Abstract
Ehrlichia chaffeensis is an obligately intracellular gram negative bacterium with a small genome that thrives in mammalian mononuclear phagoctyes by exploiting eukaryotic processes. Herein, we discuss the latest findings on moonlighting tandem repeat protein effectors and their secretion mechanisms, and novel molecular interkingdom interactions that provide insight into the intracellular pathobiology of ehrlichiae.
Keywords: Ehrlichia, human monocytotropic ehrlichiosis, zoonosis, effector, nucleomodulin, tandem repeat protein, moonlighting protein, type 1 secretion system, post translational modification
1. Introduction
Ehrlichia chaffeensis is an obligately intracellular gram-negative bacterium that demonstrates tropism for mononuclear phagocytes and is the etiologic agent of human monocytotropic ehrlichiosis (HME) [1]. HME is an emerging life-threatening tick-borne zoonosis in the United States [2], with more than 6,000 cases reported to the CDC as of 2010, and is classified as a group I NIAID emerging disease. However, a prospective investigation indicates that HME is substantially underdiagnosed and underreported [3]. HME is a seasonal disease (April-September) with a geographic distribution that coincides with that of the vector (Amblyomma americanum) [4]. E. chaffeensis is transstadially maintained in A. americanum, and the enzoonotic cycle is maintained through persistent infection of mammalian hosts, including the white tailed deer (Odocoileus virginianus), which is the primary reservoir [5]. HME presents as a flu-like illness with clinical symptoms and laboratory abnormalities that include fever, myalgia, elevated hepatic transaminases, and hematologic abnormalities, including anemia, thrombocytopenia, and leucopenia [6]. In immunocompetent patients low bacterial burden and toxic-shock like symptoms suggest immunopathologic mechanisms, such as elevated TNFα and IL-10 levels, mediate disease pathogenesis [6]. HME is a serious disease in that approximately 50% of patients are hospitalized, with severe complications, including adult respiratory distress syndrome (ARDS), intravascular coagulopathy, meningo-encephalitis, and hepatic and renal failure, associated with the 3% case fatality rate [4].
E. chaffeensis is an α-proteobacteria in the order Rickettsiales, family Anaplasmataceae, which includes genera Anaplasma, Rickettsia, Orientia, Wolbachia, and Neorickettsia [7]. E. chaffeensis has as two ultrastructurally defined forms, dense-cored cells (DC, 0.4–0.6μm) that are infectious and characterized by concentration of ribosomes and chromatin, predominate at early and late time points of infection, and reticulate cells (RC, 0.7–1.9μm) that are of pleomorphic morphology with DNA and ribosomes distributed throughout the bacterial cytoplasm and are replicate [8]. In addition to ultrastructural differences, DC and RC differentially express proteins that mediate host cell invasion, and are involved in establishment of the intracellular niche, and evasion of host innate immune response [9].
Previous reviews have focused on the host innate and adaptive immune responses as well as the cellular processes affected during E. chaffeensis infection [6, 10–12]. This review will largely focus on recent advances in our understanding of newly characterized secreted tandem repeat protein (TRP) effectors that promote intracellular survival through a large and diverse array of interactions with defined host targets and DNA, and by exploitation of host post translational pathways.
2. Intracellular development and subversion of host defense mechanisms
In mammalian cells, E. chaffeensis replication occurs in a 72 h life cycle that is initiated with DC ehrlichiae binding to DNaseX, E- and L- selectins, and other GPI-anchored proteins within caveolae at the monocyte cell surface [13–15]. This interaction is contingent on ehrlichial adhesins (ECH_1038 and TRP120), which have been shown to mediate adhesion and internalization [15, 16]. During endocytosis which is di-cyclic GMP dependent [17], DC ehrlichiae associates with caveolin 1 and phospholipase C-γ2 (PLC-γ2) and modulates host cell signaling including transglutamination, tyrosine-phosphorylation and activation of PLCγ2, inositol-(1,4,5)-triphosphate (IP3) production, and release of intracellular calcium stores [18]. Following endocytosis and for the duration of intracellular development, the bacterium resides in a membrane-bound cytoplasmic vacuole that is maintained in an exclusive caveolar endosomal recycling pathway. The ehrlichial vacuole does not fuse with lysosomes, phenotypically resembles an early-endosomal vesicle, and contains vacuolar (H+) ATPase, transferrin, transferrin receptor, and major histocompatibility molecules [19]. Within an hour of endocytosis, the DC cell transitions to RC form and divides via binary fission, doubling every 8 h for the next 48 h [8, 20], The resulting microcolony (morula) within the endosomal-like compartment, which contains as many as 400 individual bacterium [19]. By 72 h post infection, the RC transitions into the DC morphology, and the bacteria exit the host cell through undefined mechanisms including direct cell lysis, exocytosis, or cell-cell transfer via filopodia [21].
Throughout its intracellular life cycle, E. chaffeensis employs mechanisms to avoid and subvert host innate and adaptive immune responses. In contrast to related Rickettsia spp. and other gram negative bacteria, E. chaffeensis genome does not encode genes for enzymes required to synthesize pathogen-associated molecular patterns (PAMPs), lipopolysaccharide (LPS) or peptidoglycan. However, Ehrlichia does acquire cholesterol from the host cell for structural integrity of the outer membrane [22]. Following endocytosis, the bacterium actively prevents maturation and acidification of the endosomal vacuole as a means of blocking phagolysosomal fusion [10]. E. chaffeensis also expresses and secretes effector proteins, including TRP32, TRP47, TRP120, and a 200 kDa ankyrin repeat protein (Ank200) that appear to be involved in modulating host cell signaling and host gene transcription to avoid innate immune responses [11]. Early in infection Ehrlichia inhibit reactive oxygen species (ROS) production and apoptosis, and disrupt Jak/STAT signaling to prevent innate immune signaling responses [9]. Cytokine production is repressed, including IL-12, IL-15, and IL-18, thereby preventing cell-mediated immune response mechanisms, such as activation of TH1, NK cells, and cytotoxic T lymphocytes, and subsequent IFNγproduction [6, 12].
3.E. chaffeensis genome and differential gene expression
Similar to other rickettsial agents, E. chaffeensis has evolved mechanisms to survive in both arthropod and mammalian hosts. E. chaffeensis has a small genome (1.18Mbp) with low GC-content (~30%), encoding roughly 1000 genes, about half of which have predicted functions [23]. These bacteria utilize complex molecular strategies to adapt to intracellular niches in both vertebrate and invertebrate hosts, one of which involves differential bacterial gene/protein expression in order to adapt to these distinct host environments [24]. E. chaffeensis demonstrates significantly greater global transcriptional activity in tick cells (A. americanum, AAE2 cells; and Ixodes scapularis, ISE6 cells). Expression of genes involved in cellular processes, particularly metabolism and translation, is greater in tick cells suggesting E. chaffeensis is metabolically more active in the invertebrate host [24]. Expression of ehrlichial genes involved in translation and post translational modification in addition to short hypothetical proteins (30–80aa) also varies greatly between mammalian and tick cells. In contrast, significant differences in gene expression between mammalian and tick hosts were not observed in studies with R. rickettsii [25], suggesting that E. chaffeensis has evolved divergent adaptation mechanisms for intracellular survival in vertebrate and invertebrate hosts.
Differential regulation of major immunoreactive proteins that are now known to be effector proteins has also been observed. E. chaffeensis Ank200, TRP32, TRP47, and TRP120 transcripts were detected in both host cell types; however, neither TRP32 or TRP47 protein was detected in tick cells suggesting significant post transcriptional regulation occurs in the invertebrate host [24]. Conversely, TRP47 and TRP32 are hyperexpressed (10–15-fold greater than other highly expressed genes) in mammalian cells and protein is readily detected.
In human cells, TRP32 and TRP47 interact with host proteins involved in immune response, cell signaling, vesicular trafficking, cytoskeletal reorganization, and apoptosis [26, 27]. Host-dependent transcriptional regulation of these TRP effectors may reflect the ability of E. chaffeensis to establish a unique intracellular niche and survive in both mammalian macrophages and tick cells. Moreover, differential expression of the p28 family of outer membrane genes (OMP-1) occurs, with OMP-1b (p28-14) upregulated in tick cells, and p28 (p28-19), an integral membrane protein with porin activity [28] that is upregulated in mammalian cells [24].
4. Two-component systems
Ehrlichia differential gene expression, to some extent, is controlled by a series of two-component response regulator (RR) systems, three of which have been identified in the E. chaffeensis genome. These systems mediate gene expression based on queues from the host cell environment and are composed of the histidine kinase (HK), present as a transmembrane protein with a sensor domain in the periplasm and a kinase domain extending into the cytosol, as well as the RR, which is phosphorylated by the HK in the cytoplasm and subsequently binds genomic DNA to modulate bacterial gene expression. The ehrlichial two-component systems include PleC-PleD, NtrY-NtrX, and CckA-CtrA (RR-HK, respectively), which do not crosstalk [29, 30]. The most well characterized system, CckA-CtrA, regulates bacterial expression late in infection and confers stress-response mechanisms, including physical and chemical resistance to sonication and ROS, to the DC cell type in preparation for host cell evasion and release [31]. Activated CtrA binds the bolA promoter which regulates transcription of TRP120 and ECH_1038 (a highly conserved OMP) [31].
5.Ehrlichia secretion systems
E. chaffeensis genome encompasses multiple mechanisms for secretion and delivery of effector proteins to the bacterial extracellular space. Orthologs of Sec-dependent twin arginine translocase (TatA), Sec-independent TatC, and SecF genes, have been identified in the genome. Such secretion systems deliver bacterial proteins across the inner membrane into the periplasm, and these genes are expressed during both invertebrate and vertebrate host infections, with higher expression levels reported in tick cells [24].
Type 4 secretion system (T4SS) components ((VirB2, B3, B4, B6, B8, B9, B10, B11, and D4) of the VirB/D ancestral type, largely characterized in plant pathogen Agrobacterium tumefaciens, have been identified in the E. chaffeensis genome [32, 33]. These genes are expressed in both mammalian and tick host cells, with select components demonstrating >2-fold greater expression in the tick cell [24]. Ehrlichial VirB/D genes are present on two operons and operon expression is regulated by the ehrlichial DNA binding protein EcxR [34]. VirB/D loci are upregulated during bacterial replication, and downregulated during bacterial evasion and release from the mammalian host cell [33]. E. chaffeensis proteins (ECH_0261, ECH_0767, and EC_0825) have been identified as T4S substrates demonstrated to be secreted via the T4S machinery [35]. EC_0825 translocates across the ehrlichial vacuolar membrane and localizes to host cell mitochondria where it induces expression of the mitochondrial matrix protein manganese superoxide dismutase (MnSOD) to curb ROS production and inhibit mitochondrial-mediated apoptosis [35].
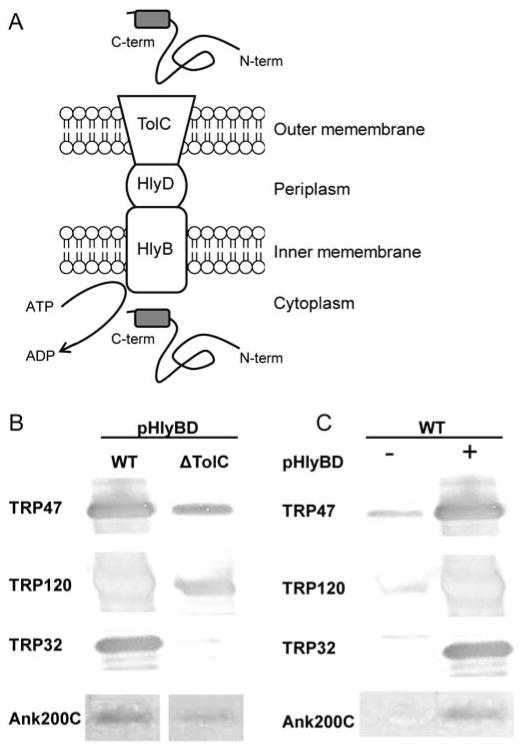
The E. chaffeensis genome also encodes three proteins (ECH_0383, ECH_0970, and ECH_1020) with homology to Escherichia coli type 1 secretion system (T1SS) components HlyB, HlyD, and TolC. T1SS mediates translocation of unfolded proteins from the cytosol, through the periplasm, and into the bacterial extracellular space. HlyD forms a homodimeric pore that spans the entire periplasm and interacts with the TolC heterotrimer that assembles in the outer membrane [36, 37] (Fig. 1a). HlyB is present as a homodimer in the inner membrane, where it recognizes C-terminal T1S signals in unfolded bacterial proteins and hydrolyses ATP to mediate translocation of the substrate across the inner membrane into the HlyD pore.
Figure 1. Type-1 secretion of E. chaffeensis effectors.
(A), E. chaffeensis orthologs of E. coli T1SS components HlyB (ECH_0383) HlyD (ECH_0970) and TolC (ECH_1020) have been identified. Schematic depicts secretion of T1SS substrates when acidic C-terminal T1S-signals in unfolded cytoplasmic proteins are recognized by inner membrane protein HlyB and in an ATP-dependent manner the substrate is translocated across the inner membrane, then the periplasm via HlyD Substrates pass through TolC, an outer membrane channel and into the extracellular space. (B), Secretion of Ehrlichia TRPs and Ank200 by E. coli in the presence or absence of TolC, and (C), and without (−) and with (+) HlyBD [38]. E. chaffeensis effector secretion is significantly enhanced in the presence of TolC or HlyB/D components of the T1SS.
Features of T1SS substrates include acidic pI, few cysteine residues, and tandem repeats, with the best characterized T1SS substrates being the repeats in toxins (RTX) family of exoproteins [36]. An uncleaved C-terminal secretion signal enriched in LDAVTSIF is observed in T1S substrates, suggesting unfolded proteins are translocated following completion of protein translation, and folded upon exit of TolC. Recently, Ank200, and TRP ehrlichial effectors were demonstrated to be T1SS substrates [38]. LDAVTSIF-enriched motifs were identified in the C-terminal 50 amino acids of these effectors. Secretion of TRP32, TRP47, TRP120, and Ank200 (a C-terminal construct) was demonstrated using a heterologous E. coli T1SS mutant strains. In the absence of the T1SS components TolC (Fig. 1b) or HlyB and HlyD (Fig. 1c), ehrlichial effector secretion was diminished [38]. Ehrlichial protein homology with T1SS components and the recent demonstration that immunoreactive TRPs and Ank200 are secreted in a type-1 dependent manner, establishes the importance of the T1SS in the pathobiology of ehrlichial infection, a first for an intracellular bacterium.
6. Characteristics of E. chaffeensis ankyrin and tandem repeat proteins
Ehrlichia proteins with ankyrin and tandem repeats have been identified as the major immunoreactive proteins that elicit strong antibody responses during infection [39–41]. Ehrlichia TRP effectors share domain architecture and molecular characteristics despite differences in protein length and absence of homology (Fig. 2). Consistent with other T1S substrates, these proteins are highly acidic and contain tandem repeats [38]. TRP32, TRP47, and TRP120 TRs are serine-rich and acidic (pI ~4), and this characteristic is responsible for the anomalous protein migration observed during gel electrophoresis [42]. In contrast, the TRP75 TR is lysine-rich with a basic pI [43]. In TRP32, TRP47, and TRP120 the acidic C-terminal region contains the T1S signal, facilitating effector secretion [38]. TRPs contain major continuous species-specific antibody epitopes [43–47], and antibodies directed at these epitopes are protective against infection [41].
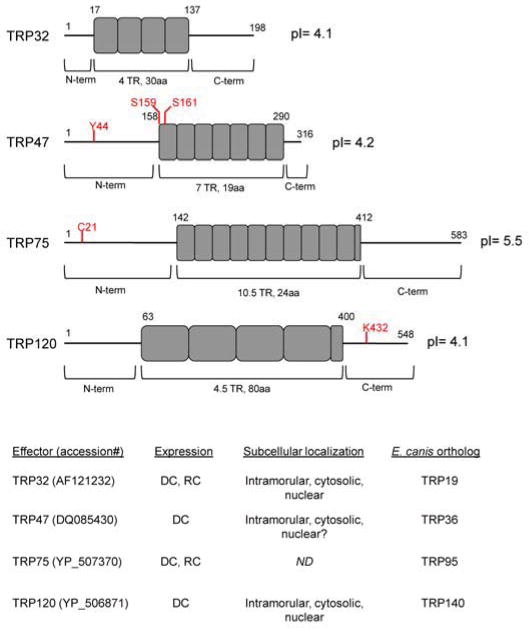
Figure 2. Illustration of TRP effector domains, including details of expression and host subcellular localization.
TR protein domains are represented by grey boxes, and locations of confirmed post translational modifications are noted in red. TRP47 is phosphorylated at Y44, S159 and/or S161[42]. TRP75 is acylated at C21 following proteolysis within the lipobox (aa 11–24) [49]. TRP120 is SUMOylated at K432 [51]. (DC, expressed by dense-core cell type; RC, expressed by reticulate cell type; ND, not determined).
In human monocytes, TRPs are differentially expressed dependent on morphological cell type. TRP32 and TRP75 are constitutively expressed by DC and RC types, while TRP47 and TRP120 are exclusively expressed by DC cell type [16, 43]. Upon T1S into the intramorular space, these effectors translocate across the vacuolar membrane via an undefined mechanism, and localize to different host subcellular compartments including the nucleus [27, 48]. However, these proteins do not contain a classic NLS and the mechanism of nuclear translocation remains undefined.
Following secretion, TRPs are substrates of host-mediated post translational modification pathways. TRP47 is phosphorylated at tyrosine 44 and serine 159 and/or serine 161 [42]. TRP75 contains an N-terminal lipobox motif and is proteolyzed and acylated at cysteine 21 [49]. Anti-phosphotyrosine immunoprecipitation demonstrated TRP75 is also tyrosine phosphorylated, although the specific modified residues remain undefined [43]. TRP120 is covalently modified by ubiquitin (Ub) at multiple lysine residues [50], as well as the small ubiquitin-like modifier (SUMO) at lysine 432 in the C-terminus [51]. The functional consequences for TRP post translational modification have not been defined, but there is evidence suggesting effector subcellular localization is affected [50] and some host protein interactions are enhanced with modified effector [51]. Ank200 is a T1S effector that contains at least 21 ankyrin repeats flanked by acidic N- and C-terminal regions that harbor major linear antibody epitopes [52]. Ank200 is secreted, escapes the ehrlichial inclusion, and translocates to the host nucleus where it directly binds host genomic DNA [53]. The mechanism of nuclear translocation remains undefined as Ank200 does not harbor a classic nuclear localization sequence (NLS).
7. TRP-host protein interactions
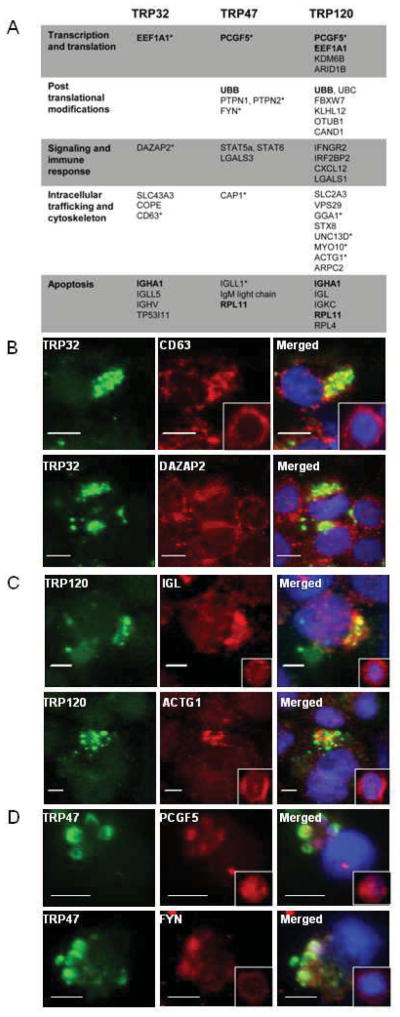
Our understanding of the role of TRPs as effectors in ehrlichial pathobiology has significantly advanced in recent years with characterization of TRP-host interactions. Yeast two-hybrid (Y2H) studies performed with a human bone marrow cDNA library have identified host target proteins that interact with TRPs [26, 27, 54]. These studies identified TRP interactions with a diverse network of host proteins involved in many host cellular processes including transcriptional and translational regulation, post translational modification, signaling and immune response, intracellular trafficking and cytoskeletal organization, and apoptosis (Fig. 3a), demonstrating that TRPs are moonlighting proteins.
Figure 3. Diverse interactions between E. chaffeensis TRP effectors and host proteins.
A, Selection of mammalian host proteins that interact with ehrlichial TRP effectors, as determined by Y2H,and classified by biological function. (*) denotes Y2H hits that were confirmed for TRP localization and interaction with immunofluorescence microscopy and coimmunoprecipitation. Common host proteins that interact with multiple TRPs are highlighted in boldface. (B–D), Microscopy of THP-1 cells 72 h post infection, fixed and probed with epitope-specific anti-TRP antibody (green), anti-specific host protein (red), and DAPI (DNA, blue) to confirm colocalization (merged, yellow) of TRP32 (B), TRP120 (C), and TRP47 (D) with interacting host proteins identified in Y2H screens. Insets demonstrate localization of host proteins in uninfected THP-1 cells and highlight redistributed host proteins in E. chaffeensis infected cells. Figures are modified reprints from [26, 27, 54].
Y2H studies for TRP47 and TRP120 utilized the full TRP open-reading frame and yielded roughly 100 host protein interactions each [27, 54]. Such interaction studies with TRP32 required use of a truncated C-terminal construct (as the TR region elicited strong autoactivation in this assay) and yielded 22 host protein interactions [26] (Fig. 3a). Immunofluorescence (Fig. 3b, c) and confocal microscropy (Fig. 3d) demonstrated colocalization of select TRP-host proteins during ectopic expression and during E. chaffeensis infection. During infection, host protein redistribution from the cytosol to the ehrlichial vacuole was observed, particularly for CD63, immunoglobulin lambda (IGL) and -actin (ACTG1) compared to healthy cells (Fig. 3b, c). TRP75 yeast two-hyrbrid studies did not reveal host interactions (Luo, Zhang, and McBride, unpublished data); however this protein encompasses an N-terminal lipobox motif that is proteolyzed and acylated [49] suggesting TRP75 may be membrane associated. The format of the yeast two-hybrid technique may not be suitable for identifying TRP75 interacting proteins.
Interestingly, several host proteins were demonstrated to interact with more than one TRP effector. TR47 and TRP120 both robustly interact with PCGF5 [27, 54]. In the nucleus, PGCF5 is a component of the polycomb repressive complex 1 (PRC1), which mediates epigenetic regulation [55]. However, during E. chaffeensis infection, PCGF5 redistributes to the cytosol and colocalizes with ehrlichial inclusions (Fig. 3d) [27], suggesting there may be a cytosolic role for PCGF5 or effector interaction may trigger clearing of the nucleus. TRP120 interaction with PCGF5 is mediated through the TR domain, but evidence suggests a conformational change is necessary to expose this domain as full length TRP120 must be SUMO conjugated to yield interaction with PCGF5 [51].
TRP47 and TRP120 interact with components of the Ub post translational modification pathways, including E3 ligases, FBXW7 and KLHL12, as well as Ub isoforms UBB and UBC. These interactions suggest the effectors are either themselves targets of Ub conjugation, which was recently observed for TRP120 [50], or the TRP interaction modulates this pathway to enhance Ub-mediated signaling changes such as receptor-mediated endocytosis, Ub-mediated subcellular localization, or Ub-mediated proteasomal degradation.
Ehrlichia and related species demonstrate delayed host cell apoptosis through expression of cell surface effectors [56] and stabilization of host cell mitochondria [57]. Microarray studies demonstrate E. chaffeensis upregulates expression of apoptosis inhibitors early in infection (1–7 h post infection) and upregulate apoptosis inducers at later time points (>7 h post infection) [9]. TRP Y2H studies suggest E. chaffeensis may also modulate programmed cell death responses through host protein interactions. TRP47 and TRP120 interact with ribosomal proteins L4 and L11 (RPL4 and RPL11), which are associated with ribosomal assembly and p53-mediated apoptosis [58, 59]. TRP effectors also interact with a range of immunoglobulins, which were recently reported to stabilize protein expression of anti-apoptotic Bcl-xL [60]. Further characterization of these interactions is necessary to determine the role of TRPs in regulating host programmed cell death; however recent host interaction studies suggest these effectors participate in the mechanism.
8.E. chaffeensis effectors function as nucleomodulins
Characterization of TRP-host protein interactions has contributed to our understanding of the role of these effectors in E. chaffeensis pathobiology. Recently, ehrlichial effectors Ank200 and TRP120 were also determined to reprogram host cell processes through directly binding host chromatin and modulating host transcriptional response [48, 53]. These nuclear effectors contribute to changes in expression levels observed for 5% of the host genes four hours post E. chaffeensis infection [9]. Observation that ehrlichial proteins function as nuclear effectors is not unique to E. chaffeensis. In fact, there is an emerging appreciation that several pathogenic bacteria, including Listeria monocytogenes [61], Xanthomonas campestris [62], and Anaplasma phagocytophilum [63], secrete effectors that access the host nucleus and modulate gene expression through interactions with host chromatin or specific DNA motifs that mediate epigenetic changes or function as transcription factors [61]. This newly defined class of bacterial effectors has been termed “nucleomodulins” [64].
Ank200 localizes to the host nucleus during ehrlichial infection, where it has been demonstrated to directly bind host genomic DNA at an adenine-rich motif in Alu-Sx elements [53]. Alu-Sx elements are the most abundant repetitive elements in the mammalian genome, and are frequently found in promoter and intronic regions. Whole genome chromatin immunoprecipitation (ChIP) and DNA sequencing demonstrated Ank200 binds DNA motifs within 456 host genes [53] which range in function from transcriptional and translational regulation, post translational modification, immune response and cell signaling, intracellular trafficking and cytoskeletal rearrangement, and transmembrane transport (Table 1). Many of these target genes are strongly upregulated during ehrlichial infection, including TNFα, STAT1, CD48 [53], suggesting the mechanism of enhanced gene expression may involve Ank200 interaction.
Table 1.
Selection of host genes that TRP120 and Ank200 bind, as determined by ChlP, classified by ontology.
| TRP120 | Ank200 | |
|---|---|---|
| Transcription and translation | EEF1A1 DNMT |
PCGF3 HDAC3, HDAC6 Histone 1, 2, 3 components EIF4E2 |
| Post translational modification | PTK2 | SUMO1 pseudogene UBE2J2, UBE2Q1 DUB3 |
| Immune response and cell signaling | NOTCH1 WNT11 PPARA RXRA IKBKB TLR5 |
NOTCH2 WNT2B WISP3 RXRB STAT1, STAT5b JAK2 MYD88 TNF TRAF7 IRF2BP2** |
| Intracellular trafficking and cytoskeleton | SNX14 TSNARE1 SNAP47 |
SNX11, SNX17 CAP1* COPA CLTA RHOA MYL6 TUBG1 ACTBL1 ARPC1B |
| Transmembrane transport |
SLC2A1 SLC26A9 KCNQ4 |
SLC2A1 SLC26A9 KCNQ4 |
Gene was also identified in Y2H as TRP47 interacting protein
Gene was also identified in Y2H as TRP120 interacting protein
Bold highlights genes bound by both TRP120 and Ank200
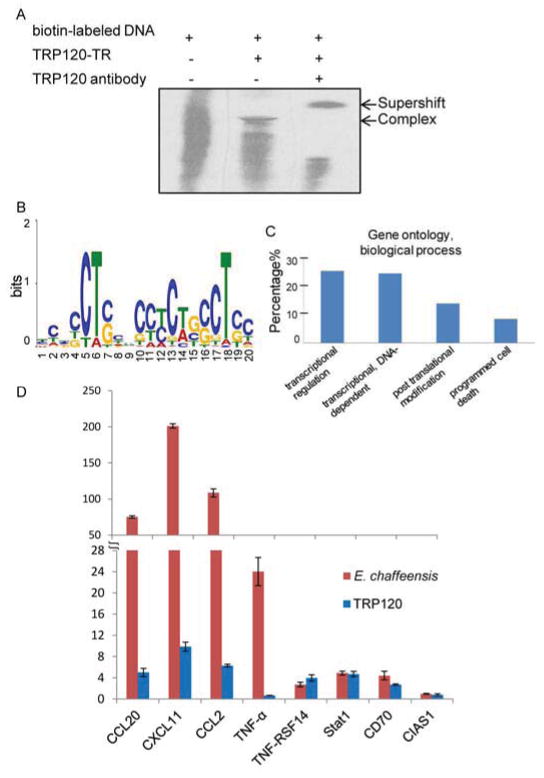
Recently, TRP120 was reported to directly interact with host genomic DNA. In these studies a recombinant TRP120-TR construct encoding two 80-amino acid TRP120-TRs was shown to directly bind host genomic DNA by EMSA (Fig. 4a) [48]. The TRP120-TR-DNA complex was confirmed following incubation with anti-TRP120 antibody and an observed supershift (Fig. 4a). DNA sequence motif analysis identified the consensus TRP120 DNA binding motif as a GC-rich sequence motif (Fig. 4b). These DNA motifs are enriched in promoter regions and serve as recognition sequences for some classes of transcription factors [65, 66]. Genome-wide TRP120 targets were identified using ChIP and DNA sequencing (Table 1) [48]. Genomic TRP120 binding sites were mapped to promoter regions as well as introns and exons within more than 2000 host genes. Gene ontology (GO) analysis classified TRP120 target genes into functionally related gene groups based on biological process, where the most highly represented categories include transcriptional regulation, protein modification, and apoptosis (Fig. 4c) [48].
Figure 4. TRP120 directly binds host genomic DNA at a specific motif and modulates host gene expression.
A, EMSA demonstrates recombinant TRP-TR directly binds biotin-labeled mammalian genomic DNA (purified from THP-1 human monocytes). Incubation with epitope-specific TRP120 antibody results in a supershift of the TRP120-TR-DNA complex. B, GC-rich consensus binding motif identified from TRP120 ChIPSeq peak analysis using WebLogo. C, Gene ontology biological process classification of host genes identified from TRP120-TR ChIP analysis. D, Induction of expression for select TRP120 target genes is observed in THP-1 cells 72 h post E. chaffeensis infection (relative to uninfected THP-1 cells) or recombinant TRP120 transfection (relative to thioredoxin control protein), as determined by qPCR analysis. Figure is a modified reprint from [48].
Following identification of TRP120 host genomic targets, TRP120-mediated modulation of transcription levels of representative gene targets was demonstrated (Fig. 4d). TRP120 targets immune response genes, including CXCL11, CCL2, and STAT1, and upregulates expression [48]. Consistent with increased CXCL11, CCL2, and STAT1 protein expression observed during E. chaffeensis infection, transfection of recombinant TRP120 protein enhances expression levels for these gene targets. This recent study suggests TRP120 contributes to the mechanism of upregulation of target gene expression during ehrlichial infection.
8.1 Ehrlichial effectors subvert host immune responses
As a potential immune subversion mechanism, Ank200 and TRP120 target the genes for nearly all major components of the Jak-Stat pathway (Jak2, Stat1, Stat3, Stat5 and IFNR2) (Table 1) [48, 53]. Transcription factors involved in host signaling pathways are also gene targets for Ank200 and TRP120. Notch and Wnt signaling molecules are targets, as well as peroxisome proliferator-activated receptor alpha (PPAR) and a retinoid x receptor alpha (RXRA). Protein products of these genes ultimately regulate cell growth, proliferation, differentiation, and immune inflammatory responses. Ehrlichial effector nucleomodulins may target these genes as a mechanism for reprogramming the host cell and downregulating host defenses.
8.2 Ehrlichial effectors target host epigenetic machinery
Genes for epigenetic machinery are also targets of Ank200 and TRP120, including histone deacetylase 1, 2, and 8 (HDAC1, 3, and 8) and DNA methyltransferase (DNMT). Y2H studies demonstrated TRP120 also directly interacts with host epigenetic machinery, including a histone methylase (NSD1) and demethylase (KDM6B), protein components of the SWI/SNF chromatin remodeling complex (ARID1B), and PCGF5, a component of PRC1 that regulates transcriptional silencing through epigenetic modification. This suggests yet another mode of transcriptional regulation through potential effector-mediated changes in expression levels of host epigenetic machinery, or changes in histone modification and chromatin remodeling. This mechanism is similar to that described for AnkA, a T4S effector from A. phagocytophilum, which recruits HDAC1 to host chromatin to repress gene expression [63].
8.3 Ehrlichial effectors modulate vesicular trafficking and cytoskeletal components
Similar to yeast two-hybrid results, ehrlichial effectors target genes involved in intracellular trafficking and cytoskeletal rearrangement. Clathrin (CLTA), coatomer (COPA), syntaxins (SNX14, SNX11, SNX17), and TSNARE (TSNARE1) are gene targets of Ank200 and TRP120, and suggest E. chaffeensis may regulate expression of vesicular trafficking machinery to facilitate delivery of requisite protein and small molecule cargo to the ehrlichial inclusion during replication. Y2H studies also demonstrated TRP120 interacts with components of the vesicular trafficking machinery [54]. Acquisition of cholesterol, iron, and essential amino acids from the host cell has been shown to be necessary for ehrlichial replication [22], and ehrlichial effectors may modulate vesicular trafficking to facilitate this process. Likewise, components of the cytoskeleton are gene targets for Ank200 [53], as well as host proteins known to interact with TRP120 [54]. Effector-mediated changes in expression of cytoskeletal components may facilitate vesicular trafficking, exocytosis or filopodia formation, particularly late in infection for bacterial egress.
Of particular note is the frequency of solute carrier family pores (SLC) and ion channels (KCNQ4) that were identified as both Ank200 and TRP120 gene targets. Y2H studies identified SLC pores as TRP32 and TRP120 interacting proteins [26, 54]. Further characterization of the importance of these transcripts is necessary; however, their putative function suggests that, similar to modulation of vesicular trafficking, ehrlichial effectors change expression levels of these small molecule and ion channels as a means of acquiring requisite materials during bacterial replication.
9. Conclusions
E. chaffeensis demonstrates host-specific gene expression which gives insight into adaptive mechanisms employed by the bacteria for survival in diverse vertebrate and invertebrate intracellular environments. E. chaffeensis interacts with and responds to its host environment via two-component systems and expression and secretion of effector proteins via T4 and T1SS. TRPs, a novel class of ehrlichial effectors, were recently shown to be T!SS substrates. This class of ehrlichial proteins is highly immunoreactive and contains species-specific epitopes in tandem repeat regions. These epitope-specific TRP antibodies are protective against infection via extracellular and intracellular mechanisms that remain undefined.
TRPs effectors mediate diverse host protein interactions as well as directly bind host genes. This recent characterization cements the fact that these proteins are indeed effectors, and points to their moonlighting functions in modulating multiple host cellular processes and subverting host defenses to promote intracellular survival. Fig. 5 illustratively summarizes what is known thus far about the roles of these effectors in ehrlichial pathobiology. Further characterization is necessary to determine the impact of these effectors on host cellular processes. The studies reviewed herein extend our understanding of the pathobiology of intracellular pathogenic bacteria, explore mechanisms used to invade and subvert host defenses, and identify novel targets for development of bacterial and host-directed small molecule therapeutics, such as effector secretion systems and cell signaling pathways, respectively.
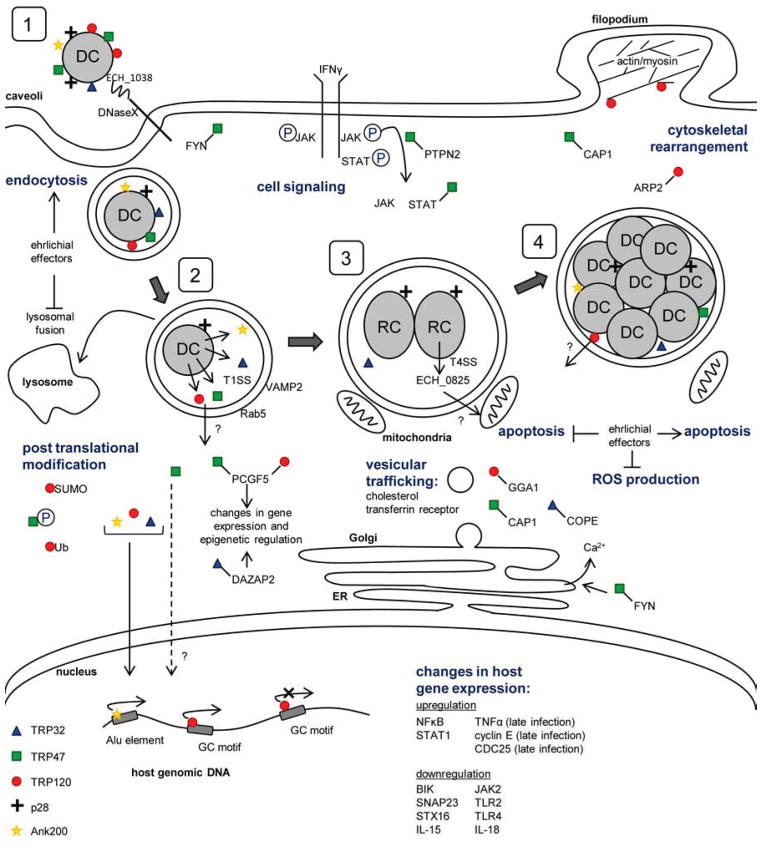
Figure 5. Depiction of effector functions during the E. chaffeensis life cycle in a mammalian monocyte.
1, DC bacteria bind DNaseX, a GPI-anchored receptor as well as other GPI-anchored proteins within caveoli at the surface of the monocyte. ECH_1038 and TRP120 mediate this interaction and facilitate bacterial endocytosis. 2, DC bacteria establish an intracellular niche in a membrane-bound vacuole, to which early-endosomal proteins, including Rab5a and VAMP2, are recruited. This inclusion does not undergo phagolysosomal fusion. Ehrlichial effectors, including TRP32, TRP47, TRP120, and Ank200 are secreted into the intramorular space via T1SS and translocate across the vacuolar membrane (via an undefined mechanism) into the host cytosol. In the cytosol these effectors are substrates of host post translational modification machinery (TRP47 is phosphorylated; TRP120 is SUMO and Ub modified), and interact with diverse host proteins to alter cell processes. Through an NLS-independent mechanism, ehrlichial effectors are also translocated to the host nucleus where Ank200 and TRP120 bind host genomic DNA at Alu-Sx elements and GC motifs, respectively, and modulate host gene expression. Changes in gene expression include upregulation of signaling genes, including NFκB, and STAT1, as well as downregulation of innate immune-related genes JAK2, TLR2, and TLR4. 3, 1 h post infection, DC bacteria transition into the RC morphology and replicate by binary fission every 8 h for the following 48 h. During this period ehrlichial effectors continue to modulate host cell processes. Host cell signaling is disrupted, including IFNγ-mediated JAK/STAT signaling, as a means of avoiding host innate immune responses. Vesicular trafficking is exploited to obtain cholesterol and iron (via transferrin and transferrin receptors) necessary for replication. Mitochondria redistribute to the periphery of the ehrlichial inclusion. T4SS effector, ECH_0825, translocates to these mitochondria to inhibit Bax-induced apoptosis and ROS production, thereby permitting intracellular survival. 4, 72 h post infection RC morphology transitions into DC cell type. Effectors, including T1SS TRP47 and TRP120 translocate across the vacuolar membrane and interact with host cytoskeletal proteins, including β-actin, γ-actin, γ-tubulin, and myosin X, as well as accessory proteins, including components of the ARP2/3 complex and CAP1, to facilitate exocytosis or filopodium formation (for direct cell-cell transfer). Late in infection ehrlichial effectors induce host apoptosis, possibly to facilitate bacterial release.
Acknowledgments
We thank all current and former laboratory members for discussions and scientific contributions towards understanding the pathobiology of Ehrlichia infections. This work was supported by grant AI105536 from the National Institute of Allergy and Infectious Diseases (NIAID) and jointly by the Clayton Foundation for Research. PSD was supported by NIH Emerging Infectious Diseases and Biodefense training grant T32 AI007536.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson BE, Dawson JE, Jones DC, Wilson KH. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. Journal of clinical microbiology. 1991;29:2838–2842. doi: 10.1128/jcm.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olano JP, Walker DH. Human ehrlichioses. The Medical clinics of North America. 2002;86:375–392. doi: 10.1016/s0025-7125(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 3.Olano JP, Masters E, Hogrefe W, Walker DH. Human monocytotropic ehrlichiosis, Missouri. Emerging infectious diseases. 2003;9:1579–1586. doi: 10.3201/eid0912.020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paddock CD, Sumner JW, Shore GM, Bartley DC, Elie RC, McQuade JG, Martin CR, Goldsmith CS, Childs JE. Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. Journal of clinical microbiology. 1997;35:2496–2502. doi: 10.1128/jcm.35.10.2496-2502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewing SA, Dawson JE, Kocan AA, Barker RW, Warner CK, Panciera RJ, Fox JC, Kocan KM, Blouin EF. Experimental transmission of Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) among white-tailed deer by Amblyomma americanum (Acari: Ixodidae) Journal of medical entomology. 1995;32:368–374. doi: 10.1093/jmedent/32.3.368. [DOI] [PubMed] [Google Scholar]
- 6.Ismail N, Bloch KC, McBride JW. Human ehrlichiosis and anaplasmosis. Clinics in laboratory medicine. 2010;30:261–292. doi: 10.1016/j.cll.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. International journal of systematic and evolutionary microbiology. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 8.Zhang JZ, Popov VL, Gao S, Walker DH, Yu XJ. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cellular microbiology. 2007;9:610–618. doi: 10.1111/j.1462-5822.2006.00812.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JZ, Sinha M, Luxon BA, Yu XJ. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infection and immunity. 2004;72:498–507. doi: 10.1128/IAI.72.1.498-507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rikihisa Y. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nature reviews. Microbiology. 2010;8:328–339. doi: 10.1038/nrmicro2318. [DOI] [PubMed] [Google Scholar]
- 11.McBride JW, Walker DH. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert reviews in molecular medicine. 2011;13:e3. doi: 10.1017/S1462399410001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansueto P, Vitale G, Cascio A, Seidita A, Pepe I, Carroccio A, di Rosa S, Rini GB, Cillari E, Walker DH. New insight into immunity and immunopathology of Rickettsial diseases. Clinical & developmental immunology. 2012;2012:967852. doi: 10.1155/2012/967852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JZ, McBride JW, Yu XJ. L-selectin and E-selectin expressed on monocytes mediating Ehrlichia chaffeensis attachment onto host cells. FEMS microbiology letters. 2003;227:303–309. doi: 10.1016/S0378-1097(03)00696-7. [DOI] [PubMed] [Google Scholar]
- 14.Lin M, Rikihisa Y. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cellular microbiology. 2003;5:809–820. doi: 10.1046/j.1462-5822.2003.00322.x. [DOI] [PubMed] [Google Scholar]
- 15.Kumar DMYM, Miura K, Los M, Coy J, Rikihisa Y. DNaseX. a GPI-anchored Nucleotidase Is a Mammalian Receptor for Entry for the Novel Ehrlichia chaffeensis Invasin ECH1038. American Society of Microbiology General Meeting; 2013. [Google Scholar]
- 16.Popov VL, Yu X, Walker DH. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microbial pathogenesis. 2000;28:71–80. doi: 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 17.Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. Journal of bacteriology. 2010;192:4122–4133. doi: 10.1128/JB.00132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin M, Zhu MX, Rikihisa Y. Rapid activation of protein tyrosine kinase and phospholipase C-gamma2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infection and immunity. 2002;70:889–898. doi: 10.1128/IAI.70.2.889-898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnewall RE, Rikihisa Y, Lee EH. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infection and immunity. 1997;65:1455–1461. doi: 10.1128/iai.65.4.1455-1461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dedonder SE, Cheng C, Willard LH, Boyle DL, Ganta RR. Transmission electron microscopy reveals distinct macrophage- and tick cell-specific morphological stages of Ehrlichia chaffeensis. PloS one. 2012;7:e36749. doi: 10.1371/journal.pone.0036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas S, Popov VL, Walker DH. Exit mechanisms of the intracellular bacterium Ehrlichia. PloS one. 2010;5:e15775. doi: 10.1371/journal.pone.0015775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infection and immunity. 2003;71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hotopp JCLM, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. Comparative genomics of emerging human ehrlichiosis agents. PloS Genetics. 2006:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuriakose JA, Miyashiro S, Luo T, Zhu B, McBride JW. Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PloS one. 2011;6:e24136. doi: 10.1371/journal.pone.0024136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Hackstadt T. Limited transcriptional responses of Rickettsia rickettsii exposed to environmental stimuli. PloS one. 2009;4:e5612. doi: 10.1371/journal.pone.0005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo T, McBride JW. Ehrlichia chaffeensis TRP32 interacts with host cell targets that influence intracellular survival. Infection and immunity. 2012;80:2297–2306. doi: 10.1128/IAI.00154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakeel A, Kuriakose JA, McBride JW. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infection and immunity. 2009;77:1734–1745. doi: 10.1128/IAI.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumagai Y, Huang H, Rikihisa Y. Expression and porin activity of P28 and OMP-1F during intracellular Ehrlichia chaffeensis development. Journal of bacteriology. 2008;190:3597–3605. doi: 10.1128/JB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumagai Y, Cheng Z, Lin M, Rikihisa Y. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infection and immunity. 2006;74:5014–5022. doi: 10.1128/IAI.00735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Z, Kumagai Y, Lin M, Zhang C, Rikihisa Y. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cellular microbiology. 2006;8:1241–1252. doi: 10.1111/j.1462-5822.2006.00704.x. [DOI] [PubMed] [Google Scholar]
- 31.Cheng Z, Miura K, Popov VL, Kumagai Y, Rikihisa Y. Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Molecular microbiology. 2011;82:1217–1234. doi: 10.1111/j.1365-2958.2011.07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao W, Kumagai Y, Niu H, Yamaguchi M, Miura K, Rikihisa Y. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. Journal of bacteriology. 2009;191:278–286. doi: 10.1128/JB.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rikihisa Y, Lin M, Niu H, Cheng Z. Type IV secretion system of Anaplasma phagocytophilum and Ehrlichia chaffeensis. Annals of the New York Academy of Sciences. 2009;1166:106–111. doi: 10.1111/j.1749-6632.2009.04527.x. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Z, Wang X, Rikihisa Y. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. Journal of bacteriology. 2008;190:2096–2105. doi: 10.1128/JB.01813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Bao W, Lin M, Niu H, Rikihisa Y. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cellular microbiology. 2012;14:1037–1050. doi: 10.1111/j.1462-5822.2012.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delepelaire P. Type I secretion in gram-negative bacteria. Biochimica et biophysica acta. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review) Molecular membrane biology. 2005;22:29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- 38.Wakeel A, den Dulk-Ras A, Hooykaas PJ, McBride JW. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Frontiers in cellular and infection microbiology. 2011;1:22. doi: 10.3389/fcimb.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride JW, Comer JE, Walker DH. Novel Immunoreactive glycoprotein orthologs of Ehrlichia spp. Annals of the New York Academy of Sciences. 2003;990:678–684. doi: 10.1111/j.1749-6632.2003.tb07443.x. [DOI] [PubMed] [Google Scholar]
- 40.McBride JW, Corstvet RE, Gaunt SD, Boudreaux C, Guedry T, Walker DH. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infection and immunity. 2003;71:2516–2524. doi: 10.1128/IAI.71.5.2516-2524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuriakose JA, Zhang X, Luo T, McBride JW. Molecular basis of antibody mediated immunity against Ehrlichia chaffeensis involves species-specific linear epitopes in tandem repeat proteins. Microbes and infection / Institut Pasteur. 2012;14:1054–1063. doi: 10.1016/j.micinf.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakeel A, Zhang X, McBride JW. Mass spectrometric analysis of Ehrlichia chaffeensis tandem repeat proteins reveals evidence of phosphorylation and absence of glycosylation. PloS one. 2010;5:e9552. doi: 10.1371/journal.pone.0009552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride JW, Zhang X, Wakeel A, Kuriakose JA. Tyrosine-phosphorylated Ehrlichia chaffeensis and Ehrlichia canis tandem repeat orthologs contain a major continuous cross-reactive antibody epitope in lysine-rich repeats. Infection and immunity. 2011;79:3178–3187. doi: 10.1128/IAI.01347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doyle CK, Nethery KA, Popov VL, McBride JW. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infection and immunity. 2006;74:711–720. doi: 10.1128/IAI.74.1.711-720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo T, Zhang X, Wakeel A, Popov VL, McBride JW. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infection and immunity. 2008;76:1572–1580. doi: 10.1128/IAI.01466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo T, Zhang X, McBride JW. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clinical and vaccine immunology: CVI. 2009;16:982–990. doi: 10.1128/CVI.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo T, Zhang X, Nicholson WL, Zhu B, McBride JW. Molecular characterization of antibody epitopes of Ehrlichia chaffeensis ankyrin protein 200 and tandem repeat protein 47 and evaluation of synthetic immunodeterminants for serodiagnosis of human monocytotropic ehrlichiosis. Clinical and vaccine immunology: CVI. 2010;17:87–97. doi: 10.1128/CVI.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu B, Kuriakose JA, Luo T, Ballesteros E, Gupta S, Fofanov Y, McBride JW. Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infection and immunity. 2011;79:4370–4381. doi: 10.1128/IAI.05422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H, Lin M, Wang X, Kikuchi T, Mottaz H, Norbeck A, Rikihisa Y. Proteomic analysis of and immune responses to Ehrlichia chaffeensis lipoproteins. Infection and immunity. 2008;76:3405–3414. doi: 10.1128/IAI.00056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunphy PSLT, McBride JW. Host Post Translational Modification Pathways Mediate Nuclear Translocation of Ehrlichia chaffeensis Type 1 Secreted Effectors. American Society of Rickettsiology Annual Meeting; 2013. [Google Scholar]
- 51.Dunphy PSLT, McBride JW. Ehrlichia Type 1 Secreted Effectors Exploit Host SUMO Pathways. American Society of Microbiology General Meeting; 2013. [Google Scholar]
- 52.Nethery KA, Doyle CK, Zhang X, McBride JW. Ehrlichia canis gp200 contains dominant speciesspecific antibody epitopes in terminal acidic domains. Infection and immunity. 2007;75:4900–4908. doi: 10.1128/IAI.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu B, Nethery KA, Kuriakose JA, Wakeel A, Zhang X, McBride JW. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infection and immunity. 2009;77:4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo T, Kuriakose JA, Zhu B, Wakeel A, McBride JW. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infection and immunity. 2011;79:4382–4391. doi: 10.1128/IAI.05608-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Junco SE, Wang R, Gaipa JC, Taylor AB, Schirf V, Gearhart MD, Bardwell VJ, Demeler B, Hart PJ, Kim CA. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure. 2013;21:665–671. doi: 10.1016/j.str.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bazzocchi C, Comazzi S, Santoni R, Bandi C, Genchi C, Mortarino M. Wolbachia surface protein (WSP) inhibits apoptosis in human neutrophils. Parasite immunology. 2007;29:73–79. doi: 10.1111/j.1365-3024.2006.00915.x. [DOI] [PubMed] [Google Scholar]
- 57.Xiong Q, Bao W, Ge Y, Rikihisa Y. Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. The Journal of infectious diseases. 2008;197:1110–1118. doi: 10.1086/533457. [DOI] [PubMed] [Google Scholar]
- 58.Kajikawa S, Nakayama H, Suzuki M, Takashima A, Murayama O, Nishihara M, Takahashi M, Doi K. Increased expression of rat ribosomal protein L4 mRNA in 5-azacytidine-treated PC12 cells prior to apoptosis. Biochemical and biophysical research communications. 1998;252:220–224. doi: 10.1006/bbrc.1998.9633. [DOI] [PubMed] [Google Scholar]
- 59.Donati G, Thomas G. Apoptosis in pluripotent stem cells: RPL11 strikes again. Cell cycle. 2012;11:840. doi: 10.4161/cc.11.5.19438. [DOI] [PubMed] [Google Scholar]
- 60.Yang SB, Chen X, Wu BY, Wang MW, Cai CH, Cho DB, Chong J, Li P, Tang SG, Yang PC. Immunoglobulin kappa and immunoglobulin lambda are required for expression of the anti-apoptotic molecule Bcl-xL in human colorectal cancer tissue. Scandinavian journal of gastroenterology. 2009;44:1443–1451. doi: 10.3109/00365520903369953. [DOI] [PubMed] [Google Scholar]
- 61.Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harbor perspectives in medicine. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voytas DF, Joung JK. Plant science. DNA binding made easy. Science. 2009;326:1491–1492. doi: 10.1126/science.1183604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rennoll-Bankert KE, Dumler JS. Lessons from Anaplasma phagocytophilum: chromatin remodeling by bacterial effectors. Infectious disorders drug targets. 2012;12:380–387. doi: 10.2174/187152612804142242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bierne H, Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cellular microbiology. 2012;14:622–633. doi: 10.1111/j.1462-5822.2012.01758.x. [DOI] [PubMed] [Google Scholar]
- 65.Medvedeva YA, Fridman MV, Oparina NJ, Malko DB, Ermakova EO, Kulakovskiy IV, Heinzel A, Makeev VJ. Intergenic, gene terminal, and intragenic CpG islands in the human genome. BMC genomics. 2010;11:48. doi: 10.1186/1471-2164-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes & development. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]