Abstract
AIM: To examine the status and clinical significance of anaplastic lymphoma kinase (ALK) gene alterations in hepatocellular carcinoma (HCC) patients.
METHODS: A total of 213 cases of HCC were examined by fluorescent in situ hybridization using dual color break-apart ALK probes for the detection of chromosomal translocation and gene copy number gain. HCC tissue microarrays were constructed, and the correlation between the ALK status and clinicopathological variables was assessed by χ2 test or Fisher’s exact test. Survival analysis was estimated using the Kaplan-Meier approach with a Log-rank test. Univariate and multivariate analyses of clinical variables were performed using the Cox proportional hazards regression model.
RESULTS: ALK gene translocation was not observed in any of the HCC cases included in the present study. ALK gene copy number gain (ALK/CNG) (≥ 4 copies/cell) was detected in 28 (13.15%) of the 213 HCC patients. The 3-year progression-free-survival (PFS) rate for ALK/CNG-positive HCC patients was significantly poorer than ALK/CNG-negative patients (27.3% vs 42.5%, P = 0.048), especially for patients with advanced stage III/IV (0% vs 33.5%, P = 0.007), and patients with grade III disease (24.8% vs 49.9%, P = 0.023). ALK/CNG-positive HCC patients had a significantly poorer prognosis than ALK/CNG-negative patients in the subgroup that was negative for serum hepatitis B virus DNA, with significantly different 3-year overall survival rates (18.2% vs 63.6%, P = 0.021) and PFS rates (18.2% vs 46.9%, P = 0.019). Multivariate Cox proportional hazards regression analysis suggested that ALK/CNG prevalence can predict death in HCC (HR = 1.596; 95%CI: 1.008-2.526, P = 0.046).
CONCLUSION: ALK/CNG, but not translocation of ALK, is present in HCC and may be an unfavorable prognostic predictor.
Keywords: Anaplastic lymphoma kinase gene, Hepatocellular carcinoma, Prognostic predictor
Core tip: This study retrospectively analyzed the status and clinical significance of anaplastic lymphoma kinase (ALK) gene alterations in a relatively large number of hepatocellular carcinoma (HCC) patients. We used HCC tissue microarrays to detect ALK transcripts by fluorescent in situ hybridization. No positive cases of ALK gene rearrangements and ALK amplification were observed. However, we found that ALK gene copy number gain (ALK/CNG) was common in HCC (13.15%, 28/213). Our findings suggest that ALK/CNG may serve as a prognostic marker for HCC, especially in patients with advanced stage, grade III pathology.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common human cancers and is the third leading cause of cancer-related death worldwide[1]. Only 30%-40% of patients who present with early-stage tumors are deemed eligible for curative intervention[2]. Although a combination of surgical resection, liver transplantation, and chemoembolization can be used to treat this disease, late-stage HCC is almost uniformly fatal[3,4]. With growing knowledge of the molecular pathways of carcinogenesis, several molecular-targeting drugs have been developed. Sorafenib, which is approved for renal cell carcinoma, is a multikinase inhibitor that targets vascular endothelial growth factor receptor (VEGFR)-2/3, platelet-derived growth factor receptor (PDGFR)-β, Raf, Flt-3 and c-Kit[4]. Phase II and III data have revealed improved overall survival (OS) in patients with advanced HCC following sorafenib treatment, establishing a new standard of care. The United States Food and Drug Administration has approved sorafenib for advanced HCC[4-6]. However, sorafenib has some limitations: the responses are not durable and its safety is questionable. Several potential novel anticancer agents are currently under investigation for the treatment of HCC, including bevacizumab, ramucirumab, sunitinib, everolimus and linifanib[6].
In 2007, Soda reported another type of tyrosine kinase with accelerated activity in a subset of patients with non-small cell lung cancer (NSCLC) resulting from a small inversion in chromosome 2p that created a fusion gene between echinoderm microtubule-associated protein-like 4 (EML4) and anaplastic lymphoma kinase (ALK)[7]. However, the prevalence of the fusion is low, and is present in approximately 5% of all NSCLC cases, depending on the population studied and screening methods used[8-10]. Simultaneous with the discovery of ALK translocation in NSCLC, crizotinib (PF-02341066, Pfizer, New York, NY, United States), originally in development as a Met inhibitor, was shown to have significant activity against EML4-ALK[11,12]. Therefore, efficient screening for the EML4-ALK fusion gene is a crucial issue in clinical practice.
In addition to NSCLC, ALK gene translocations are associated with other tumor types, including anaplastic large-cell lymphoma, neuroblastoma, and inflammatory myofibroblastic tumor[13-15]. The ALK gene copy number is also increased in NSCLC, neuroblastoma, and esophageal cancer[16-18]. Activating mutations and ALK amplification are the underlying mechanisms in sporadic tumors mainly of metastatic stages, and are associated with a subgroup of high-risk neuroblastoma[19,20].
In this retrospective study, in order to provide information for the first time on the association between ALK alterations and HCC, we examined the status of the ALK gene in 213 clinical samples using fluorescent in situ hybridization (FISH). The relationship between ALK status and HCC prognosis was investigated. Overall, our findings indicate that ALK gene copy number gain (ALK/CNG) may serve as an independent prognostic marker for predicting poor prognosis in HCC patients.
MATERIALS AND METHODS
Patients and sample collection
Two hundred and thirteen HCC patients who underwent surgical resection from 1999 to 2004 at Sun Yat-Sen University Cancer Center (Guangzhou, China) were enrolled. All patient data were from inpatient and outpatient medical records. All samples were pathologically confirmed by two pathologists. The cancer TNM stage was defined according to the 1997 American Joint Committee on Cancer staging system. Cancer histopathological classification was defined according to World Health Organization classification criteria. The study was approved by the Institutional Research Ethics Committee of the Sun Yat-Sen University Cancer Center.
Tissue microarray construction
We collected tissues from all 213 HCC cases which were then formalin-fixed and paraffin-embedded. Using hematoxylin and eosin-stained slides, a representative tumor site was chosen for tissue microarray (TMA). Details on TMA design and the staining procedure have been reported previously[21,22].
ALK FISH assay
Currently, there are three methods to detect ALK rearrangements: immunohistochemistry, FISH and reverse-transcriptase polymerase chain reaction (RT-PCR)[23,24]. FISH can be applied to formalin-fixed paraffin-embedded tissues[25] and was considered the gold standard for confirmation of ALK status in recent clinical trials of crizotinib[8].
The ALK gene copy number per cell was investigated by FISH using the Vysis ALK Break Apart FISH Probe Kit (Abbott Laboratories, Abbott Park, IL, United States) according to a published protocol with minor modifications[26]. Sections (4 μm thick) from the recipient blocks were mounted onto glass slides and stored at 4 °C for FISH staining. Signals for each locus-specific FISH probe were assessed under an Olympus BX51 TRF microscope (Olympus, Tokyo, Japan) equipped with a triple-pass filter (DAPI/Green/Orange; Vysis). FISH analysis was independently performed by pathologists who were blinded to the clinical characteristics and molecular variables of the patients.
Break-apart FISH uses a green centromeric probe and an orange telomeric probe on loci in chromosome 2. A fluorescence microscope equipped with appropriate excitation and emission filters allows visualization of the probes contained in the kit. Thus, if the ALK gene is split, the orange and green signals are separated; if the orange and green signals are adjacent or fused, yellow signals are seen under the filter. According to the manufacturer’s protocol, positivity consists of separation of these orange and green signals by two or more signal diameters, or a single orange signal without a corresponding green signal. A minimum of 50 nuclei was scored.
The criteria for ALK/CNG have not been established. A receiver operating characteristic (ROC) curve based on the detection of ALK/CNG in 213 HCC patients was used. The cutoff value was determined by optimal Youden’s index through analyzing the survivin ROC among patients. When ALK/CNG was 4, the sensitivity, specificity and Youden’s index were 35.6%, 97.1% and 0.327, respectively. The survival outcome of patients with ALK/CNG ≥ 4 was worse than in the other groups. Therefore, we defined ALK/CNG positivity as the presence of ≥ 4 copies of ALK per cell in ≥ 40 of the 100 analyzed cells.
Patient follow-up and classification of the cause of death
Clinical follow-up information was obtained from the medical records of inpatients or outpatients, as well as telephone interviews. Clinical follow-up visits with each patient were scheduled on a semi-annual basis. Follow-up information was complete for approximately 69% of all treated HCC patients. For deceased patients, we classified the underlying cause of death according to the death certificate. Local relapse was defined as the recurrence of HCC as determined by biopsy, and distant metastasis was defined by evidence on chest X-ray, computed tomography, abdominal ultrasound, or bone scan. The endpoints were OS and progression-free-survival (PFS). We calculated OS from the date of diagnosis (for non-randomized patients) or from randomization until death, or up to the last date of follow-up. For PFS, we recorded the time from diagnosis or randomization to the first locoregional or distant metastasis failure. The last follow-up date was December 6, 2012, and the median follow-up for the entire series was 42.47 mo (range, 0.87-164.93 mo). In total, 147 (69.0%) deaths had occurred at the end of the last follow-up.
Statistical analysis
The probabilities of OS and PFS were estimated using the Kaplan-Meier method, with a Log-rank test to detect differences. The correlation between ALK status and clinicopathological variables was assessed by χ2 test or Fisher’s exact test. The simultaneous prognostic effect of various factors was estimated using multivariate Cox proportional hazards regression models. The proportional hazards assumption for each covariate was tested by graphical methods, with no significant violations found. The relationships between histological classification and OS and PFS were estimated by hazard ratio with 95%CI. All statistical tests were two-sided, and P < 0.05 was considered statistically significant. All statistical analyses were performed with SPSS version 13.0 (SPSS, Chicago, IL, United States).
RESULTS
Patient characteristics
We retrospectively enrolled 213 patients with newly diagnosed, histologically confirmed, previously untreated HCC at Sun Yat-Sen University Cancer Center in Guangzhou, China, between January 1999 and December 2004. Of the 213 patients, 196 (92.0%) were male and 17 (8.0%) were female, with a median age of 49 years (range, 25-75 years). The clinicopathological characteristics of the 213 HCC cases are summarized in Table 1.
Table 1.
Clinicopathological characteristics of hepatocellular carcinoma patients n (%)
| Characteristics | Patients |
| Total | 213 (100) |
| Age (yr)a | |
| < 49 | 105 (49.3) |
| ≥ 49 | 108 (50.7) |
| Sex | |
| Female | 17 (8.0) |
| Male | 196 (92.0) |
| Grade | |
| I | 19 (8.9) |
| II | 97 (45.5) |
| III | 91 (42.7) |
| IV | 6 (2.8) |
| Stage | |
| I + II | 174 (81.7) |
| III + IV | 39 (18.3) |
| Relapse | |
| No | 83 (39.0) |
| Yes | 130 (61.0) |
| AFP (ng/mL) | |
| < 400 | 125 (58.7) |
| ≥ 400 | 88 (41.3) |
| Child-Pugh classification | |
| A | 195 (91.5) |
| B | 16 (7.5) |
| C | 2 (0.9) |
| HBV DNA | |
| Positive | 113 (53.1) |
| Negative | 100 (46.9) |
| Postoperative platelet count | |
| < 100 × 109/L | 35 (16.4) |
| ≥ 100 × 109/L | 178 (83.6) |
Median: 49, range: 25-75 years. AFP: Alpha fetoprotein; HBV: Hepatitis B virus.
Detection of ALK gene rearrangements and lack of association with patient characteristics
Among the 213 HCC patients, no positive cases of ALK gene rearrangements as detected by break-apart FISH were observed. Therefore, FISH signals were used to calculate the copy number for determination of ALK/CNG positivity. The criterion for ALK/CNG positivity was ≥ 4 copies per cell in ≥ 40 of 100 analyzed cells. Using this cutoff, 28 cases (13.15%) showed ALK/CNG positivity (Figure 1). No statistically significant association was observed between ALK/CNG and age, sex, grade, stage, relapse, α-fetoprotein (AFP) level, Child-Pugh classification, hepatitis B virus (HBV) status, or postoperative platelet count in the HCC patients (Table 2).
Figure 1.
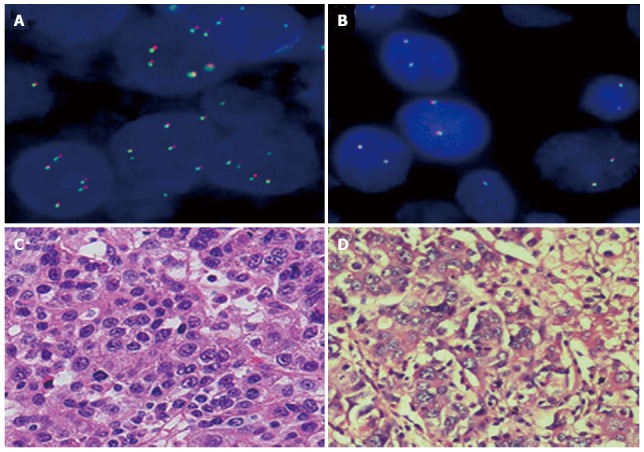
Representative fluorescent in situ hybridization images of tumors negative for anaplastic lymphoma kinase translocation. The two tumors varied in copy number, but both were negative for anaplastic lymphoma kinase (ALK) translocation, as indicated by the signals that neither had green and orange in close proximity or were fused to form a yellow locus. A: ALK/copy number gain (CNG)-positive cells (≥ 4 copies/cell); B: ALK/CNG-negative cells (< 4 copies/cell); C and D: HE staining of hepatocellular carcinoma cells from A and B (× 200).
Table 2.
Association between anaplastic lymphoma kinase copy number gain and clinicopathological characteristics of hepatocellular carcinoma patients n (%)
| Characteristics | Total cases |
ALK/CNG |
P value | |
| Positive (28 cases) | Negative (185 cases) | |||
| Age (yr) | ||||
| < 49 | 105 | 12 (11.4) | 93 (88.6) | 0.465 |
| ≥ 49 | 108 | 16 (14.8) | 92 (85.2) | |
| Sex | ||||
| Female | 17 | 3 (17.6) | 14 (82.4) | 0.843 |
| Male | 196 | 25 (12.8) | 171 (87.2) | |
| Grade | ||||
| I + II | 116 | 12 (10.3) | 104 (89.7) | 0.186 |
| III + IV | 97 | 16 (16.5) | 81 (83.5) | |
| Stage | ||||
| I + II | 174 | 21 (12.1) | 153 (87.9) | 0.326 |
| III + IV | 39 | 7 (17.9) | 32 (82.1) | |
| Relapse | ||||
| No | 83 | 13 (15.7) | 70 (84.3) | 0.385 |
| Yes | 130 | 15 (11.5) | 115 (88.5) | |
| AFP (ng/mL) | ||||
| < 400 | 125 | 19 (15.2) | 106 (84.8) | 0.290 |
| ≥ 400 | 88 | 9 (10.2) | 79 (89.8) | |
| Child-Pugh- classification | ||||
| A | 195 | 25 (12.8) | 170 (87.2) | 0.922 |
| B + C | 18 | 3 (16.7) | 15 (83.3) | |
| HBV DNA | ||||
| Positive | 113 | 17 (15.0) | 96 (85.0) | 0.383 |
| Negative | 100 | 11 (11.0) | 89 (89.0) | |
| Postoperative platelet count | ||||
| < 100 × 109/L | 35 | 2 (5.7) | 33 (94.3) | 0.250 |
| ≥ 100 × 109/L | 178 | 26 (14.6) | 152 (85.4) | |
ALK/CNG: Anaplastic lymphoma kinase copy number gain; AFP: Alpha fetoprotein; HBV: Hepatitis B virus.
ALK/CNG and HCC patient survival analyses and prognostic determinants
To determine the potential impact on survival of ALK aberrations, we compared the ALK/CNG with the OS and PFS rates. Among the 213 patients, the 3-year OS and PFS rates were 53.0% and 40.8%, respectively. The 3-year OS rates did not differ significantly between subgroups of ALK/CNG-positive and ALK/CNG-negative HCC patients (38.1% vs 57.6%, P = 0.089) (Figure 2A). However, the 3-year PFS rates showed a significant difference between ALK/CNG-positive and ALK/CNG-negative HCC patients (27.3% vs 42.5%, P = 0.048) (Figure 2B).
Figure 2.
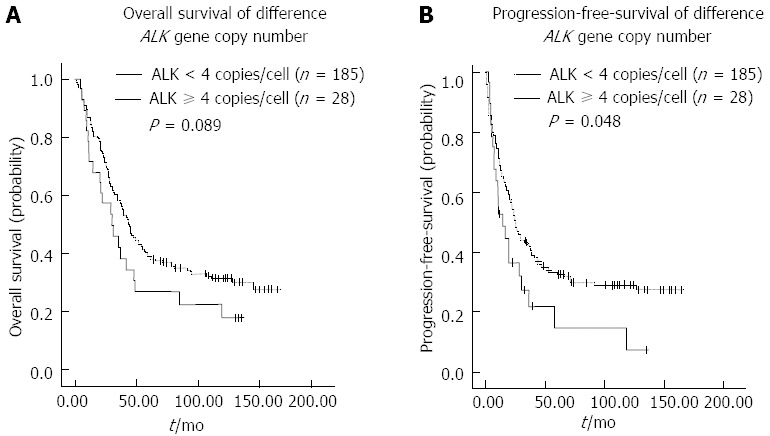
Kaplan-Meier survival curves of hepatocellular carcinoma patients. A: Overall survival of hepatocellular carcinoma (HCC) patients with ≥ 4 anaplastic lymphoma kinase (ALK) gene copies/cell did not differ significantly compared to patients with < 4 ALK gene copies/cell; B: Progression-free survival of HCC patients with ≥ 4 ALK gene copies/cell was significantly different from patients with < 4 ALK gene copies/cell.
To assess the prognostic ability of ALK/CNG at different stages of disease, we stratified the patients by clinical stage. In advanced stage (III and IV), the 3-year OS and PFS rates for patients were 33.3% and 27.4%, respectively. The 3-year OS rates did not differ significantly between subgroups of ALK/CNG-positive and ALK/CNG-negative HCC patients (0% vs 40.6%, P = 0.054) (Figure 3A). The 3-year PFS rates for ALK/CNG-positive patients was significantly poorer than that of the ALK/CNG-negative HCC patients (0% vs 33.5%, P = 0.007) (Figure 3B). However, in early stage (I and II) cancers, the 3-year OS (56.7% vs 61.8%, P = 0.394) and PFS rates (36.9% vs 44.4%, P = 0.314) were not significantly different between subgroups of ALK/CNG-positive and ALK/CNG-negative HCC patients (Figure 3C and D). These results suggest that ALK/CNG may serve as a reliable predictor for advanced, but not early stage disease.
Figure 3.
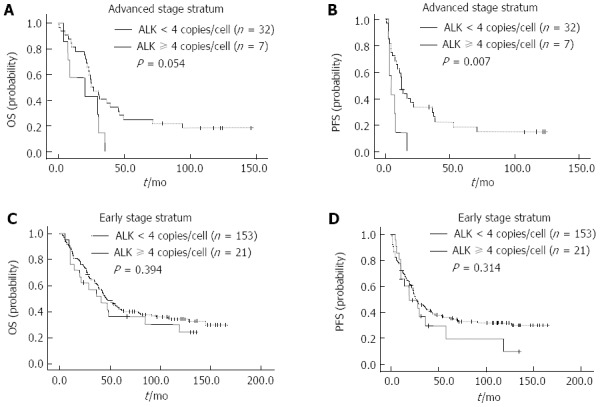
Kaplan-Meier survival curves of hepatocellular carcinoma patients classified according to stage stratum. A and B: In the advanced stage stratum ( IIIand IV), overall survival (OS) (A) and progression-free survival (PFS) (B) of hepatocellular carcinoma (HCC) patients with anaplastic lymphoma kinase copy number gain (ALK/CNG)-positivity and ALK/CNG-negativity; C and D: In the early stage stratum (I and II), OS (C) and PFS (D) of HCC patients with ALK/CNG-positivity and ALK/CNG-negativity.
We also assessed the prognostic ability of ALK/CNG for different pathological grades. In grade III, the 3-year OS and PFS rates for patients were 46.9% and 29.9%, respectively. The 3-year OS rates for ALK/CNG-positive patients was significantly poorer than that of ALK/CNG-negative patients (24.8% vs 49.9%, P = 0.023) (Figure 4A). The 3-year PFS rates did not differ significantly between subgroups of ALK/CNG-positive and ALK/CNG-negative HCC patients (0% vs 31.4%, P = 0.174) (Figure 4B). However, the 3-year OS (45.5% vs 58.8%, P = 0.440) and PFS rates (0% vs 43.6%, P = 0.109) did not differ significantly between subgroups of ALK/CNG-positive and ALK/CNG-negative HCC patients with grade II pathology (Figure 4C and D), suggesting that ALK/CNG status has greater prognostic ability for more advanced histological grades.
Figure 4.
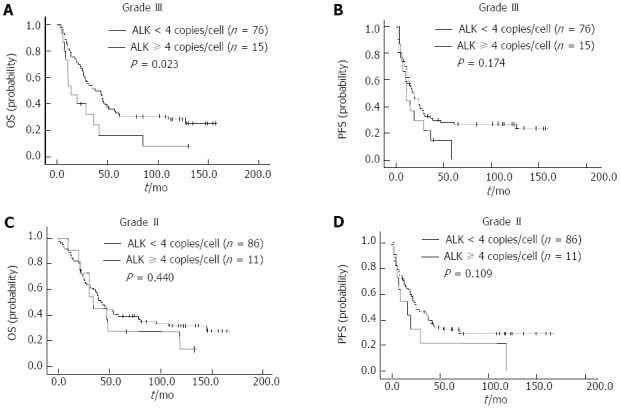
Kaplan-Meier survival curves of hepatocellular carcinoma patients classified according to pathological grade. A and B: In grade III disease, overall survival (OS) (A) and progression-free survival (PFS) (B) of hepatocellular carcinoma (HCC) patients with anaplastic lymphoma kinase copy number gain (ALK/CNG)-positivity and ALK/CNG-negativity; C and D: In grade II disease, the OS (C) and PFS (D) of HCC patients with ALK/CNG-positivity and ALK/CNG-negativity.
After stratification by HBV status, ALK/CNG remained a significant predictor of OS and PFS in HCC patients who were negative for HBV DNA. In the HBV-DNA-negative group, 3-year OS and PFS rates were 58.5% and 43.6%, respectively. The 3-year OS and PFS rates for ALK/CNG-positive patients was significantly poorer than that of the ALK/CNG-negative HCC patients (18.2% vs 63.6%, P = 0.021 and 18.2% vs 46.9%, P = 0.019, respectively) (Figure 5A and B). However, the 3-year OS (52.3% vs 51.0%, P = 0.977) and PFS rates (22.8% vs 36.3%, P = 0.813) did not differ significantly between subgroups of ALK/CNG-positive and ALK/CNG-negative HCC patients with HBV DNA (Figure 5C and D).
Figure 5.
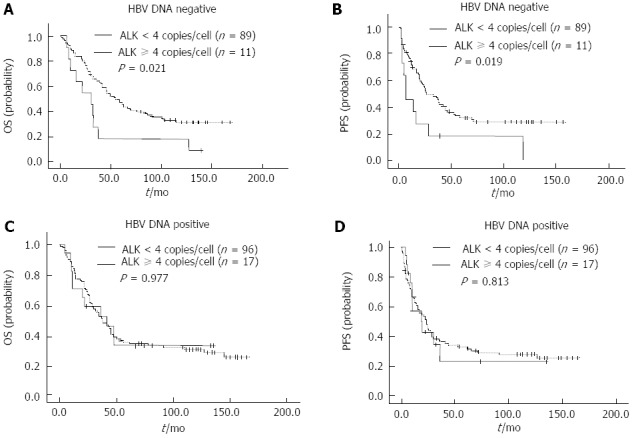
Kaplan-Meier survival curves of hepatocellular carcinoma patients classified according to hepatitis B virus status. A and B: In hepatocellular carcinoma (HCC) patients negative for serum hepatitis B virus (HBV) DNA, the overall survival (OS) (A) and progression-free survival (PFS) (B) of HCC patients with anaplastic lymphoma kinase copy number gain (ALK/CNG)-positivity and ALK/CNG-negativity; C and D: The OS (C) and PFS (D) in HCC patients positive for serum HBV DNA.
When stratified by AFP level, Child-Pugh classification and postoperative platelet count, ALK/CNG was not related to HCC patient survival.
Univariate and multivariate Cox proportional hazard regression analysis for OS and PFS were conducted using age, sex, AFP, grade, stage, Child-Pugh classification, HBV status, postoperative platelet count and ALK/CNG as variables. Univariate analysis for OS suggested that ALK/CNG was not a significant predictive factor for HCC (HR = 1.479, 95%CI: 0.939-2.329, P = 0.091) (data not shown). However, in the univariate analysis for PFS, ALK/CNG (HR = 1.598, 95%CI: 1.001-2.552, P = 0.049), Child-Pugh classification (HR = 2.165, 95%CI: 1.300-3.608, P = 0.002) and stage (HR = 1.926, 95%CI: 1.304-2.845, P = 0.001) were predictive factors for HCC patients. Furthermore, multivariate analysis supported a role for ALK/CNG (HR = 1.596, 95%CI: 1.008-2.526, P = 0.046), Child-Pugh classification (HR = 1.744, 95%CI: 1.021-2.979, P = 0.042) and stage (HR = 1.775, 95%CI: 1.179-2.673, P = 0.006) as independent predictors for PFS in HCC patients (Table 3).
Table 3.
Cox regression analysis of progression-free-survival in hepatocellular carcinoma patients
| Variables | Subset | HR (95%CI) | P value |
| Univariate analysis (n = 213) | |||
| Age (yr) | < 49 vs ≥ 49 | 1.153 (0.834-1.596) | 0.389 |
| Sex | Female vs male | 1.464 (0.770-2.783) | 0.245 |
| AFP (ng/mL) | < 400 vs ≥ 400 | 1.219 (0.879-1.690) | 0.236 |
| Grade | I + II vs III + IV | 1.335 (0.966-1.847) | 0.080 |
| Stage | I + II vs III + IV | 1.926 (1.304-2.845) | 0.001 |
| ALK/CNG | Yes vs no | 1.598 (1.001 -2.552) | 0.049 |
| Child-Pugh classification | A vs B + C | 2.165 (1.300-3.608) | 0.002 |
| HBV DNA | Positive vs negative | 1.067 (0.771-1.476) | 0.696 |
| Postoperative plateletcount | < 100 × 109/L vs ≥ 100 × 109/L | 1.267 (0.839-1.914) | 0.260 |
| Multivariate analysis (n = 213) | |||
| Stage | I + II vs III + IV | 1.775 (1.179-2.673) | 0.006 |
| ALK/CNG | Yes vs no | 1.596 (1.008-2.526) | 0.046 |
| Child-Pugh classification | A vs B + C | 1.744 (1.021-2.979) | 0.042 |
ALK/CNG: Anaplastic lymphoma kinase copy number gain; AFP: Alpha fetoprotein; HBV: Hepatitis B virus.
DISCUSSION
HCC has a high therapeutic failure rate and a low median survival rate due to the aggressive nature of the disease[27]. To improve patient survival, it is important to seek new therapeutic methods. Targeted therapies provide hope for the future of cancer therapy. The key signaling pathways that have been found in the pathogenesis of HCC include those mediated by epidermal growth factor receptor, VEGFR, PDGFR, insulin-like growth factor receptor, and the Ras/Raf/MEK/PTEN/mTOR pathways[4]. Therefore, it is important to select targeted therapy strategies according to the specific clinicopathological features of the tumors[28,29]. In the present study, we investigated, for the first time, ALK gene aberrations in a large series of HCC tissue samples.
ALK belongs to the insulin receptor superfamily of tyrosine kinase receptors and is activated by fusion with a variety of other genes, such as NPM, EML4, TMP2 and KIF5B[30]. In our study, we used TMAs to detect ALK transcripts in a large number of clinical samples by FISH. TMA is a practical and valuable tool, particularly in performing large-scale analyses. Compared with conventional approaches, this technology can facilitate the standardization of FISH and has been proven to markedly decrease the time and cost involved[31]. As a disadvantage, TMA may be limited to tumor materials of sufficient size and quality, and therefore, genetic aberrations may be missed. In addition, samples in which the cells harboring increased gene copy numbers are scattered on TMA sections may be difficult to detect.
We did not detect ALK rearrangement in HCC patients. It has been reported that the EML4-ALK fusion transcript is not detected in liver and gastrointestinal cancer by RT-PCR[32]. Therefore, the target proportion may be limited. However, we found that ALK/CNG was common in HCC. This is believed to be the first study to detect ALK/CNG using FISH in a large number of HCC tissue samples. We used ≥ 4 copies per cell in ≥ 40 of 100 cells analyzed as a cut-off for ALK/CNG positivity. Our findings showed that the survival rate of ALK/CNG-positive patients was lower than that of ALK/CNG-negative patients (27.3% vs 42.5%, P = 0.048). In the present study, ALK/CNG was significantly associated with tumor progression of HCC. After stratification by clinical stage, ALK/CNG remained a significant predictor of HCC prognosis in the advanced stage (III and IV) (0% vs 33.5%, P = 0.007), but not a significant predictor in the early stage (I and II). Univariate and multivariate Cox proportional hazards survival analysis suggested that ALK/CNG had a significantly worse prognostic impact on PFS in HCC patients (HR = 1.596, 95%CI: 1.008-2.526, P = 0.046). These results indicate that ALK/CNG may serve as a prognostic marker for survival of HCC patients.
ALK amplification as an oncogenic event was reported first in neuroblastoma in 2002[33,34], and subsequent investigations suggested that neuroblastoma may benefit from ALK inhibitors[14,35,36]. Recently, ALK was also reported to be amplified in esophageal cancer (11.1%) and brain metastases of NSCLC (11.0%)[18,37]. Additionally, ALK gene amplification was recently demonstrated in inflammatory breast cancer, and low doses of the ALK inhibitor, crizotinib, were shown to promote tumor shrinkage in mouse xenograft models[38]. Compared with mutation or amplification, ALK/CNG appears to be more common, and in fact may occur in 15%-20% of cases of neuroblastoma[32,39]. ALK gene amplifications and copy number gains were initially reported at frequencies as high as 74% in NSCLC, but there was no associated prognostic relevance[17], and the influence on the levels of ALK protein expression for NSCLC remains to be elucidated. Shao et al[40] reported that expression of ALK gene was examined in two cases of HCC by immunohistochemistry, and that ALK gene may be involved in the origin and development of HCC. Future studies will be needed to investigate ALK expression level and correlate it to ALK gene copy number in HCC. Therefore, although this report is believed to be the first to describe ALK/CNG in HCC, there is a basis for the prominence and role of ALK/CNG in other cancers, which further supports the potential use of ALK/CNG as both a prognostic marker and therapeutic target for HCC. Recent findings on neuroblastoma suggest that aberrant activation of the ALK gene may contribute to tumor development through copy number alterations, and specific ALK-targeting drugs inhibit cell proliferation and induce apoptosis[14,41]. We speculated that the mechanism of ALK/CNG that affect prognosis may be associated with tumor cell proliferation and apoptosis in HCC. Hopefully, this hypothesis will be studied in the future.
In conclusion, ALK/CNG, but not ALK translocation, was frequently detected in HCC patients. ALK/CNG may be a potential prognostic marker for HCC patients with advanced stage, grade III, tumors or serum HBV DNA negativity. We will continue to investigate whether patients with ALK/CNG can be candidates for ALK inhibitor therapy in the future.
COMMENTS
Background
Hepatocellular carcinoma (HCC) is one of the most common human cancers and is the third leading cause of cancer-related death worldwide. Targeted therapies provide hope for the future of cancer therapy. The anaplastic lymphoma kinase (ALK) inhibitor, crizotinib, has significant activity in non-small cell lung cancer (NSCLC). ALK gene translocations are also associated with other tumor types. Therefore, it is important to select targeted therapy strategies according to the specific clinicopathological features of the tumors.
Research frontiers
ALK belongs to the insulin receptor superfamily of tyrosine kinase receptors. The prevalence of the fusion was found in approximately 5% of all NSCLC cases. In this study, the authors investigated the status and clinical significance of ALK gene alterations in HCC patients by fluorescent in situ hybridization (FISH).
Innovations and breakthroughs
In addition to NSCLC, ALK gene translocations were recently demonstrated in anaplastic large-cell lymphoma, neuroblastoma, and inflammatory myofibroblastic tumor. The ALK gene copy number is also increased in NSCLC, neuroblastoma, and esophageal cancer. In the present study, the authors investigated ALK gene aberrations in a large series of HCC tissue samples using FISH. The results suggest that ALK gene copy number gain (≥ 4 copies) may serve as a potential prognostic marker for HCC patients, especially for those with advanced stage, grade III, tumors.
Applications
The results suggested that ALK gene copy number gain, but not ALK translocation, was frequently detected in HCC patients, and may serve as an independent prognostic marker for predicting poor prognosis in HCC patients.
Terminology
ALK gene copy number gain (ALK/CNG): The criteria for ALK/CNG have not been established, and a receiver operating characteristic curve based on the detection of ALK/CNG in HCC patients was analyzed. Therefore, authors defined ALK/CNG positivity as the presence of ≥ 4 copies of ALK per cell in ≥ 40 of the 100 analyzed cells.
Peer review
The authors examined the status and clinical significance of ALK gene alterations in HCC patients. Unfortunately, there were no positive cases for ALK gene rearrangements as detected by break-apart FISH. However, ALK/CNG had a significantly worse prognostic impact on progression-free survival of HCC patients. The findings may be relevant to clinical practice. Overall, this manuscript is highly relevant and interesting.
Footnotes
P- Reviewers: Baba H, Iwasaki Y, Jin S S- Editor: Gou SX L- Editor: Webster JR E- Editor: Wu HL
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Lu Y, Wang C, Bai W, Qu J, Chen Y, Chang X, An L, Zhou L, Zeng Z, et al. Cryotherapy is associated with improved clinical outcomes of sorafenib for the treatment of advanced hepatocellular carcinoma. Exp Ther Med. 2012;3:171–180. doi: 10.3892/etm.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek YH, Kim KT, Lee SW, Jeong JS, Park BH, Nam KJ, Cho JH, Kim YH, Roh YH, Lee HS, et al. Efficacy of hepatic arterial infusion chemotherapy in advanced hepatocellular carcinoma. World J Gastroenterol. 2012;18:3426–3434. doi: 10.3748/wjg.v18.i26.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cervello M, McCubrey JA, Cusimano A, Lampiasi N, Azzolina A, Montalto G. Targeted therapy for hepatocellular carcinoma: novel agents on the horizon. Oncotarget. 2012;3:236–260. doi: 10.18632/oncotarget.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka S, Arii S. Molecular targeted therapies in hepatocellular carcinoma. Semin Oncol. 2012;39:486–492. doi: 10.1053/j.seminoncol.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Frenette C, Gish R. Targeted systemic therapies for hepatocellular carcinoma: clinical perspectives, challenges and implications. World J Gastroenterol. 2012;18:498–506. doi: 10.3748/wjg.v18.i6.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 8.Murakami Y, Mitsudomi T, Yatabe Y. A Screening Method for the ALK Fusion Gene in NSCLC. Front Oncol. 2012;2:24. doi: 10.3389/fonc.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vincent MD, Kuruvilla MS, Leighl NB, Kamel-Reid S. Biomarkers that currently affect clinical practice: EGFR, ALK, MET, KRAS. Curr Oncol. 2012;19:S33–S44. doi: 10.3747/co.19.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki T, Rodig SJ, Chirieac LR, Jänne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010;46:1773–1780. doi: 10.1016/j.ejca.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou SH. Crizotinib: a novel and first-in-class multitargeted tyrosine kinase inhibitor for the treatment of anaplastic lymphoma kinase rearranged non-small cell lung cancer and beyond. Drug Des Devel Ther. 2011;5:471–485. doi: 10.2147/DDDT.S19045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mossé YP, Wood A, Maris JM. Inhibition of ALK signaling for cancer therapy. Clin Cancer Res. 2009;15:5609–5614. doi: 10.1158/1078-0432.CCR-08-2762. [DOI] [PubMed] [Google Scholar]
- 13.Kutok JL, Aster JC. Molecular biology of anaplastic lymphoma kinase-positive anaplastic large-cell lymphoma. J Clin Oncol. 2002;20:3691–3702. doi: 10.1200/JCO.2002.12.033. [DOI] [PubMed] [Google Scholar]
- 14.George RE, Sanda T, Hanna M, Fröhling S, Luther W, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulford K, Lamant L, Espinos E, Jiang Q, Xue L, Turturro F, Delsol G, Morris SW. The emerging normal and disease-related roles of anaplastic lymphoma kinase. Cell Mol Life Sci. 2004;61:2939–2953. doi: 10.1007/s00018-004-4275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Łastowska M, Viprey V, Santibanez-Koref M, Wappler I, Peters H, Cullinane C, Roberts P, Hall AG, Tweddle DA, Pearson AD, et al. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene. 2007;26:7432–7444. doi: 10.1038/sj.onc.1210552. [DOI] [PubMed] [Google Scholar]
- 17.Salido M, Pijuan L, Martínez-Avilés L, Galván AB, Cañadas I, Rovira A, Zanui M, Martínez A, Longarón R, Sole F, et al. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J Thorac Oncol. 2011;6:21–27. doi: 10.1097/JTO.0b013e3181fb7cd6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoppmann SF, Streubel B, Birner P. Amplification but not translocation of anaplastic lymphoma kinase is a frequent event in oesophageal cancer. Eur J Cancer. 2013;49:1876–1881. doi: 10.1016/j.ejca.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carén H, Abel F, Kogner P, Martinsson T. High incidence of DNA mutations and gene amplifications of the ALK gene in advanced sporadic neuroblastoma tumours. Biochem J. 2008;416:153–159. doi: 10.1042/bj20081834. [DOI] [PubMed] [Google Scholar]
- 21.Holm C, Rayala S, Jirström K, Stål O, Kumar R, Landberg G. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98:671–680. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 22.Wang HY, Li YY, Shao Q, Hou JH, Wang F, Cai MB, Zeng YX, Shao JY. Secreted protein acidic and rich in cysteine (SPARC) is associated with nasopharyngeal carcinoma metastasis and poor prognosis. J Transl Med. 2012;10:27. doi: 10.1186/1479-5876-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thunnissen E, Bubendorf L, Dietel M, Elmberger G, Kerr K, Lopez-Rios F, Moch H, Olszewski W, Pauwels P, Penault-Llorca F, et al. EML4-ALK testing in non-small cell carcinomas of the lung: a review with recommendations. Virchows Arch. 2012;461:245–257. doi: 10.1007/s00428-012-1281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bang YJ. The potential for crizotinib in non-small cell lung cancer: a perspective review. Ther Adv Med Oncol. 2011;3:279–291. doi: 10.1177/1758834011419002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soda M, Isobe K, Inoue A, Maemondo M, Oizumi S, Fujita Y, Gemma A, Yamashita Y, Ueno T, Takeuchi K, et al. A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin Cancer Res. 2012;18:5682–5689. doi: 10.1158/1078-0432.CCR-11-2947. [DOI] [PubMed] [Google Scholar]
- 26.Li YH, Wang F, Shen L, Deng YM, Shao Q, Feng F, An X, Wang FH, Wang ZQ, Xu RH, et al. EGFR fluorescence in situ hybridization pattern of chromosome 7 disomy predicts resistance to cetuximab in KRAS wild-type metastatic colorectal cancer patients. Clin Cancer Res. 2011;17:382–390. doi: 10.1158/1078-0432.CCR-10-0208. [DOI] [PubMed] [Google Scholar]
- 27.Shiraha H, Yamamoto K, Namba M. Human hepatocyte carcinogenesis (review) Int J Oncol. 2013;42:1133–1138. doi: 10.3892/ijo.2013.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi T, Sonobe M, Kobayashi M, Yoshizawa A, Menju T, Nakayama E, Mino N, Iwakiri S, Sato K, Miyahara R, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17:889–897. doi: 10.1245/s10434-009-0808-7. [DOI] [PubMed] [Google Scholar]
- 29.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 30.Motegi A, Fujimoto J, Kotani M, Sakuraba H, Yamamoto T. ALK receptor tyrosine kinase promotes cell growth and neurite outgrowth. J Cell Sci. 2004;117:3319–3329. doi: 10.1242/jcs.01183. [DOI] [PubMed] [Google Scholar]
- 31.Jawhar NM. Tissue Microarray: A rapidly evolving diagnostic and research tool. Ann Saudi Med. 2009;29:123–127. doi: 10.4103/0256-4947.51806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuyoshi Y, Inoue H, Kita Y, Utsunomiya T, Ishida T, Mori M. EML4-ALK fusion transcript is not found in gastrointestinal and breast cancers. Br J Cancer. 2008;98:1536–1539. doi: 10.1038/sj.bjc.6604341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyake I, Hakomori Y, Shinohara A, Gamou T, Saito M, Iwamatsu A, Sakai R. Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene. 2002;21:5823–5834. doi: 10.1038/sj.onc.1205735. [DOI] [PubMed] [Google Scholar]
- 34.Azarova AM, Gautam G, George RE. Emerging importance of ALK in neuroblastoma. Semin Cancer Biol. 2011;21:267–275. doi: 10.1016/j.semcancer.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 36.Grande E, Bolós MV, Arriola E. Targeting oncogenic ALK: a promising strategy for cancer treatment. Mol Cancer Ther. 2011;10:569–579. doi: 10.1158/1535-7163.MCT-10-0615. [DOI] [PubMed] [Google Scholar]
- 37.Preusser M, Berghoff AS, Ilhan-Mutlu A, Magerle M, Dinhof C, Widhalm G, Dieckmann K, Marosi C, Wöhrer A, Hackl M, et al. ALK gene translocations and amplifications in brain metastases of non-small cell lung cancer. Lung Cancer. 2013;80:278–283. doi: 10.1016/j.lungcan.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Tuma RS. ALK gene amplified in most inflammatory breast cancers. J Natl Cancer Inst. 2012;104:87–88. doi: 10.1093/jnci/djr553. [DOI] [PubMed] [Google Scholar]
- 39.De Brouwer S, De Preter K, Kumps C, Zabrocki P, Porcu M, Westerhout EM, Lakeman A, Vandesompele J, Hoebeeck J, Van Maerken T, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res. 2010;16:4353–4362. doi: 10.1158/1078-0432.CCR-09-2660. [DOI] [PubMed] [Google Scholar]
- 40.Shao CK, Su ZL, Feng ZY, Rao HL, Tang LY. [Significance of ALK gene expression in neoplasms and normal tissues] Ai Zheng. 2002;21:58–62. [PubMed] [Google Scholar]
- 41.Bagci O, Tumer S, Olgun N, Altungoz O. Copy number status and mutation analyses of anaplastic lymphoma kinase (ALK) gene in 90 sporadic neuroblastoma tumors. Cancer Lett. 2012;317:72–77. doi: 10.1016/j.canlet.2011.11.013. [DOI] [PubMed] [Google Scholar]


