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr. Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–8. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper MP, Machwe A, Orren DK, Brosh RM, Jr., Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–12. [PMC free article] [PubMed] [Google Scholar]
- 26.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–54. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112:1413–23. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins C, Kan J, Hoatlin ME. Targeting the fanconi anemia pathway to identify tailored anticancer therapeutics. Anemia. 2012;2012:481583. doi: 10.1155/2012/481583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moeller BJ, Pasqualini R, Arap W. Targeting cancer-specific synthetic lethality in double-strand DNA break repair. Cell Cycle. 2009;8:1872–6. doi: 10.4161/cc.8.12.8743. [DOI] [PubMed] [Google Scholar]
- 30.Damerla RR, Knickelbein KE, Strutt S, Liu FJ, Wang H, Opresko PL. Werner syndrome protein suppresses the formation of large deletions during the replication of human telomeric sequences. Cell Cycle. 2012;11:3036–44. doi: 10.4161/cc.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Brosh RM., Jr. G-quadruplex nucleic acids and human disease. FEBS J. 2010;277:3470–88. doi: 10.1111/j.1742-4658.2010.07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lachapelle S, Oesterreich S, Lebel M. The Werner syndrome helicase protein is required for cell proliferation, immortalization, and tumorigenesis in Scaffold attachment factor B1 deficient mice. Aging (Albany NY) 2011;3:277–90. doi: 10.18632/aging.100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–33. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, Shimamura A, D’Andrea AD. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest. 2007;117:1440–9. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CC, Kennedy RD, Sidi S, Look AT, D’Andrea A. CHK1 inhibition as a strategy for targeting Fanconi Anemia (FA) DNA repair pathway deficient tumors. Mol Cancer. 2009;8:24. doi: 10.1186/1476-4598-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]