Abstract
AIM: To investigate the effect of bile acid on the expression of histidine decarboxylase (HDC), which is a major enzyme involved in histamine production, and gene expression of gastric transcription factors upon cooperative activation.
METHODS: HDC expression was examined by immunohistochemistry, reverse transcriptase polymerase chain reaction, and promoter assay in human gastric precancerous tissues, normal stomach tissue, and gastric cancer cell lines. The relationship between gastric precancerous state and HDC expression induced by bile acid was determined. The association between the expression of HDC and various specific transcription factors in gastric cells was also evaluated. MKN45 and AGS human gastric carcinoma cell lines were transfected with farnesoid X receptor (FXR), small heterodimer partner (SHP), and caudal-type homeodomain transcription factor (CDX)1 expression plasmids. The effects of various transcription factors on HDC expression were monitored by luciferase-reporter promoter assay.
RESULTS: Histamine production and secretion in the stomach play critical roles in gastric acid secretion and in the pathogenesis of gastric diseases. Here, we show that bile acid increased the expression of HDC, which is a rate-limiting enzyme of the histamine production pathway. FXR was found to be a primary regulatory transcription factor for bile acid-induced HDC expression. In addition, the transcription factors CDX1 and SHP synergistically enhanced bile acid-induced elevation of HDC gene expression. We confirmed similar expression patterns for HDC, CDX1, and SHP in patient tissues.
CONCLUSION: HDC production in the stomach is associated with bile acid exposure and its related transcriptional regulation network of FXR, SHP, and CDX1.
Keywords: Histamine, Histidine decarboxylase, Bile acid, Farnesoid X receptor
Core tip: Histamine production and secretion in the stomach play critical roles in gastric acid secretion and in the pathogenesis of gastric diseases. Bile acids are tumor promoters, and higher levels of bile acids are found in patients with atrophic chronic gastritis and intestinal metaplasia. Increased histidine decarboxylase (HDC), which is a histamine producing enzyme, has been detected in intestinal-type gastric carcinoma. In this study, we provide new evidence that bile acids induce HDC expression in gastric cells and a series of specific transcription factors (farnesoid X receptor, small heterodimer partner, and caudal-type homeodomain transcription factor 1) play critical roles in bile acid-mediated HDC induction.
INTRODUCTION
Histidine decarboxylase (HDC) is the only known enzyme that converts histidine to histamine[1], a bioamine that plays important roles in many physiological processes, including allergies, inflammation, neurotransmission, and gastric acid secretion[2-4]. In stomach cells, HDC promoter activity is upregulated by a variety of stimuli, including gastrin[5], phorbol 12-myristate 13-acetate[6], oxidative stress[7], thrombopoietin[8], Helicobacter pylori (H. pylori) infection[9], and pituitary adenylate cyclase-activating polypeptide[10].
HDC expression and histamine production play important roles in a number of physiological processes in many tissues; however, the functional regulation of these processes in the stomach is not yet defined. Gastric disease pathogenesis results from a combination of genetic alterations and various environmental factors, such as H. pylori infection, excessive salt intake, and low intake of vegetables and fruits[11,12]. Intestinal metaplasia is a premalignant condition of the stomach and is associated with chronic atrophic gastritis and gastric carcinoma[13]. In addition, an association between histamine production and intestinal metaplasia has been reported, and histamine deregulation is considered to be a risk factor for intestinal metaplasia of the stomach as well as an initiator of gastric carcinogenesis[14,15].
Caudal-related homeobox family (CDX1), is essential for the control of normal intestine differentiation and maintenance of the intestinal phenotype[16]. Although CDX1 is not expressed in normal stomach tissue, aberrant expression of these proteins causes gastric inflammation and intestinal metaplasia of the stomach, which induces gastric carcinoma[17-19]. Hence, CDX1 is the most likely candidate to be involved in the induction of intestinal metaplasia in the stomach.
Bile acids, such as deoxycholic acid (DCA) and chenodeoxycholic acid (CDCA), are tumor promoters, and higher levels of bile acids are found in patients with atrophic chronic gastritis and intestinal metaplasia in gastric carcinoma. Increased HDC expression has been detected in intestinal-type gastric carcinoma and dysplastic precursor lesions in the sequence of gastric carcinogenesis, suggesting a role for HDC in the early events associated with gastric carcinogenesis[20]. Here, we provide the first evidence that bile acids induce HDC expression by activating farnesoid X receptor (FXR), small heterodimer partner (SHP), and CDX1.
MATERIALS AND METHODS
Endoscopy and histological examination
We reviewed the upper gastrointestinal endoscopy records in our institution from June 2006 to December 2007. During this period, two biopsy specimens were taken from each of 33 patients for the evaluation of gastric mucosal inflammation status. Experienced endoscopists performed gastrointestinal endoscopy. Biopsy specimens were taken from the antrum and greater curvature of the middle corpus of all patients to evaluate the gastric mucosal status, including intestinal metaplasia. All endoscopic procedures were performed without sedation. Biopsy specimens were examined with hematoxylin and eosin staining. The specimens from the antrum and corpus were assessed for the presence of intestinal metaplasia, which was recognized morphologically by the presence of goblet cells, absorptive cells, and cells resembling colonocytes.
Cell culture and reagents
The MKN45 and AGS human gastric carcinoma cell lines were maintained in RPMI 1640 supplemented with 10% heat inactivated fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, United States) and 1% (v/v) penicillin-streptomycin (Gibco) at 37 °C in a humidified atmosphere containing 5% CO2 and fed every 2 d. DCA and CDCA were purchased from Sigma (St Louis, MO, United States). The cells were treated with 5 to 100 μmol/L DCA and CDCA in medium containing 20% FBS and 0.2% lactoalbumin hydrolysate for 24 h to avoid the confounding variable of serum-induced signaling.
Transient transfection assay and luciferase assay
MKN45 and AGS cells were seeded in a 24-well culture plate and transfected with reporter vector and β-galactosidase expression plasmids, along with each indicated expression plasmid using the Polyfect reagent (Qiagen, Valencia, CA, United States) and JetPEI (Polyplus-transfection, Illkirch, France), according to the manufacturer’s instructions. The total amount of plasmids was maintained by adding 100 ng of pcDNA3.1/His C. The cells were incubated with DNA precipitates for 18-24 h, washed or recovered in fresh media before treatment with bile acids, and maintained for 48 h before harvest. The basal luciferase reporter promoter activity in the presence of an empty expression vector was normalized to 1.0, and the activities of the remaining transfection reactions were expressed as fold activation of the basal promoter.
RNA isolation and reverse transcription-polymerase chain reaction
Total RNA from MKN45 and AGS cells was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s recommendation. Total RNA was converted into single-strand cDNA by Moloney murine leukemia virus reverse transcriptase (Promega) with random hexamer primers. A one-tenth aliquot of cDNA was subjected to polymerase chain reaction (PCR) amplification using gene-specific primers. The reverse transcription (RT)-PCR bands were quantified and normalized relative to the β-actin mRNA control band with ImageJ, version 1.35d (National Institutes of Health, Bethesda, MD, United States).
Statistical analysis
Statistical analyses were conducted with unpaired or paired t tests, as appropriate. All data are reported as mean ± SD. A P value of < 0.05 was considered significant.
RESULTS
Relative mRNA expression levels of HDC, cyclooxygenase 2, CDX1, SHP, and mucin 2 in gastritis patients with or without intestinal metaplasia
To compare the in vivo expression levels of intestinal metaplasia markers and transcription factors with that of HDC, human gastritis tissues were collected via endoscopic resection from each of 33 patients following informed consent. Patients were defined by an aberrant high level of bile acid in the bottom of their stomachs. The intestinal metaplasia region and gastritis mucosa were extracted via endoscopy, and pathological identities were verified via tissue examination. RT-PCR was conducted to determine the mRNA expression levels of HDC, cyclooxygenase 2 (COX-2), CDX1, SHP, and mucin (MUC)2 in mucosal tissues with or without intestinal metaplasia. Figure 1A shows that the mRNA expression levels of HDC, COX-2, CDX1, and SHP were significantly higher in intestinal metaplasia than in gastritis mucosa. As shown in Figure 1B, CDX1 and HDC were abundantly expressed in the intestinal metaplasia region. MUC2 expression, which is detected in goblet cells and considered to be an intestinal differentiation marker, was evaluated in cells displaying intestinal metaplasia. These results indicate that the concomitant expression of CDX1, SHP, and HDC may actually induce intestinal metaplasia via transcriptional regulation of precancerous target genes in addition to being a diagnostic marker.
Figure 1.
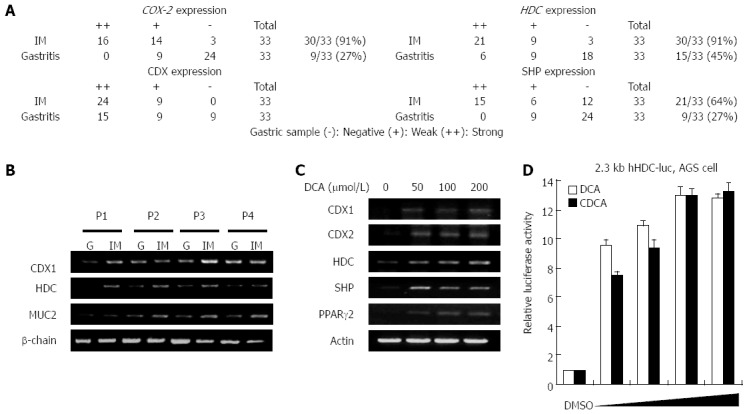
Relative expression levels of histidine decarboxylase in gastritis patients with or without intestinal metaplasia and bile acid-treated cells. A: mRNA expression patterns of cyclooxygenase 2 (COX-2), caudal-related homeobox family (CDX1), small heterodimer partner (SHP), and histidine decarboxylase (HDC) in 11 patients. Tissue samples for molecular analysis were collected from patients undergoing gastritis surgery or endoscopy for diagnosis of gastric disease. Numbers represent patient numbers showing the same intensity of mRNA expression; B: Total RNA was prepared from four gastric mucosa samples representing gastritis and intestinal metaplasia, after which CDX1, HDC, and mucin (MUC)2 mRNA levels were measured by reverse transcription-polymerase chain reaction (RT-PCR) with β-actin as a loading control; C: MKN45 cells were maintained either under control conditions (DMSO) or in the presence of various concentrations of bile acids (deoxycholic acid, DCA) for 24 h. Total RNA was prepared from the cells, after which mRNA levels of CDX, HDC, SHP, and peroxisome proliferator-activated receptor (PPAR)γ2 were assessed by RT-PCR; D: Human gastric adenocarcinoma (AGS) cells were transfected with the 2.3 kb HDC-luc promoter construct and maintained either under control conditions (DMSO) or in the presence of various concentrations (10, 20, 50, and 100 μmol/L) of bile acids (DCA; chenodeoxycholic acid, CDCA) for 24 h. Cells were harvested and analyzed for luciferase activity. The experiment was repeated three times in triplicate.
Bile acids increase histidine decarboxylase gene expression in gastric cells
To confirm bile acid-mediated HDC induction, we used RT-PCR to examine mRNA expression of HDC in the presence of increasing doses of DCA in MKN45 gastric cells. As shown in Figure 1C, DCA increased mRNA expression of HDC in a dose-dependent manner together with CDX1, SHP, and peroxisome proliferator-activated receptor (PPAR)-2. The results in Figure 1A-C show that a high level of bile acid induced HDC expression in gastric cells of intestinal metaplasia lesions as well as in the bile acid-treated gastric cell line.
To evaluate the effects of bile acids on HDC gene transcription in gastric cells, we compared human HDC gene promoter activity in the absence or presence of bile acids (DCA and CDCA) in AGS cells. Both bile acids significantly increased HDC promoter activation in a dose-dependent manner (Figure 1D). We observed the same results in MKN45 cells, another gastric cell line (data not shown). The effect of bile acids on HDC gene transcription was further verified by measuring mRNA expression via RT-PCR (data not shown). These results suggest that bile acids, which can induce intestinal metaplasia in the stomach, elevate HDC gene expression in gastric cells.
FXR mediates bile acid-induced HDC expression
Bile acids are known to act as a potent ligand for FXR. To confirm whether FXR plays a critical role as a transcriptional activator in the elevation of HDC gene transcription by bile acids, FXR-1 and FXR-2 were transfected into gastric cells along with a native hHDC promoter reporter. As shown in Figure 2A, both FXRs significantly increased transactivation of hHDC promoter activity. CDCA treatment with ectopic FXRα expression synergistically enhanced promoter activity as well as mRNA expression (Figure 2B). To further determine whether FXRα is involved in bile acid-mediated hHDC expression, we used the FXRα inhibitor z-guggulsterone. As predicted, z-guggulsterone inhibited FXRα and bile acid-enhanced transactivation of hHDC promoter activity (Figure 2C). The specific effects of the bile acids and z-gugglusterone were confirmed via RT-PCR assay (Figure 2D). These results suggest that FXRα is a major transcriptional regulator of bile acid-mediated hHDC gene induction.
Figure 2.
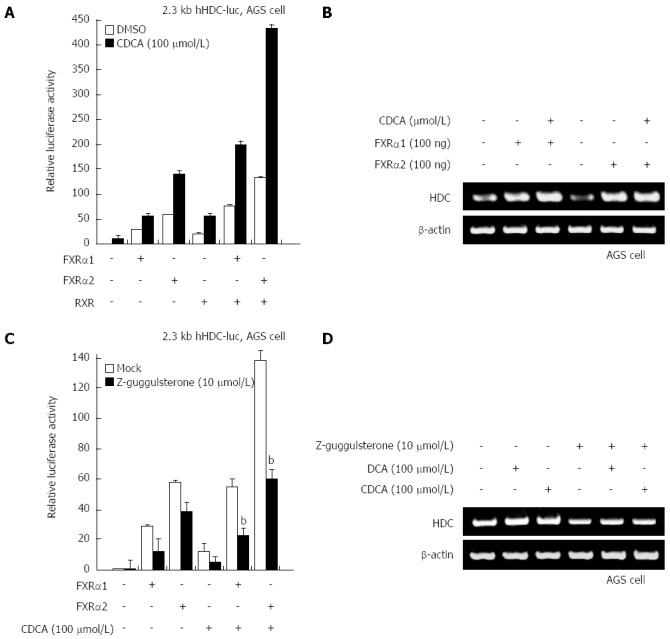
Farnesoid X receptor mediates bile acid-induced increase in histidine decarboxylase in gastric cancer cells. A: Human gastric adenocarcinoma (AGS) cells were cotransfected with the 2.3 kb histidine decarboxylase (HDC)-luc promoter construct and the indicated plasmids and then maintained either under control conditions or in the presence of chenodeoxycholic acid (CDCA). Cells were harvested and analyzed for luciferase activity; B: AGS cells were transfected with farnesoid X receptor (FXR)α1/2 expression plasmid and maintained either under control conditions or in the presence of CDCA. Total RNA was prepared from the cells, after which HDC mRNA levels were evaluated by reverse transcription-polymerase chain reaction (RT-PCR); C: AGS cells were cotransfected with the 2.3 kb HDC-luc construct and FXRα1/2 expression plasmid or in the presence of CDCA. Cells were then maintained either under control conditions or in the presence of z-guggulsterone. Cells were harvested and analyzed for luciferase activity. bP < 0.01 vs mock transfectants; D: AGS cells were treated with deoxycholic acid (DCA) or CDCA. Cells were maintained either under control conditions or in the presence of z-guggulsterone. Total RNA was prepared from the cells, after which HDC mRNA levels were determined by RT-PCR.
SHP is associated with bile acid-induced HDC expression
FXRα regulates transcriptional activation of SHP, which is indirectly dependent on ligand-binding of bile acids. To assess whether SHP increases HDC gene expression induced by bile acids, we examined hHDC promoter transactivation induced by ectopic expression of SHP in the absence and presence of DCA and CDCA. According to the results, higher expression of SHP significantly increased hHDC promoter activity both with and without bile acids (Figure 3A). DCA and CDCA treatment synergistically enhanced SHP-mediated HDC gene transactivation and mRNA expression (Figure 3B). Additional transfection of FXRα 1/2 with SHP also dramatically increased transactivation (Figure 3C) and mRNA expression (Figure 3D). A knock-down experiment using SHP siRNA confirmed that SHP increased bile acid- or FXRα 1/2-mediated transactivation of the hHDC gene promoter (Figure 3E) and mRNA expression (Figure 3F). These results suggest that SHP plays a critical role as a major transcriptional regulator of bile acid/FXRα-stimulated hHDC gene expression.
Figure 3.
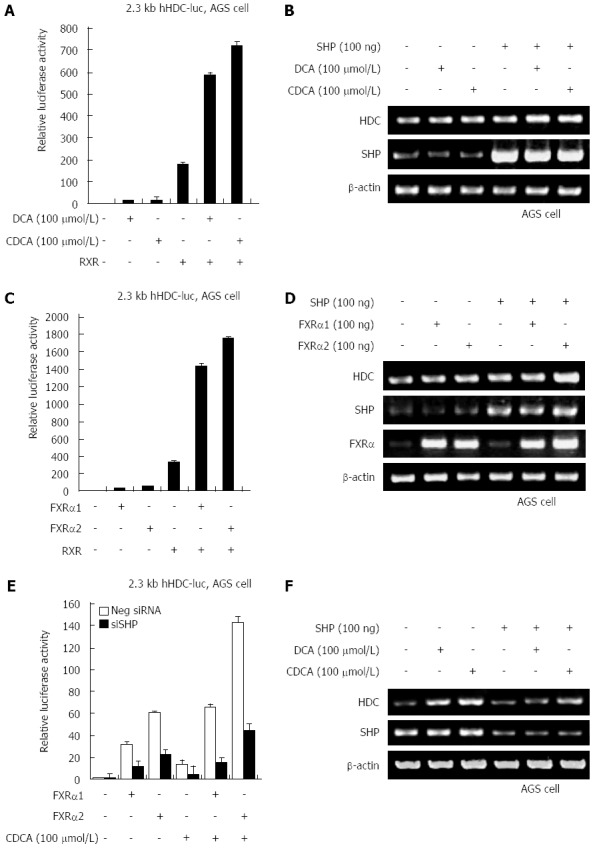
Small heterodimer partner is associated with bile acid-induced histidine decarboxylase transactivation. A: Human gastric adenocarcinoma (AGS) cells were cotransfected with the 2.3 kb histidine decarboxylase (HDC)-luc promoter construct and small heterodimer partner (SHP) expression plasmid in the presence of deoxycholic acid (DCA) or chenodeoxycholic acid (CDCA). Cells were analyzed for luciferase activity; B: AGS cells were cotransfected with the 2.3 kb HDC-luc promoter construct and expression plasmids for farnesoid X receptor (FXR)α1/2 and SHP. Cells were analyzed for luciferase activity; C: AGS cells were cotransfected with the 2.3 kb HDC-luc promoter construct and FXRα1/2 expression plasmid in the presence of CDCA. Cells were analyzed for luciferase activity; D: AGS cells were transfected with SHP expression plasmid in the presence of DCA or CDCA. Total RNA was prepared from the cells, after which HDC mRNA levels were determined by reverse transcription-polymerase chain reaction (RT-PCR); E: AGS cells were transfected with expression plasmids for FXRα1/2 and SHP. Total RNA was prepared from the cells, after which HDC mRNA levels were determined by RT-PCR; F: AGS cells were transfected with SHP siRNA construct in the presence of DCA or CDCA. Total RNA was prepared from the cells, after which HDC mRNA levels were determined by RT-PCR.
Bile acids induce HDC expression by CDX1/2, which is involved in the induction of intestinal metaplasia
In our previous report, SHP increased mRNA and protein expression of the CDX1 transcriptional regulator in gastric cells[21]. As bile acids increased SHP and HDC gene expression, we examined whether bile acids regulate CDX1/2 expression and transactivation activity. As shown in Figure 4A, ectopic expression of CDX1 and CDX2 significantly increased transactivation of the hHDC promoter induced by bile acid (DCA and CDCA) treatment. In particular, CDX1 was more responsible for bile acid-mediated hHDC gene transactivation. We further confirmed the CDX1/2 effect on hHDC mRNA induction via RT-PCR assay (Figure 4B). Additionally, knock-down of endogenous CDX1/2 expression by siRNA verified that CDX1/2 affected bile acid-mediated hHDC gene expression (Figure 4C and D). Furthermore, CDX1/2-mediated HDC induction was attenuated by expression of SHP siRNA, as shown in Figure 4E. These results suggest that CDX1/2 regulates bile acid-induced HDC expression in gastric cells.
Figure 4.
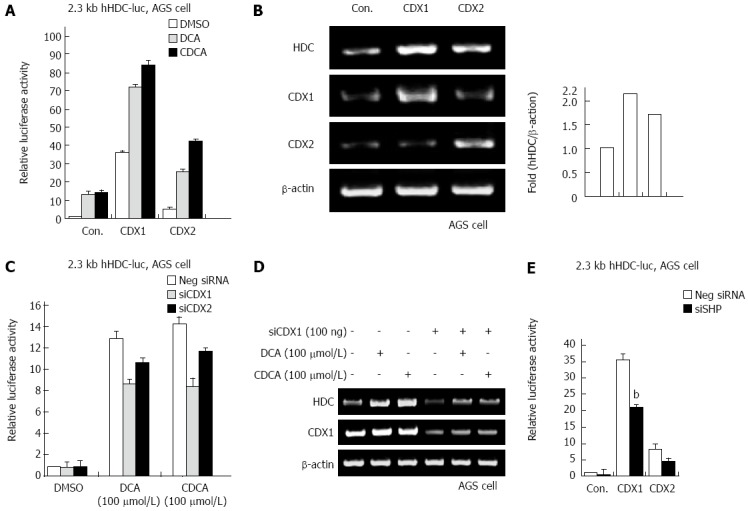
Bile acid induces histidine decarboxylase expression by caudal-related homeobox 1, which is mediated by small heterodimer partner. A: Human gastric adenocarcinoma (AGS) cells were cotransfected with the 2.3 kb histidine decarboxylase (HDC)-luc promoter construct and caudal-related homeobox (CDX)1 expression plasmid in the presence of deoxycholic acid (DCA) or chenodeoxycholic acid (CDCA). Cells were analyzed for luciferase activity; B: AGS cells were transfected with CDX1 or CDX2 expression plasmid for 48 h. Total RNA was prepared from the cells, after which HDC mRNA levels were determined by reverse transcription-polymerase chain reaction (RT-PCR); C: AGS cells were cotransfected with the 2.3 kb HDC-luc promoter construct and siCDX1/2 constructs and then maintained either under control conditions or in the presence of DCA or CDCA. Cells were analyzed for luciferase activity; D: AGS cells were transfected with siCDX1 construct in the presence of DCA or CDCA. Total RNA was prepared from the cells, after which HDC mRNA levels were determined by RT-PCR; E: AGS cells were cotransfected with the 2.3 kb HDC-luc promoter construct and CDX1/2 expression plasmid or siSHP. Cells were harvested and analyzed for luciferase activity. The experiment was repeated three times in triplicate. bP < 0.01 vs mock transfectants.
DISCUSSION
Increased expression of HDC has been observed in adults during growth or regeneration, including in rat liver tissue after partial hepatectomy[22], healing skin wound tissue[23], the small intestine after ischemia-reperfusion injury[24], and the gastric tissues surrounding restraint and cold-induced stress ulcerations[25]. Given the importance of histamine in normal physiology, it is not surprising that HDC gene expression is under tight regulatory control.
Histamine has been postulated as an autocrine growth factor for tumors. HDC is expressed in a number of cancers and tumor cell lines, and high concentrations of histamine can be detected in primary tumors such as colorectal[26] and breast cancers[27]. Elucidation of the steps leading to metaplasia is important for identifying early prognostic or diagnostic markers of tumor development. Chronic gastritis and associated intestinal metaplasia are common precancerous conditions of gastric carcinoma[28], and intestinal metaplasia of the stomach is a risk factor for developing intestinal-type gastric cancer. The most commonly recognized gut metaplasia is characterized by transdifferentiation of gastric epithelial cells to an intestinal phenotype. The intestinal metaplasia phenotype is also associated with expression of intestine-specific genes, including MUC2, villin, sucrase/isomaltase, carbonic anhydrase I, intestinal trefoil factor, and intestinal fatty acid-binding protein, within the gastric mucosa. Several other transcription factors, such as CDX1, CDX2, PDX1, and OCT1 have been detected in tissues with intestinal metaplasia.
Bile reflux into the stomach is significantly associated with intestinal metaplasia. Furthermore, bile reflux has been shown to directly induce intestinal metaplasia and progression to neoplasia in the gastrointestinal tract in animal models. Other studies have shown that bile reflux is the most important causative factor and initiator of gastric carcinogenesis. The presence of the nuclear bile acid receptor, FXR, has recently been reported in gastric cancer. Bile acids activate this receptor and increase SHP gene expression, resulting in the activation of a variety of signaling cascades along with promotion of cell proliferation. Previous studies on SHP expression have demonstrated that FXR acts as a nuclear factor-κB (NF-κB) transcription coactivator and is essential for NF-κB transactivation[28]. By altering the chemical environment of the mucosal surface, bile acids and histamine affect the pathogenic patterns of other damage-causing factors such as gastric acid and H. pylori, thereby potentiating their activities. Consequently, bile reflux and abnormal histamine production exert harmful effects on gastric mucosa.
COMMENTS
Background
Histamine production and secretion in the stomach play critical roles in gastric acid secretion and in the pathogenesis of gastric diseases. Histidine decarboxylase (HDC) is the only known enzyme that converts histidine to histamine. Increased HDC expression has been detected in intestinal-type gastric carcinoma. Bile acids are tumor promoters, and higher levels of bile acid occur in patients with atrophic chronic gastritis and intestinal metaplasia in gastric carcinoma.
Research frontiers
In this study, the authors identified a series of specific transcription factors for bile acid-mediated HDC induction. The activation of these transcription factors can be used as molecular markers for early diagnosis and evaluation of prognosis in histamine-related gastric diseases.
Innovations and breakthroughs
HDC expression and histamine production play important roles in a number of physiological processes in many tissues; however, the functional regulation in the stomach is not yet defined. Here, the authors provide the first evidence that bile acids induce HDC expression by activating farnesoid X receptor, small heterodimer partner, and caudal-type homeodomain transcription factor 1.
Applications
Histamine has been postulated to be an autocrine growth factor for some tumors. HDC is expressed in a number of cancers and tumor cell lines, and high concentrations of histamine can be detected in primary tumors such as colorectal and breast cancers. Understanding the steps leading to metaplasia is important for identifying early prognostic or diagnostic markers of gastric tumor development.
Terminology
Intestinal metaplasia is the transformation (metaplasia) of epithelium, usually of the stomach or the esophagus, to a type that bears some resemblance to the intestine as seen in Barrett’s esophagus.
Peer review
This paper describes the effect of bile acid on HDC gene expression (and other important genes involved in bile acid action) in biopsy specimens taken from antrum and corpus of normal stomach and gastric precancerous state as well as in gastric cell lines. The main thrust of the paper is that the authors claim to have demonstrated the positive association between HDC gene expression in the stomach and bile acid exposure. This paper well written and reports a potentially interesting and an important study.
Footnotes
Supported by National Research Foundation of Korea grant funded by the Korea government, No. 2009-0093193
P- Reviewers: Khurana S, Swierczynski JT, Wang DS S- Editor: Gou SX L- Editor: Webster JR E- Editor: Liu XM
References
- 1.Medina MA, Quesada AR, Núñez de Castro I, Sánchez-Jiménez F. Histamine, polyamines, and cancer. Biochem Pharmacol. 1999;57:1341–1344. doi: 10.1016/s0006-2952(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 2.Barocelli E, Ballabeni V. Histamine in the control of gastric acid secretion: a topic review. Pharmacol Res. 2003;47:299–304. doi: 10.1016/s1043-6618(03)00009-4. [DOI] [PubMed] [Google Scholar]
- 3.Gelfand EW. Role of histamine in the pathophysiology of asthma: immunomodulatory and anti-inflammatory activities of H1-receptor antagonists. Am J Med. 2002;113 Suppl 9A:2S–7S. doi: 10.1016/s0002-9343(02)01431-6. [DOI] [PubMed] [Google Scholar]
- 4.Rangachari PK, Prior T, Bell RA, Huynh T. Histamine potentiation by hydroxylamines: structure-activity relations; inhibition of diamine oxidase. Am J Physiol. 1992;263:G632–G641. doi: 10.1152/ajpgi.1992.263.5.G632. [DOI] [PubMed] [Google Scholar]
- 5.Lappin D, Moseley HL, Whaley K. Effect of histamine on monocyte complement production. II. Modulation of protein secretion, degradation and synthesis. Clin Exp Immunol. 1980;42:515–522. [PMC free article] [PubMed] [Google Scholar]
- 6.Höcker M, Zhang Z, Koh TJ, Wang TC. The regulation of histidine decarboxylase gene expression. Yale J Biol Med. 1996;69:21–33. [PMC free article] [PubMed] [Google Scholar]
- 7.Höcker M, Rosenberg I, Xavier R, Henihan RJ, Wiedenmann B, Rosewicz S, Podolsky DK, Wang TC. Oxidative stress activates the human histidine decarboxylase promoter in AGS gastric cancer cells. J Biol Chem. 1998;273:23046–23054. doi: 10.1074/jbc.273.36.23046. [DOI] [PubMed] [Google Scholar]
- 8.Pacilio M, Debili N, Arnould A, Machavoine F, Rolli-Derkinderen M, Bodger M, Arock M, Duménil D, Dy M, Schneider E. Thrombopoietin induces histidine decarboxylase gene expression in c-mpl transfected UT7 cells. Biochem Biophys Res Commun. 2001;285:1095–1101. doi: 10.1006/bbrc.2001.5296. [DOI] [PubMed] [Google Scholar]
- 9.Wessler S, Rapp UR, Wiedenmann B, Meyer TF, Schöneberg T, Höcker M, Naumann M. B-Raf/Rap1 signaling, but not c-Raf-1/Ras, induces the histidine decarboxylase promoter in Helicobacter pylori infection. FASEB J. 2002;16:417–419. doi: 10.1096/fj.01-0766fje. [DOI] [PubMed] [Google Scholar]
- 10.McLaughlin JT, Ai W, Sinclair NF, Colucci R, Raychowdhury R, Koh TJ, Wang TC. PACAP and gastrin regulate the histidine decarboxylase promoter via distinct mechanisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G51–G59. doi: 10.1152/ajpgi.00169.2002. [DOI] [PubMed] [Google Scholar]
- 11.Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003;3:592–600. doi: 10.1038/nrc1141. [DOI] [PubMed] [Google Scholar]
- 12.Walker MM. Is intestinal metaplasia of the stomach reversible? Gut. 2003;52:1–4. doi: 10.1136/gut.52.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller MS, Ezaki T, Guo RJ, Lynch JP. Cdx1 or Cdx2 expression activates E-cadherin-mediated cell-cell adhesion and compaction in human COLO 205 cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:G104–G114. doi: 10.1152/ajpgi.00484.2003. [DOI] [PubMed] [Google Scholar]
- 14.Dixon MF, Mapstone NP, Neville PM, Moayyedi P, Axon AT. Bile reflux gastritis and intestinal metaplasia at the cardia. Gut. 2002;51:351–355. doi: 10.1136/gut.51.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobala GM, O’Connor HJ, Dewar EP, King RF, Axon AT, Dixon MF. Bile reflux and intestinal metaplasia in gastric mucosa. J Clin Pathol. 1993;46:235–240. doi: 10.1136/jcp.46.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck F. The role of Cdx genes in the mammalian gut. Gut. 2004;53:1394–1396. doi: 10.1136/gut.2003.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukamoto T, Inada K, Tanaka H, Mizoshita T, Mihara M, Ushijima T, Yamamura Y, Nakamura S, Tatematsu M. Down-regulation of a gastric transcription factor, Sox2, and ectopic expression of intestinal homeobox genes, Cdx1 and Cdx2: inverse correlation during progression from gastric/intestinal-mixed to complete intestinal metaplasia. J Cancer Res Clin Oncol. 2004;130:135–145. doi: 10.1007/s00432-003-0519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutoh H, Sakurai S, Satoh K, Osawa H, Hakamata Y, Takeuchi T, Sugano K. Cdx1 induced intestinal metaplasia in the transgenic mouse stomach: comparative study with Cdx2 transgenic mice. Gut. 2004;53:1416–1423. doi: 10.1136/gut.2003.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silberg DG, Sullivan J, Kang E, Swain GP, Moffett J, Sund NJ, Sackett SD, Kaestner KH. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122:689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- 20.Saukkonen K, Rintahaka J, Sivula A, Buskens CJ, Van Rees BP, Rio MC, Haglund C, Van Lanschot JJ, Offerhaus GJ, Ristimaki A. Cyclooxygenase-2 and gastric carcinogenesis. APMIS. 2003;111:915–925. doi: 10.1034/j.1600-0463.2003.1111001.x. [DOI] [PubMed] [Google Scholar]
- 21.Park MJ, Kim KH, Kim HY, Kim K, Cheong J. Bile acid induces expression of COX-2 through the homeodomain transcription factor CDX1 and orphan nuclear receptor SHP in human gastric cancer cells. Carcinogenesis. 2008;29:2385–2393. doi: 10.1093/carcin/bgn207. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa E, Toki A, Moriyama T, Matsuoka Y, Aikawa T, Suda M. A study on the induction of histidine decarboxylase in tumor-bearing rat. J Biochem. 1970;68:347–358. doi: 10.1093/oxfordjournals.jbchem.a129365. [DOI] [PubMed] [Google Scholar]
- 23.Kahlson G, Rosengren E. New approaches to the physiology of histamine. Physiol Rev. 1968;48:155–196. doi: 10.1152/physrev.1968.48.1.155. [DOI] [PubMed] [Google Scholar]
- 24.Fujimoto K, Imamura I, Granger DN, Wada H, Sakata T, Tso P. Histamine and histidine decarboxylase are correlated with mucosal repair in rat small intestine after ischemia-reperfusion. J Clin Invest. 1992;89:126–133. doi: 10.1172/JCI115552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouclier M, Jung MJ, Gerhart F. Histamine receptor blockade (H2) versus inhibition of histamine synthesis in stress ulceration in rats. Eur J Pharmacol. 1983;90:129–132. doi: 10.1016/0014-2999(83)90224-8. [DOI] [PubMed] [Google Scholar]
- 26.Boér K, Darvas Z, Baki M, Kaszás I, Pál Z, Falus A. Expression of histidine decarboxylase in human colonic cancer cells and adenomatous polyps. Inflamm Res. 2003;52 Suppl 1:S76–S77. doi: 10.1007/s000110300062. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Caballero M, Neugebauer E, Rodriguez F, Nuñez de Castro I, Vara-Thorbeck C. Histamine synthesis and content in benign and malignant breast tumours. Its effects on other host tissues. Surg Oncol. 1994;3:167–173. doi: 10.1016/0960-7404(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 28.Kim YS, Han CY, Kim SW, Kim JH, Lee SK, Jung DJ, Park SY, Kang H, Choi HS, Lee JW, et al. The orphan nuclear receptor small heterodimer partner as a novel coregulator of nuclear factor-kappa b in oxidized low density lipoprotein-treated macrophage cell line RAW 264.7. J Biol Chem. 2001;276:33736–33740. doi: 10.1074/jbc.M101977200. [DOI] [PubMed] [Google Scholar]


