The most basic aspects of hepatitis C virus (HCV) biology remain a mystery in part because studying HCV in its native environment, the human hepatocyte in the liver, has been fraught with technical challenges. Autofluorescence, low levels of virus replication and viral protein per infected cell, and limited availability of high quality tissue are just a few of the challenges complicating in situ detection of HCV RNA and antigen in the liver1, 2. The study by Kandathil et al., published in this issue, describes a clever and unique approach to circumvent these technical issues that have historically plagued efforts to understand HCV biology in the liver at single cell resolution.
Laser capture microdissection (LCM) was first reported in 1996 as a tool for selectively isolating cells of interest from a complex mixture of cells3. The technology originated at the National Cancer Institute’s Laboratory of Pathology as a means of cleanly capturing tumor or adjacent healthy tissue in patient biopsies for genomic analysis, thus melding classical pathology with the dawning era of genomics to create a new discipline often called “genomic diagnostics”. Using LCM, Kandathil et al. captured single hepatocytes (scLCM) from sections of liver biopsies from patients with chronic HCV (CHC) and probed each cell’s HCV RNA status retrospectively by qRT-PCR rather than attempting to identify infected cells prior to capture. Because the cells were acquired in a uniform grid-like array, “viroscapes” could be created charting the location and viral load of captured cells in their original spatial orientation. To maximize the number of hepatocytes tested, RNA from pairs of adjacent hepatocytes was combined such that the total amount of RNA assessed for viral load was equivalent to one cell with each pair of cells contributing one half. Additionally, HCV infection frequency was estimated by comparing the numbers of HCV RNA positive and negative cells per grid. With this approach, the authors confirmed earlier results by Liang et al. obtained using 2-photon immunofluorescence microscopy 2. Reassuringly, these two studies, using completely different methodologies, came to the same conclusion: HCV infected cells in the liver tend to be found in clusters2. The proportion of HCV RNA positive cells reported by Kandathil et al. (21–45%) was similar to that seen with antigen staining (7–20%)2. Due to the small amount of tissue sampled during biopsy in relation to the size of the liver, one would expect significant biopsy-to-biopsy variation within the same patient, and even greater differences between patients when performing high resolution studies like those of Kandathil et al. and Liang et al2. This makes the convergent results of the two studies even more striking. The fact that several studies1, 2 have now reported the focal nature of HCV RNA in patient liver suggests that HCV spread in vivo is likely from cell to adjacent cell or to cells in very close proximity. Given the high viral loads seen in chronically infected patients, it is somewhat surprising that this “inoculum” coursing through the blood in the liver does not create a more uniformly infected tissue. Thus, it will be very interesting to determine the biology behind the nature of HCV RNA clustering.
In addition to estimating infection frequency, the authors also quantitated the HCV copy number per cell (2–94.6 copies/cell). While the authors demonstrated their HCV qRT-PCR assay limit of quantitation to be 1 IU, the authors did not perform the exact same scLCM process on healthy donor liver sections to demonstrate that these samples were routinely below the limit of HCV RNA quantitation. Given the potential for captured cell contamination by viremic blood distributed throughout the tissue, one could imagine every cell evaluated would be positive for HCV RNA to varying degrees if the RNA isolation procedure was 100% efficient in recovery. A complementary approach, single molecule RNA fluorescence in situ hybridization (smRNA FISH), which does not require RNA isolation, could be used to confirm the HCV RNA clustering phenotype and potentially the viral load per cell in the very same tissue sections analyzed via scLCM. But even with very sensitive molecular techniques like qPCR and smFISH, it is difficult to accurately define cells at the limit of HCV genome quantitation. Are cells with one copy of HCV RNA truly infected or is this signal a result of contamination from blood borne HCV? Might a cell with one copy of HCV RNA today have 100 copies tomorrow? These questions highlight the difficulties associated with the study of HCV in patients. Hopefully with time, these questions can be addressed with new technologies.
Previous studies have characterized the host response in CHC patients via microarray of liver biopsy material4, 5. Since biopsy tissue contains a mixture of infected hepatocytes, non-infected hepatocytes and various non-parenchymal cells, these studies accurately reported the molecular signatures of the tissue but the signals originating from HCV infected cells were unclear. The molecular pathways influencing clearance or chronicity are likely to be found by determining host responses in HCV-infected and uninfected cells in the liver. Thus, defining key differences in the molecular pathways modulated within infected and uninfected cells may yield new angles for antiviral therapies. To this end, Kandathil et al. quantified gene expression levels in the LCM material in an attempt to draw a correlation between viral RNA load and the expression of interferon stimulated genes (ISGs) which are believed to affect HCV6. While studies using LCM and molecular approaches to determine HCV RNA copy number and inflammatory gene transcription in patient biopsy samples have been reported, the Kandathil study has greatly improved resolution (1 cell vs. >100 cells) and hence provides more detailed spatial and molecular information1. IFITM3 was the only ISG that could routinely be detected at the single cell level under their optimized PCR conditions. Interestingly, IFITM3 positive cells tended to be HCV RNA negative and HCV RNA positive cells tended to be IFITM3 negative though these associations were not deemed statistically significant. Unlike HCV RNA, IFITM3 expression was not clustered and appeared to be randomly dispersed.
Given that the Kandathil study was performed with a small, homogenous patient cohort (N = 4, 100% African American, 75% Male, 100% CHC, all HCV genotype 1), one can envision several reasons for the inefficient detection of ISG expression at the single cell level. First, the list of ISG targets measured, although exhibiting anti-HCV activity, may not be those most highly upregulated in these patients making their detection difficult. Secondly, RNA integrity in the samples, or the techniques used for RNA isolation and PCR may not have been sensitive enough to detect biologically relevant ISG expression at the single cell level. As new sequencing technologies improve, it may be possible to perform similar studies with less bias and greater sensitivity. Thirdly, due to the cohort size and homogeneity, the lack of ISG induction at the single cell level may be a function of the patients analyzed. Clinically unfavorable single nucleotide polymorphisms in interferon lambda (IFNL) loci are enriched in people of African heritage and are associated with poor clinical outcomes 7–9. Although patients with clinically unfavorable IFNL alleles have been reported to have higher intrahepatic ISG expression than patients with favorable alleles7, 8, it is not known what cell type(s) is primarily responsible for this phenotype (i.e. infected hepatocytes, bystander hepatocytes, non-parenchymal cells, inflammatory cells, etc.). The techniques developed by Kandathil et al. provide the field with an opportunity to gain deeper insight into these clinical phenotypes at the single cell level in hepatocytes and in non-parenchymal cells in the liver.
This study is a harbinger of a new genomic era where deep transcriptional profiling of HCV infected cells from patients at the resolution of a single cell seems increasingly less like science fiction. In the current issue of Gastroenterology, Kandathil et al. provide a proof of principle study demonstrating that HCV RNA and host gene expression can be measured at the resolution of the single cell in patient biopsy material. As the sensitivity of molecular techniques increases, similar translational studies in humans infected with HCV will hopefully become more common, as humans are the best but perhaps also the most challenging model for the study of HCV. Even though new directly acting antiviral drugs show immense promise in achieving cure in HCV infected patients, HCV is likely to remain a global public health concern due to drug resistance, cost and availability of treatment. Thus, studies to elucidate basic aspects of HCV pathogenesis should continue since they hold great promise for revealing both HCV-specific and more general liver disease mechanisms.
Future directions
The techniques described by Kandathil et al. can now be applied to more heterogeneous cohorts of patients. Similar studies on patients infected with other HCV genotypes will be especially interesting given viral genotype dependent differences in disease presentation. It will also be interesting to determine possible differences in HCV and ISG viroscapes in patients with low and high viral loads. Using serial sections from a single biopsy, it should be possible to construct a three dimensional view of the infection landscape in the organ simply by stacking viroscapes from each section. Capturing infected and bystander hepatocytes as well as non-parenchymal cells via scLCM on liver biopsies from patients of known favorable and unfavorable IFNL3 genotypes should provide clues as to the determinants that govern treatment response. As molecular techniques advance, data gleaned using techniques like Kandathil’s will increase exponentially to yield reliable transcriptional profile data from patient materials at single cell resolution. These techniques have the potential to revolutionize all disciplines of gastroenterology by allowing more precise molecular analyses of pathology yielding new insights and avenues for treatment.
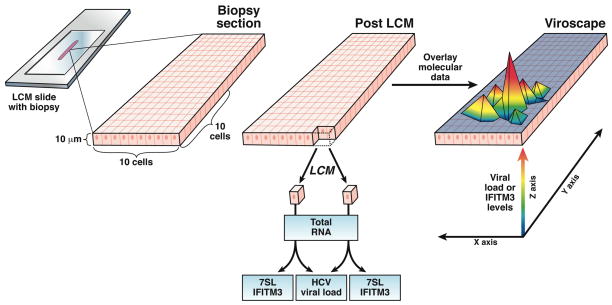
Figure. A schematic of the scLCM process.
10μm sections of fresh frozen liver biopsies were cut onto a PEN membrane LCM slide. Ten by ten grid-like arrays of single hepatocytes were cut from the section via LCM. Total RNA was extracted from each single cell. Levels of the housekeeping gene, 7SL, and the interferon stimulated gene, IFITM3, were measured via qPCR in each single cell. To measure HCV viral load, RNA from pairs of adjacent hepatocytes was combined such that the total amount of RNA assessed for viral load was equivalent to one cell with each pair of cells contributing one half. The molecular data was combined with the spatial information for each cell within a grid to create a “viroscape”.
Acknowledgments
Funding Sources: This work was supported by National Research Service Award F32 AI084448-01 (T.P.S.), National Institute for Diabetes, Digesting and Kidney Diseases 5R01DK085713-03 (C.M.R), and Clinical Translational Science Award (CTSA, RUCCTS Grant#8 UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS, NIH)).
Footnotes
Conflict of interest. The authors disclose no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stiffler JD, Nguyen M, Sohn JA, Liu C, Kaplan D, Seeger C. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, Vargas G, Lemon SM. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–58. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 3.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 4.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7034–9. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, Schweigler LM, Theodore D, Zacks SL, Liang TJ, Fried MW. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–63. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O’Brien TR. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nature genetics. 2013;45:164–71. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 9.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]