Abstract
Colletotrichum species have been rarely implicated in human disease. We describe a case of deep soft tissue mycosis following a penetrating injury with a lemon tree thorn. Direct Blankophor BA (Bayer) stain from intraoperative tissue showed fungal elements. Pure growth fungus was apparent at 2–4 days. Morphological features provisionally identified the isolate as a coelomycetous fungus, likely Colletotrichum species. This was confirmed with molecular analysis of the internal transcribed spacer region (ITS) region.
Keywords: Colletotrichum, Deep soft tissue mycosis, Internal transcribed spacer region
1. Introduction
Colletotrichum Corda is a complex form genus of the Coleomycetes class of fungi. Colletotrichum was first reported by Tode (1790) in the genus Vermcularia. Colletotrichum was later described by Corda (1831) and comprises Coelomycetes with a Glomerella teleomorph stage. Coleomycetes are asexual fungi which produce conidia from conidiophores compacted into structures known as pycnidia, acervuli or sporodochia (visible to the naked eye as dense dots in the mycelium). Colletotrichum species are commonly described in the agricultural literature as either plant commensal, plant pathogen or a saprophyte. Colletotrichum species are rarely implicated in human disease, with occasional reports of keratitis, subcutaneous infection following penetrating injury, and systemic infection in immunocompromised patients [1,2].
We describe deep soft tissue mycosis in an immunocompetent man following penetrating injury with a lemon tree thorn. Highlighted is the importance of optimising fungal culture in a high-risk patient and challenges in morphological and molecular mycology diagnostics. Deep soft tissue mycoses are associated with considerable morbidity and mortality, particularly in the immunocompromised [3].
2. Case
A 57-year-old man presented with a painful right foot 3-weeks after a lemon tree thorn penetrated his foot through a gardening boot. The patient had no response from treatment with empiric broad-spectrum oral antibiotics. The patient was previously healthy with mild oesophageal reflux managed with esomeprazole. Plain radiograph showed no evidence of osteomyelitis. T1 weighted Gadolinium enhanced MRI showed phlegmonous change in the periosseous region of the 3rd proximal phalanx, MTP joint and the adjacent 3–4th phalanx interspace, with no intra-cortical oedema or evidence of osteomyelitis (see Fig. 1). Intraoperative findings included: vegetative matter extending to but not penetrating cortical bone and black deposits in the penetration track. The patient required substantial operative debridement and washout. The patient was commenced on voriconazole after receiving the direct microscopy results. Voriconazole was ceased on day 4 as the patient suffered severe visual hallucinations, nausea and diarrhoea. The drug interaction between voriconazole and proton pump inhibitor may have contributed to these side effects. In view of the clear intraoperative margins achieved, the patient was monitored clinically without further antifungal therapy and there was no clinical or radiological evidence of recurrence at 12 months.
Fig. 1.
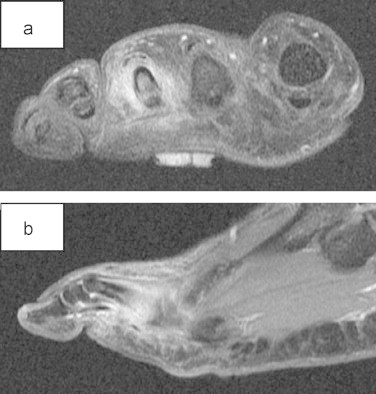
MRI showing phlegmonous change periosseous region proximal phalanx 3rd toe. (a) T1-weighted coronal MRI post Gadolinium and (b) T1-weighted sagittal MRI post Gadolinium contrast.
Histopathology showed fibroadipose tissue with reactive and foreign body changes.
The microbiology laboratory received 2 intraoperative specimens: 1 tissue (including embedded thorn remnants) and 1 swab. Direct Gram stain showed scant polymorphs and no organisms. A direct Blankophor BA (Bayer) fluorescent stain on the tissue sample (but not the swab) showed numerous irregular septate hyphae with irregular swollen round bodies. Routine mycology agar plates were inoculated: including antibiotics (chloramphenicol 0.16 g/L and gentamicin 0.16 g/L); BBL Sabouraud dextrose agar Emmons agar (SDA) incubated at 28 °C and 37 °C, BBL Mycosel agar (with cycloheximide 0.4 g/L) 28 °C and BBL brain heart infusion agar (37 °C). A pure growth of fungus was present on all plates for 2–4 days. The colony was initially pale coloured, fluffy, slightly raised and spreading. With age, the fungal cultures became darker and mottled with brown and grey colours and also produced some orange pigment. Reverse pigmentation was dark. After 3 weeks, fungal growth filled the petri dish. Initial microscopy of the isolate showed septate, pigmented hyphae, with swollen parts up to 4 μm in diameter.
The isolate was subcultured onto various agars including potato dextrose (PDA), Czapek-Dox and malt extract agars at 28 °C, 37 °C, 46 °C and 2% water agar at 28 °C (see Fig. 2a,b). Extended incubation of greater than 30 days showed the appearance of black spots in the mycelium. These appeared microscopically to be acervuli fruiting bodies. Also present were appressoria that were lobulated, had an entire margin and 2–3 guttulae (Fig. 2c). Conidia were straight, cylindrical with an obtuse apex and a truncated base (Fig. 2d). Conidial dimensions were 6–25×4–6 μm. Setae up to 200 μm long were produced and appeared septate, dark, thick-walled with pointed ends.
Fig. 2.
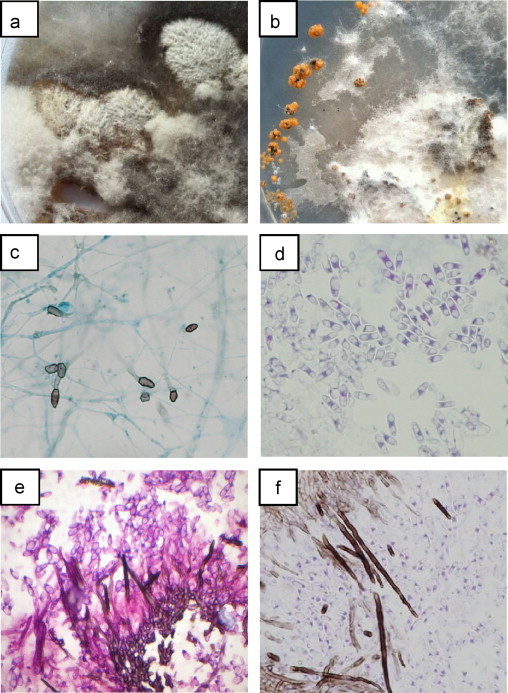
(a) Pure growth fungus on potato dextrose agar plate. (b) Orange pigmented bodies on Czapek-Dox agar plate. (c) Appressoria darkly pigmented with a complete margin, irregular edge, not deeply lobed (lactophenol cotton blue stain) Mag X788. (d) Conidia hyaline, straight, cylindrical, obtuse at the apex (H & E stain) Mag X788. (e) Vertical section of an acervular conidiomata with setae having pointed ends and conidia that are hyaline, straight, cylindrical, obtuse at the apex (PAS stain) Mag X 788. (f) As above with H&E stain.
Histological sections of the acervuli were prepared by fixation in 10% buffered formalin, then later embedded in paraffin blocks and 4 μm sections were microtomed and stained with Grocott's methenamine silver nitrate stain (GMS), periodic acid and Schiff's reagent (PAS) and haematoxylin and eosin (H&E) stains (Fig. 2e–f). Morphological features identified the isolate as a coelomycetous fungus, likely Colletotrichum. Bacterial cultures yielded no growth with extended incubation.
Molecular studies were utilised to assist with identification. DNA extraction was performed using Qiagen M48 Bio Robot in conjunction with Qiagen MagAttract DNA Mini M48 Kit. ITS region (ITS1 and ITS2) were amplified using the primers ITS 1 (TCCGTAGGTGAACCTGCGG) and ITS 2 (TCCTCCGCTTATTGATATG). PCR was carried out in the 25 μL reaction mixture containing PuReTaq bead (GE Healthcare), 18 μL H2O, 2.0 μL of each primer and 5 μL of DNA. 40 cycles of PCR amplification was performed using the G-Storm thermocycler. The amplification products (10 μL) were visualised after gel electrophoresis and staining with ethidium bromide under UV light in a transilluminator. The amplicon was purified with the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer's instruction for sequencing. Sequence analysis was performed using Applied Biosystem 3130 genetic analyser for sequencing incorporating forward and reverse sequencing reaction using the big dye terminator kits.
The fungus was identified as Glomerella cingulata (teleomorph stage of the anamorph Colletotrichum gloeosporioides) using the Basic Local Alignment Search BLAST search program www.ncbi.nlm.nih.gov/BLAST. The nucleotide sequence showed 96% homology match with accession number jq619481.1.
Susceptibility testing using SENSITITRE® YeastOne® Trek Diagnostics tray resulted in M.I.C levels (mg/L) of amphotericin B 0.5 mg/L, 5-fluorocytosine 16 mg/L, posaconazole 1 mg/L, voriconazole 0.12 mg/L, itraconazole 0.5 mg/L and fluconazole >256 mg/L. Results were difficult to interpret as the fungus was slow growing, endpoints were difficult to read with confidence, and there is no clinical correlation or validation in the literature.
3. Discussion
Deep soft tissue mycoses are relatively rare, but are an important consideration when there is a history of penetrating trauma with vegetative matter. Diagnosis can be optimised by the combination of clinical history, intraoperative findings, histopathology and microbiology.
Microbiological diagnosis can be optimised by the collection of multiple intraoperative tissue specimens. The value of intraoperative tissue specimens over swabs is illustrated by this case—the isolate only being isolated from the tissue specimen. Swabs are considered inferior to tissue or fluid/pus aspiration due to insufficient volume of specimen collected, inability to release specimen during processing, reduced microbial survival compared to aspirated fluid or pus [4].
Direct fungal stain using Blankophor BA (Bayer) provided a rapid diagnosis of subcutaneous mycosis, prompt initiation of antifungal therapy and close surgical review in this case. Blankophor and Calcofluor are white staining techniques which bind to chitin and cellulose and fluoresce on exposure to UV light facilitating rapid and reliable fungal detection in a variety of specimens [5,6].
Fungal identification relies on a combination of morphological features and molecular techniques. Identification of Colletrotrichum species in the past has utilised morphological features such as cultural characteristics, microscopic features including: conidial and appressoria appearance, presence of setae, scletoria, acervuli (size and shape), teleomorph state and host specificity in plants (see Table 1) [7]. These features have proven to be inadequate and unreliable due to the presence of few and variable characters, variability in morphology and phenotype due to environmental differences (such as agar types, incubation temperatures) and extensive host range. These challenges have led to inaccuracies in the literature, including incorrect identification, misnamed isolates, and variations in species concepts [8]. Susceptibility testing in these unusual isolates is also challenging, as there is a paucity of data available.
Table 1.
Morphological characteristics of the most clinically important Colletrotrichum species [7].
| Species | Sclerotia | Appressoria | Conidia |
|---|---|---|---|
| C. gloeosporoiodes | Absent | Margin entire, may be lobed | Straight, cylindrical, obtuse apex, base truncate 6–25×4–6 μm |
| C. coccodes | Present—globose | Margin entire | Straight, fusiform, each end abruptly tapered 16–22×3–4 μm |
| C. crassipes | Absent | Margin crenate or deeply lobed | Straight, cylindrical, obtuse apex, base truncate 11–20×6–8 μm |
| C. dematium | Present—conical | Margin entire or slightly irregularly lobed | Fusiform, falcate each end gradually tapered 19–25×2.5–3.5 μm |
| C. graminicola | Present—irregular | Margin very irregular | Fusiform, falcate, gradually tapered to the apex and base 24–28×4–6 μm |
Molecular sequencing also poses numerous challenges, including: poor standardisation of technique, variation of target region chosen, database quality, and changing taxonomy. Type strains of Colletotrichum have been found to be missing or in poor condition while many rDNA-ITS sequences have been wrongly named [9,10,11]. Cai et al. analysed rDNA-ITS sequences of 343C. gloeosporioides in GenBank and found conflicting results with 86% of these results.
Divergences of rDNA-internal transcribed spacer region (ITS) are also challenging for species distinction, hence C. gloeosporioides is more correctly under the umbrella C. gloeosporioides sensu lato. The ITS target is probably too conservative to accurately differentiate between taxa compared to other target genes [8]. Taxonomy of this genus is therefore under constant revision as molecular knowledge increases [8,12].
Fortunately, the patient described was cured with extensive initial debridement and has not required further surgery or recommencement of antifungal therapy at 12-month follow-up.
Conflict of interest
The authors declare no financial or personal conflicts of interest.
Acknowledgments
Kimiko Blendell and Nathan Hall from Anatomical Pathology Dept RNSH PaLMS for preparing the paraffin embedding, sections and stains of the fungi.
References
- 1.Castro L.G., da Silva Lacaz C., Guarro J., Gené J., Heins-Vaccari E.M., de Freitas Leite R.S. Phaeohyphomycotic cyst caused by Colletotrichum crassipes. Journal of Clinical Microbiology. 2001;39:2321–2324. doi: 10.1128/JCM.39.6.2321-2324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guarro J., Svidzinski T.E., Zaror L., Forjaz M.H., Gené J., Fischman O. Subcutaneous hyalohyphomycosis caused by Colletotrichum gloeosporioides. Journal of Clinical Microbiology. 1998;36:3060–3065. doi: 10.1128/jcm.36.10.3060-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinz T., Perfect J., Schell W., Ritter E., Ruff G., Serafin D. Soft tissue fungal infections: surgical management of 12 immunocompromised patients. Plastic and Reconstructive Surgery. 1996;97:1391–1399. doi: 10.1097/00006534-199606000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Wilson M.L., Winn W. Laboratory diagnosis of bone, joint, soft-tissue and skin infections. Clinical Infectious Diseases. 2008;46:453–457. doi: 10.1086/525535. [DOI] [PubMed] [Google Scholar]
- 5.Rüchel R., Margraf S. Rapid microscopic diagnosis of deep-seated mycoses following maceration of fresh specimens and staining with optical brighteners. Mycoses. 1993;36(239-42):6. doi: 10.1111/j.1439-0507.1993.tb00757.x. [DOI] [PubMed] [Google Scholar]
- 6.Monod M., Jaccoud S., Stirnimann R., Anex R., Villa F., Balmer S. Economical microscope configuration for direct mycological examination with fluorescence in dermatology. Dermatology. 2000;201:246–248. doi: 10.1159/000018496. [DOI] [PubMed] [Google Scholar]
- 7.Cano J., Guarro J., Gené J. Molecular and morphological identification of Colletotrichum species of clinical interest. Journal of Clinical Microbiology. 2004;42:2450–2454. doi: 10.1128/JCM.42.6.2450-2454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon P.F., Damm U., Johnston P.R., Weir B.S. Colletotrichum—current status and future directions. Studies in Mycology. 2012;73:181–213. doi: 10.3114/sim0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyde K.D., Cai L., McKenzie E.H.C., Yang Y.L., Zhang J.Z., Prihastuti H. Colletotrichum: a catologue of confusion. Fungal Diversity. 2009;39:1–17. [Google Scholar]
- 10.Hyde K.D., Cai L., Cannon P.F., Crouch J.A., Crous P.W., Damm U. Colletotrichum—names in current use. Fungal Diversity. 2009;39:147–182. [Google Scholar]
- 11.Cai L., Hyde K.D., Taylor P.W.J., Weir B.S., Waller J.M., Abang M.M. A polyphasic approach for studying Colletotrichum. Fungal Diversity. 2009;39:183–204. [Google Scholar]
- 12.Crouch J.A., Clarke B.B., Hillman B.I. What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate-spored graminicolous Colletotrichum group. Mycologia. 2009;101:648–656. doi: 10.3852/08-231. [DOI] [PubMed] [Google Scholar]


