Abstract
The Etruscan civilisation originated in the Villanovan Iron Age in the ninth century BC and was absorbed by Rome in the first century BC. Etruscan tombs, many of which are subterranean, are one of the best representations of this culture. The principal importance of these tombs, however, lies in the wall paintings and in the tradition of rich burial, which was unique in the Mediterranean Basin, with the exception of Egypt. Relatively little information is available concerning the biodeterioration of Etruscan tombs, which is caused by a colonisation that covers the paintings with white, circular to irregular aggregates of bacteria or biofilms that tend to connect each other. Thus, these colonisations sometimes cover extensive surfaces. Here we show that the colonisation of paintings in Tomba del Colle is primarily due to bacteria of the order Rhizobiales (Alphaproteobacteria), which were likely influenced by the neighbouring rhizosphere community and the availability of nutrients from root exudates.
In recent years, the study of subterranean environments has attracted the attention of microbiologists. This attraction can be attributed to the increasing biodeterioration observed in rock art caves1,2,3 and in mural paintings from catacombs and tombs4,5,6. It remains unknown whether the white colonisations found in caves and tombs, which exhibit apparent morphological similarities, are constituted by comparable microbial communities, or whether they are influenced by environmental factors and geological/geographical location.
Relatively little information is available concerning the biodeterioration of paintings in Etruscan tombs. In the 1980s and 90s, a small number of reports were published in congresses and rarely in journals7,8,9,10. These reports emphasised the involvement of Streptomyces in biodeterioration processes and described the application of biocides with questionable success. Recently, Diaz-Herraiz et al.11 reported that the paintings from Tomba della Scimmia were heavily colonised by Actinobacteria, primarily Nocardia and Pseudonocardia, by a process similar to that reported in caves1,12.
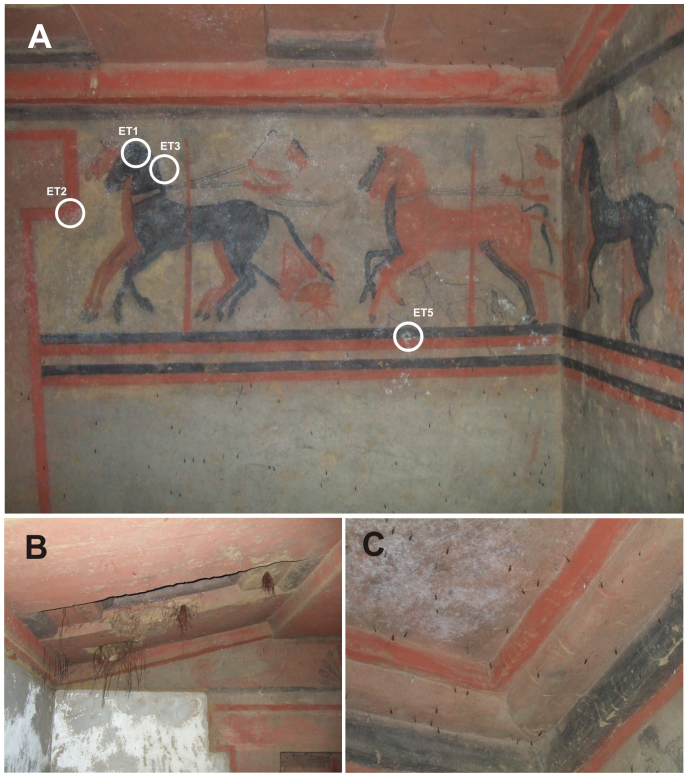
Many Etruscan tombs can be found near Chiusi, in the Tuscany region of Italy. The Tomba del Colle (i.e., Tomb of the Hill), which is also known as Tomba Casuccini, is slightly east on this town. This tomb dates to the early fifth century B.C. This tomb is interesting because the original door and its two travertine door knockers have been retained and because it consists of two decorated chambers with paintings on the walls that depict figurative scenes of games, dances, musicians, athletes, chariot races (Fig. 1A), fighters, and a banquet scene. At the time of sampling, the ceiling was invaded by roots from an acacia that was growing above the tomb (Fig. 1B) and the tomb chambers were populated by hundreds of mosquitoes (Fig. 1C). In addition, pines, olive trees and gramineous plants composed the vegetation above the tomb.
Figure 1. Paintings in Tomba del Colle.
(A). Sampling points on white colonisations. (B). Roots on the ceiling and wall efflorescences. (C). Ceiling efflorescences. Photographs taken by the authors.
The main deterioration pathologies observed in the tomb were extensive areas of efflorescence on some walls and on the ceiling (Fig. 1B, C) and biodeterioration (Figs. 1A and 2). The paintings were colonised by bacteria that formed white, circular to irregular aggregates or biofilms, which extended throughout the paintings, regardless of the pigment used (Fig. 1A). Due to the bioreceptivity of some pigmented areas that facilitates the colonisation of certain bacteria11, a study on the different pigmented areas was performed to determine whether the minerals used in the paints selected for specific bacteria and to investigate the metabolic activity of the bacteria colonising the walls.
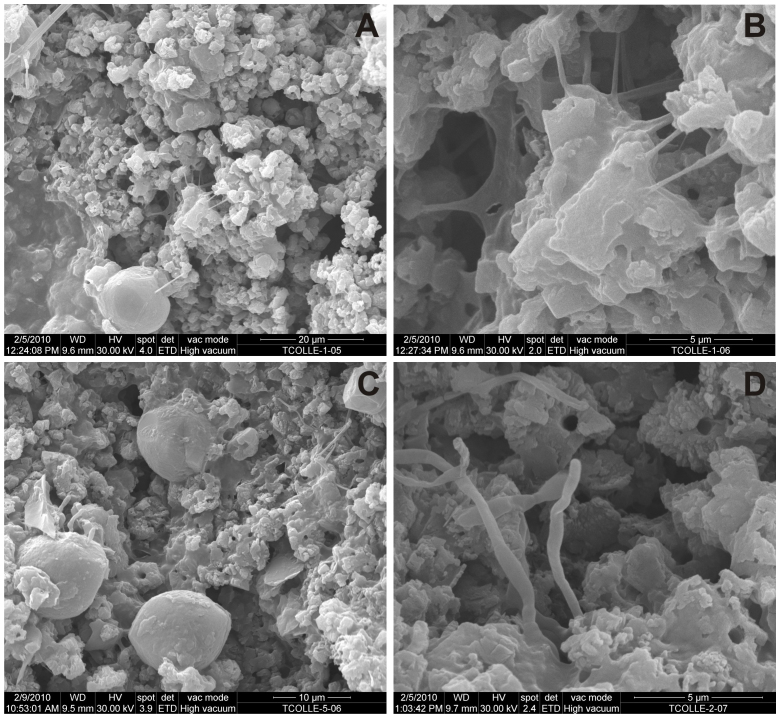
Figure 2. Scanning electron micrographs of the bacterial colonisations in Tomba del Colle.
(A). A colonisation on black pigment (i.e., ET1) with a continuous bed formed of CaCO3 nest-like aggregates and dispersed spheroidal elements. (B). Detail of A at the point where aggregates are attached by filaments and extracellular polymeric substances. (C). A colonisation on black pigment (i.e., ET5) with similar composition to ET1 and more spheroidal elements. (D). A colonisation on red pigment (i.e., ET2) with nest-like aggregates and microbial filaments emerging from the mineral substratum.
Results
Site and Tomb description
Like other Chiusi tombs Tomba del Colle was excavated in thick-bedded, massive host-rock characterised by Pliocene clastic sediments, which consisted of weakly cemented sands with intercalations of clay layers and pebble beds. The rough rocks were smoothed, and the paintings were made on a thin clay coating that was applied directly to the walls. The tomb is connected directly to the exterior by an original travertine door and is located immediately below the soil. Therefore, the tomb is directly affected by plant roots, seasonal temperatures and soil moisture variations. These factors resulted in marked degradation that was aggravated by the presence of trees directly above the tomb. The roots of these trees affected the left side of the atrium and the ceiling, producing cracks and areas with high moisture gradients (Figs. 1B, C). Humidity variations in the sandy surfaces that were caused by external weather events produced bulges, sand disaggregation and powdering of the rock surface, with gravitational shedding of particles and occasional large detachments, which affected most of the walls and the ceiling. Stone blocks, which were detached as a result of the action of the roots, were previously clamped using metal belts.
In addition, one of the characteristic effects of the weathering processes was the crystallisation of abundant salts on the walls (Fig. 1B, C) due to cyclical wetting–drying processes. Like other Etruscan tombs in the Chiusi area, white bacterial colonisations, morphologically similar to those reported for Tomba della Scimmia11, were observed.
Scanning electron microscopy
To investigate the biodeterioration of the paintings, representative samples of the white colonisations on black (i.e., ET1 and ET5), red (i.e., ET2) and ochre (i.e., ET3) pigments were analysed using a scanning electron microscope. Figure 2 shows an example of the microbial colonisations, which were primarily composed of filamentous microorganisms, CaCO3 deposits and extracellular polymeric substances (EPS).
Microbial growth was associated with a type of CaCO3 deposits that exhibited a morphology of nest or rosette-like fabrics that contain aggregates of subeuhedral to euhedral calcite crystals (2–4 μm in size). The crystals usually have a central hole that is 0.5–0.7 μm in diameter, and they generally rest on the substratum. In addition, a second, less frequent type of CaCO3 deposit was represented by spheroidal or hemispheroidal elements averaging 8–10 μm in diameter (Fig. 2A, C).
Microbial growth was associated with CaCO3 deposits. The image presented in Figure 2B demonstrates that extensive EPS and filaments held together the CaCO3 crystals and nest-like fabrics. Bacterial filaments with a diameter of 0.5–0.7 μm were often observed immersed and emerging from the substratum (Fig. 2D).
Previous scanning electron micrographs of actinobacterial colonisations from Tomba della Scimmia, Italy11 and Altamira Cave, Spain12 revealed similar biological structures and mineral patterns. Bioinduced minerals coated the rock surfaces of the caves and tombs, and CaCO3 nest-like aggregates and spheroidal elements were predominant11. We hypothesised that the nest or rosette-like fabrics were bioinduced around bacterial filaments because the diameter of the central hole and the filaments are the same. Thus, the hole might indicate the location that was previously occupied by lysed bacterial structures, in a similar manner to that described for the calcite or manganese oxide sheaths of reticulated filaments13.
While similar mineral biofabrics have been observed in association with actinobacterial colonisations in other subsurface environments11,12 the micrographs of the white colonisations covering the surface of the paintings in Tomba del Colle are different from those previously reported for Tomba della Scimmia. In this latter tomb, the surface of the paintings is coated by abundant nest-like aggregates and spheroidal elements, which are held together by abundant actinobacterial filaments and other biological structures. In contrast, in Tomba del Colle, the bacteria develop beneath the mineral layer, and sometimes emerged to the surface (Fig. 2C, D). The minerals are occasionally connected by 0.25 μm filaments and abundant EPS, while emerging filaments reach 0.5–0.7 μm in diameter (Fig. 2B). It appears that in Tomba del Colle, bioinduction and mineral precipitation on the painted surfaces was more extensive than in Tomba della Scimmia, for which micrographs from the painted surface reveal a predominance of actinobacterial structures11.
Hirsch14 reported that budding may be a characteristic of bacteria that live in habitats that undergo frequent and sudden changes, which is suggested by the extensive efflorescence area. These bacteria have a tendency to attach to surfaces in habitats that are encrusted with mucilage and minerals. It is believed that polar growth allows the bacterial cell to emerge and facilitates bacteria survival in oligotrophic environments by increasing nutrient uptake. The image presented in Figure 2D supports this hypothesis.
Microbial communities
Previous reports indicated that subterranean environments are mainly populated by Actinobacteria11,12. The quasi-extremophile subsurface environment appears to posses ideal conditions for actinobacterial development. The low temperatures, high relative humidity and dissolved organic carbon (DOC) from dripping waters appear to favour actinobacterial growth in Tomba della Scimmia11, but this was not the case in Tomba del Colle where Alphaproteobacteria (order Rhizobiales) predominated over Actinobacteria, as indicated by the 16S rRNA gene libraries.
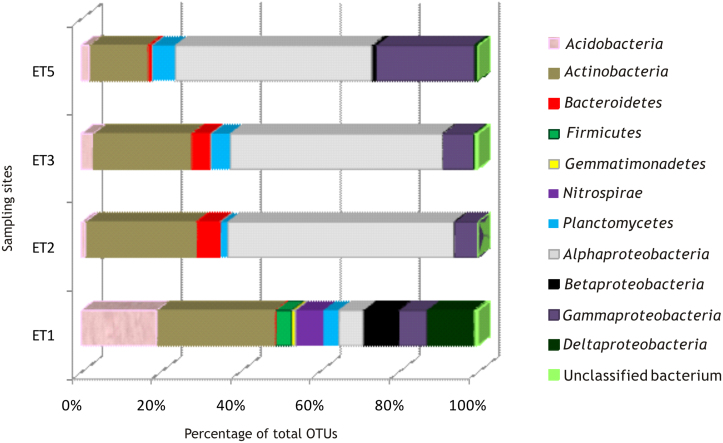
A total of 456 non-chimeric bacterial sequences were obtained from four samples from different area and pigments. Amplification of archaea and fungi was unsuccessful from all of the samples. The bacterial sequences are reported in Supplementary Tables S1–S4. The distribution of different phyla in the samples is summarised in Figure 3. The major phylogenetic groups were Alphaproteobacteria and Actinobacteria. Alphaproteobacteria represented between 49 and 57% of the DNA sequences in three samples (i.e., ET2, ET3 and ET5), but only 6% in sample ET1. Actinobacteria represented 25 to 30% of the clones in three of the samples and only 15% of the clones in sample ET5. In total, Alphaproteobacteria and Actinobacteria comprised between 64 and 85% of all of the clones in samples ET2, ET3 and ET5, but only 36% of the clones in sample ET1.
Figure 3. Percentage of shared and specific OTUs in the Tomba del Colle samples.
Among the Actinobacteria, several sequences, which were tentatively affiliated to the genera Catelliglobosispora, and Streptosporangium (Supplementary Tables S2–S4), were common and abundant in three of the samples (i.e., ET2, ET3 and ET5). The clones identified as Jiangella muralis, reached similarities as high as 99%, facilitating their assignment to this particular species.
The distinct abundance of sequences assigned to Alphaproteobacteria (order Rhizobiales) was of great interest. The order Rhizobiales includes 13 families, and genera from seven of these families were found in the tomb: the Hyphomicrobiaceae family, with the genera Filomicrobium, Hyphomicrobium, Pedomicrobium, Prosthecomicrobium, Rhodoplanes and Blastochlorys, the Phyllobacteriaceae family, with the genera Phyllobacterium and Mesorhizobium, the Xanthobacteraceae family, with the genera Xanthobacter and Pseudolabrys, the Bradyrhizobiaceae family, with the genus Afipia, the Methylobacteriaceae family, with the genus Microvirga, the Rhodobiaceae family, with the genus Rhodobium and the Rhizobiaceae family with the genus Rhizobium. The order Rhizobiales comprised 36% of the clones from the four samples. However, this order comprised 50% of the clones when we excluded sample ET1, which was atypical due to the high percentage of Actinobacteria and Acidobacteria.
The family Hyphomicrobiaceae alone represented 29% of all of the clones in the four samples but this family represented 41% of the clones when considering only samples ET2, ET3 and ET5. The most abundant genera were Hyphomicrobium, Pedomicrobium, and Prosthecomicrobium. Species assignment was only possible in the case of Hyphomicrobium aestuarii (99% similarity). The genus Hyphomicrobium consists of restricted facultative methylotrophs that are found in soils, aquatic environments and sewage sludges15. Hyphomicrobium spp. are also endophytic bacteria in roots16.
Sample ET1 contained a high number of actinobacterial clones (30%), and 25% of these clones corresponded to Pseudonocardia. This sample is the only sample that was similar to Tomba della Scimmia11 with respect to actinobacterial abundance. In sample ET1, 55% of the clones shared a similarity below 90% to previously described species, including all of the Acidobacteria, Firmicutes, Gemmatimonadetes, Planctomycetes and Deltaproteobacteria clones, suggesting an unusual contribution of unknown bacteria to the clones in this particular sample.
In addition, we investigated sample ET3 to assess the potentially active fraction of bacteria in the painting using RNA analysis17. We obtained a total of 115 non-chimeric sequences (Supplementary Table S5). A comparison of the DNA (Supplementary Table S3) and RNA data (Supplementary Table S5) indicated that the bacteria present in sample ET3, as indicated by DNA analysis, were primarily composed of Alphaproteobacteria (54%) and Actinobacteria (25%), while the potentially active bacteria, as indicated by RNA analysis, were primarily composed of Alphaproteobacteria (68%) and Actinobacteria (22%).
The DNA and RNA clone libraries both contained the genera Catelliglobosispora, Jiangella, Streptosporangium and Marmoricola among the Actinobacteria; Hyphomicrobium, Pedomicrobium, Prosthecomicrobium, Afipia, Azospirillum, Dongia, Filomicrobium and Xanthobacter among the Alphaproteobacteria, and Flavitalea and Chitinophaga among the Bacteroidetes, although some differences in clone percentages were observed.
It is interesting that the most abundant members of the Alphaproteobacteria phyla that were retrieved in the DNA library from the ET3 sample were shown to be potentially active and were also common to the ET2 and ET5 samples. The number and size of samples, which was imposed by cultural heritage preservation guidelines, limited our capacity to assign a general distribution of Rhizobiales to every tomb wall and painting. However, the abundant presence of potentially active members of this order, particularly the family Hyphomicrobiaceae, in the samples suggests an important and active role of this family of bacteria in this tomb, as well as an involvement of bacteria of the order Rhizobiales in biogeochemical and biodeterioration processes (Fig. 2).
Discussion
We assessed the bacterial community colonising the Etruscan paintings from Tomba del Colle using DNA analysis, which reflected the bacteria present in the community, and using RNA analysis, which was indicative of the potentially active bacteria in the community. RNA-based analyses have been used frequently to identify the active fraction of the total bacterial community11,17. It was recently suggested that these analyses may have some limitations with respect to the relationships between RNA and growth and activity18. However, our previous data from another Etruscan tomb11 showed a high degree of confidence in this respect.
In Tomba del Colle the most abundant phyla were Alphaproteobacteria and Actinobacteria, in both DNA- and RNA-based analyses. Snider19 found that roots were abundant inside lava tube caves and that both roots and wall samples had their closest relatives within the Acidobacteria, Alphaproteobacteria and Actinobacteria. In addition, she found a small number of clones closely related to Hyphomicrobium and Pedomicrobium.
Invasive woody legumes, such as Acacia spp., form a symbiosis with nitrogen-fixing bacteria, which increase the nitrogen in the invaded areas20. Nitrogen-fixing species of the genera Mesorhizobium, Phyllobacterium, Azospirillum and Xanthobacter were found in this tomb, as well as denitrifying bacteria, such as the hyphomicrobia. In addition, Andreote et al.21 reported that one of the most frequently observed families in the rhizoplane of Eucalyptus was Hyphomicrobiaceae, with the genera Hyphomicrobium, Blastochloris and Rhodoplanes, which was also found in Tomba del Colle. Other Rhizobiales genera, such as Afipia, Phyllobacterium and Rhizobium, were detected in both the Eucaliptus rhizoplane and this tomb.
The high abundance of members of the family Hyphomicrobiaceae in this tomb was not previously reported for other tombs, catacombs or caves1,4,11,12, although some clones from Hyphomicrobium spp. were found in Movile Cave22. The abundance of Rhizobiales observed in Tomba del Colle appears to be consistent with previous data from nitrogen-fixing bacteria and rhizosphere studies. Therefore, we suggest that roots can act as a conduit for microorganisms from the rhizosphere.
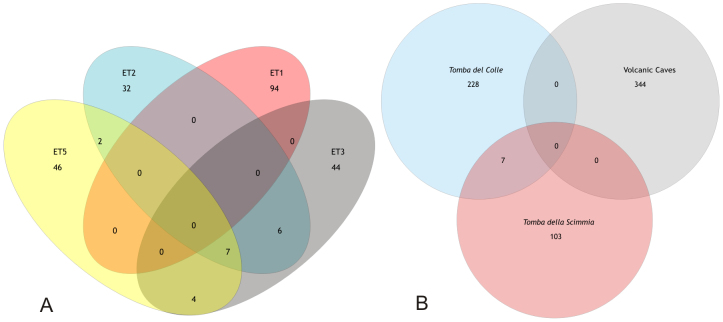
To determine overlapping diversity, the sequences obtained from Tomba del Colle were compared to previously studied sequences from Tomba della Scimmia11 and to other white colonisations from lava caves23 (Fig. 4).
Figure 4. Venn diagram of the OTUs in the Tomba del Colle samples (A), and the Tomba del Colle, Tomba della Scimmia and volcanic caves samples (B) at a distance of 0.03.
The comparison of the four samples obtained from Tomba del Colle revealed that the community structures were likely to be significantly different, although the test was not convincing for the comparison between samples ET2 and ET3 or samples ET2 and ET5 (data not shown). A Venn diagram demonstrated that the ET2, ET3 and ET5 samples shared only seven operational taxonomic units (OTUs) and that these samples shared no OTUs with sample ET1. These findings indicate that the bacterial communities were different. When the white colonisations from Tomba del Colle, Tomba della Scimmia and volcanic caves23 were compared, only seven OTUs from a total richness of 682 OTUs were shared between the Etruscan tombs and no OTUs were shared with the volcanic caves. These results demonstrate that the microbial communities from the two tombs have little in common with each other or with volcanic caves.
In an attempt to correlate phyla with environmental factors, we observed that Tomba del Colle was invaded by tree roots (Fig. 1B). The roots can bring microorganisms into the cave, as well as organic carbon (OC) that occurs primarily as root litter and root exudates and that may enhance microbial activity24, selecting for distinct microbial communities in response to nutrient availability at or near the rhizosphere.
Small scale variability of OC should have an important influence on subterranean microbial communities. Condensation of water drops on the painted surface and local input of OC from volatile organic compounds, root exudates or percolating DOC can promote the heterogeneous spatial biodiversity observed in the tomb (e.g. sample ET1 vs the rest of samples). We did not observed a direct effect of the pigmented minerals used in the paintings on the selection of communities, as indicated by the similarity of the bacteria phyla identified in samples with red, black and ochre pigments (Supplementary Tables S2–S4). In contrast, local variations appear to occur within the same pigment (e.g., ET1 vs. ET5) due to microenvironmental factors (see Supplementary Table S1 vs. S4).
The high diversity between Tomba del Colle and Tomba della Scimmia11 is striking. The tombs are approximately 1.7 km apart, located on a straight line, and excavated in the same geological formation (i.e., Pliocene clastic sediments) with the same climatic conditions. Our hypothesis was that these two factors (i.e., climate and parent rock) have less influence on the microbial community composition than other environmental factors, such as vegetation and the availability of organic matter. The belowground diversity in Tomba del Colle appears to be largely influenced by the rooting system and root-associated microorganisms that are invading the tomb. These factors are highly specific for promoting a Rhizobiales colonisation. These factors may also be the reason why the bacterial communities from Tomba del Colle were different from those reported for Tomba della Scimmia, which lies under soil with grasses and is dominated by the genera Nocardia and Pseudonocardia11.
It is worth noting that Tomba del Colle was cleaned and restored from 1994–1996. In an additional treatment performed from 2003–2006, the walls were cleaned sequentially with 2% and 5% solutions of the biocide Preventol R80 (i.e., dodecyldimethyl dichlorobenzyl ammonium chloride) before consolidation with ethyl silicate. The cleaning did not prevent further colonisation, as indicated by the presence of potentially active bacteria in this study. Similar results were obtained in other Etruscan and Roman tombs, where re-colonisation was observed shortly after cleaning with biocides and restoration9,25. In Tomba del Colle, after cleaning, rhizosphere microbial communities that were established around the tomb would have been transported by water infiltration, promoting the re-colonisation of the communities observed on the painted walls. Cleaning of subterranean environments with similar biocides has been shown to be detrimental in many cases3,9,25,26. In contrast, preventive conservation to control environmental and microclimate factors was successful in another case1.
Methods
Sample collection
Sample ET1, ET2, ET3 and ET5 were collected from Tomba del Colle in July 2009. All of these samples represented macroscopic white colonisations that were located on the paintings. ET1 was collected from the head and mouth of the black horse, ET2 was collected from the lower angle of the red frame in front of the two horses, ET3 was collected from the yellow zone behind the neck of the black horse and ET5 was collected from the black line just below the right leg of the dog (Fig. 1). More extensive sampling was not possible due to the limitations imposed by the protection of cultural heritage site guidelines. The samples were obtained by scraping off the colonisations with a sterile scalpel. One millilitre of RNA-later (Life Technologies) RNA stabilisation reagent was added to the set of tubes. After collection, the samples were stored on ice and processed or frozen when they arrived in the laboratory.
Nucleic acid extraction and PCR amplification
The employed method of total nucleic acids extraction was described in detail by Griffiths et al.15 and Diaz-Herraiz et al.11. Briefly, complementary DNA (cDNA) was synthesised from the RNA using Superscript II Reverse Transcriptase (Invitrogen) with the following single specific primers: 907R for bacterial 16S rRNA gene, Arch 1000R for the archaeal 16S rRNA gene and ITS4 for fungal internal transcribed spacer regions. Amplification of the cDNA was performed using the bacteria-specific primers 616F and 907R, the archaea-specific primers Arch 340F and Arch 1000R, and fungi-specific primers ITS1 and ITS4, as described by Diaz-Herraiz et al.11. In addition, the FastDNA Spin kit for soil (MP Biomedicals) was used to extract the genomic DNA.
Cloning and sequencing
DNA libraries of PCR-amplified products were constructed using the pGEM-T Easy Vector (Promega) and subsequently transformed into One Shot Max Efficiency DH5a-T1 chemically competent Escherichia coli (Invitrogen), according to manufacturer's instructions. Transformants were picked randomly, transferred to multiwell plates containing Luria–Bertani medium supplemented with 100 μg.mL−1 ampicillin and 15% w/v glycerol and stored at −80°C. We constructed four libraries. On average 100 clones from each library were sequenced at Macrogen Inc., Seoul, Korea, using the universal bacterial primer 616F.
Sequence analyses
Sequences were checked for chimera using chimera.slayer, as implemented in the Mothur software package27. Putative chimeric sequences were excluded from further analysis, and 571 sequences were included in phylogenetic analyses. The sequences were aligned using the Mothur program. After this analysis, all sequences were compared to the non-redundant database of sequences deposited at the National Center for Biotechnology (NCBI) and EzTaxon28 using the BLASTN algorithm29. The aligned sequences were clustered into OTUs using Mothur with a 97% sequence identity cut-off. Rarefaction curves (Supplementary Fig. S1) and diversity indices were also obtained using Mothur.
Nucleotide sequence accession
The nucleotide sequences generated in this study were deposited into the NCBI GenBank database under the accession numbers HG379790-HG380020.
Environmental scanning electron microscopy
Textural and microestructural characterisation of different white colonisations were performed using an Inspect-S50 low-vacuum environmental scanning electron microscope (FEI Co., Japan) that includes an energy-dispersive spectroscopy probe. The samples were first observed and described under controlled low-vacuum conditions. The samples were then covered with gold sputter using an Emitech K550Y gold coater and observed under high-vacuum conditions to improve the photographic quality and EDS microanalysis.
Author Contributions
M.D.-H. and V.J. contributed equally to this work. C.S.-J. conceived the study; C.S.-J., V.J., S.C., S.S.-M., P.T. and P.P. participated in the field survey and collected samples; P.T. and P.P. contributed information concerning the tomb; M.D.-H. and V.J. performed the experiments, V.J. developed bioinformatic tools; L.L. supervised the laboratory work of M.D.-H.; C.S.-J. was responsible for the interpretation of the data and wrote the paper.
Supplementary Material
Deterioration of an Etruscan tomb by bacteria from the order Rhizobiales
Acknowledgments
This work was funded through the projects CGL2010-17183, 201030E011 and Consolider 2007-00058. M.D.H. was supported by a JAE Research Fellowship from CSIC, and S.C. was supported by a Juan de la Cierva contract.
Accession codes Nucleotide sequences have been deposited in GenBank under the accession codes HG379790-HG380020.
References
- Saiz-Jimenez C. et al. Paleolithic art in peril: Policy and science collide at Altamira Cave. Science 334, 42–43 (2011). [DOI] [PubMed] [Google Scholar]
- Saiz-Jimenez C., Miller A. Z., Martin-Sanchez P. M. & Hernandez-Marine M. Uncovering the origin of the black stains in Lascaux Cave in France. Environ. Microbiol. 14, 3220–3231 (2012). [DOI] [PubMed] [Google Scholar]
- Martin-Sanchez P. M., Nováková A., Bastian F., Alabouvette C. & Saiz-Jimenez C. Use of biocides for the control of fungal outbreaks in subterranean environments: The case of the Lascaux Cave in France. Environ. Sci. Technol. 46, 3762–3770 (2012). [DOI] [PubMed] [Google Scholar]
- Sanchez-Moral S. et al. Deterioration of building materials in Roman Catacombs: The influence of visitors. Sci. Total Environ. 349, 260–276 (2005). [DOI] [PubMed] [Google Scholar]
- Laiz L. et al. Isolation of Rubrobacter strains from biodeteriorated monuments. Naturwissenschaften 96, 71–79 (2009). [DOI] [PubMed] [Google Scholar]
- Vasanthakumar A., DeAraujo A., Mazurek J., Schilling M. & Mitchell R. Microbiological survey for analysis of the brown spots on the walls of the tomb of King Tutankhamun. Int. Biodeterior. Biodegr. 79, 56–63 (2013). [Google Scholar]
- Agarossi G., Ferrari R., Monte M., Gugliandolo C. & Maugeri T. In: VIth International Congress on Deterioration and Conservation of Stone. Supplement, 82–91 (Nicholas Copernicus University, Torun, 1988). [Google Scholar]
- Agarossi G., Ferrari R. & Monte M. In: The Conservation of Monuments in the Mediterranean Basin, (ed Zezza F., ed. ), 511–517 (Grafo, Bari, 1989). [Google Scholar]
- Monte M. & Ferrari R. Biodeterioration in subterranean environments. Aerobiologia 9, 141–148 (1993). [Google Scholar]
- Agarossi G. In: Studi e Ricerche sulla Conservazione delle Opere d'Arte Dedicati alla Memoria di Marcello Paribeni. (ed Zezza, F.), 1–11 (C.N.R., Roma, 1994). [Google Scholar]
- Diaz-Herraiz M. et al. The actinobacterial colonization of Etruscan paintings. Sci. Rep. 3, 1440 | 10.1038/srep01440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuezva S. et al. The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiol. Ecol. 81, 281–290 (2012). [DOI] [PubMed] [Google Scholar]
- Miller A. Z. et al. Enigmatic reticulated filaments in subsurface granite. Environ. Microbiol. Rep. 4, 596–603 (2012). [DOI] [PubMed] [Google Scholar]
- Hirsch P. Budding bacteria. Annu. Rev. Microbiol 28, 391–440 (1974). [DOI] [PubMed] [Google Scholar]
- Osaka T. et al. Identification of acetate- or methanol-assimilating bacteria under nitrate-reducing conditions by stable-isotope probing. Microb. Ecol. 52, 253–266 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang Y. Z. et al. Effects of rhizobial inoculation, cropping systems and growth stages on endophytic bacterial community of soybean roots. Plant Soil 347, 147–151 (2011). [Google Scholar]
- Griffiths R. I., Whiteley A. S., O'Donnell A. G. & Bailey M. J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazewicz S. J., Barnard R. L., Daly R. A. & Firestone M. K. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J. R. Comparison of microbial communities on roots, ceiling and floors of two lava tube caves in New Mexico. Master Thesis, University of New Mexico, Albuquerque, 2010, http://repository.unm.edu/handle/1928/11135. [Google Scholar]
- Crisóstomo J. A., Rodríguez-Echeverría S. & Freitas H. Co-introduction of exotic rhizobia to the rhizosphere of the invasive legume Acacia saligna, an intercontinental study. Appl. Soil Ecol. 64, 118–126 (2013). [Google Scholar]
- Andreote F. D. et al. Culture-independent assessment of Rhizobiales-related Alphaproteobacteria and the diversity of Methylobacterium in the rhizosphere and rhizoplane of transgenic Eucalyptus. Microb. Ecol. 57, 82–93 (2009). [DOI] [PubMed] [Google Scholar]
- Hutchens E., Radajewski S., Dumont M. G., McDonald I. R. & Murrell J. C. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6, 111–120 (2004). [DOI] [PubMed] [Google Scholar]
- Hathaway J. J. M. et al. Comparison of bacterial diversity in Azorean and Hawaiian lava cave microbial mats. Geomicrobiol J. in press (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R. P., Finzi A. C. & Bernhardt E. S. Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long-term CO2 fumigation. Ecol. Lett. 14, 187–194 (2011). [DOI] [PubMed] [Google Scholar]
- Akatova E., Roldan M., Hernandez-Marine M., Gonzalez J. M. & Saiz-Jimenez C. In: Science and Cultural Heritage in the Mediterranean Area, 317–322 (Regione Siciliana, Palermo, 2009). [Google Scholar]
- Saiz-Jimenez C. In: Cave Microbiomes: A Novel Resource for Drug Discovery (ed Cheeptham, N.) vol. 1, 69–84 (SpringerBriefs in Microbiology, 2013). [Google Scholar]
- Schloss P. D. et al. Introducing mothur: Open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Deterioration of an Etruscan tomb by bacteria from the order Rhizobiales