Abstract
The increasing complexity of in vivo imaging technologies, coupled with the development of cell therapies, has fuelled a revolution in immune cell tracking in vivo. Powerful magnetic resonance imaging (MRI) methods are now being developed that use iron oxide- and 19F-based probes. These MRI technologies can be used for image-guided immune cell delivery and for the visualization of immune cell homing and engraftment, inflammation, cell physiology and gene expression. MRI-based cell tracking is now also being applied to evaluate therapeutics that modulate endogenous immune cell recruitment and to monitor emerging cellular immunotherapies. These recent uses show that MRI has the potential to be developed in many applications to follow the fate of immune cells in vivo.
Non-invasive cell tracking is an emerging approach for imaging cells in their native environment. The phenotype of an immune cell is partly defined by the patterns and expression levels of cell surface molecules and secreted factors. These molecules are commonly assayed in vitro using techniques such as flow cytometry and immunohistochemistry. However, determining which cell surface molecules and secreted factors are present, and at what levels, under various culture conditions or disease states, is only one piece of the puzzle. A more challenging question to answer relates to the biological roles of these molecules in vivo. Non-invasive imaging of the trafficking patterns of phenotypically defined populations of immune cells can have an important role in answering this question. Moreover, immune cells are increasingly being used as next-generation therapeutics to treat chronic conditions such as autoimmune disease and cancer. A common requirement for the development of nearly all cell therapies is a non-invasive means to visualize the biodistribution of cells following injection. Imaging of cell trafficking can provide crucial information regarding the persistence and the motility of transferred cells, as well as the optimal routes of delivery and the therapeutic doses for individuals. On the regulatory side, emerging therapies using immune cells can be slow to gain regulatory approval partly because clinical researchers are challenged to verify where the cells traffic to immediately after inoculation and where they migrate to over time. Cell tracking in vivo can potentially provide this information and might help to overcome regulatory barriers.
Inflammation is a hallmark of many of the major diseases on which biomedical research commonly focuses, such as autoimmune diseases, neurological disorders, transplant rejection and cancer. Measuring the effectiveness of treatment in terms of the inflammatory burden can be challenging. Conventional methods often include invasive biopsies and histology or imaging methods that are nonspecific for inflammation or that lack quantitative measures. There is a need for improved inflammation-specific imaging diagnostics, as well as surrogate biomarkers of inflammation, that could enable researchers to determine the efficacy of an anti-inflammatory therapy safely, quickly, quantitatively and in a longitudinal manner. There is also a need for pharmacological safety profiling to detect off-target inflammatory side effects in preclinical and clinical drug trials. Vital imaging can help to steer the decision-making process at the preclinical and clinical trial stages; it can facilitate smaller, less costly trials by enabling the enrolment of fewer patients. Imaging can potentially yield a rich data set from each patient in terms of inflammation severity and its time course in a three-dimensional anatomical context.
Given the clear need for in vivo cell tracking, much progress has been made in this area in recent years. Imaging methods using radionuclides have traditionally been used for the non-invasive imaging of leukocytes. However, technologies using magnetic resonance imaging (MRI) (BOX 1) are now emerging, and the field is experiencing a rapid expansion in the development of new imaging probes and genetically encoded reporters that enable the visualization of specific cell populations and molecular events in vivo in both animals and humans. These new capabilities have been made possible by next-generation, non-toxic cell labelling probes and by MRI methods. MRI has the advantage that it does not use ionizing radiation and can safely image deep tissues at high resolution.
Box 1. Magnetic resonance imaging.
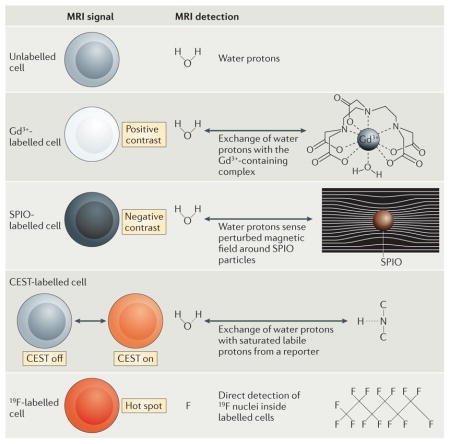
The signal used for magnetic resonance imaging (MRI) is derived from endogenous mobile water protons (1H) or fluorinated molecules (such as 19F) that are present or introduced in the subject. When the subject is placed in a large static magnetic field, the magnetic moment associated with 1H or 19F tends to align along the direction of the magnetic field. The 1H or 19F nuclei are perturbed from this equilibrium by pulsed radio-frequency radiation. Following the removal of the radio-frequency radiation, the nuclei recover to equilibrium and induce a transient voltage in a receiver antenna; this transient voltage constitutes the nuclear magnetic resonance (NMR) signal. The physical properties of a specific tissue, such as the density of nuclei, the nuclear spin–lattice relaxation time (T1) and the spin–spin relaxation time (T2), often determine the amount of signal that is available. The alignment of the nuclei along the magnetic field direction is not instantaneous, but occurs gradually over a period that is parameterized by the time constant T1. T2 is the characteristic time constant for which nuclei remain in ‘phase’ with each other, and its value is reflected in the duration of the transient NMR signal. MRI-based cell tracking involves detecting cells that exhibit a differential signal. The MRI signal can be controlled in four ways, as discussed below.
Positive contrast agents containing paramagnetic metals
Paramagnetic contrast agents primarily affect T1. Most often, T1 contrast agents contain Gd3+ that is chelated to a low-molecular-mass molecule to limit toxicity. The surrounding water protons rapidly exchange with the complex, which results in a reduction of T1 and an increase in signal intensity (positive contrast) of Gd3+-labelled cells on T1-weighted magnetic resonance images.
Negative contrast agents containing superparamagnetic iron oxides
Superparamagnetic iron oxide (SPIO) contrast agents primarily affect T2 by virtue of their iron oxide crystals, which have a strong magnetic moment. These agents commonly consist of small crystalline particles of ferrous and ferric oxides (FeO–Fe2O3) that are coated with dextran. These particulates strongly perturb the magnetic field that they are in proximity to. Surrounding water molecules subsequently experience a highly inhomogeneous magnetic field, which results in a local signal loss (negative contrast) of SPIO-labelled cells on T2-weighted magnetic resonance images.
Molecular probes that induce chemical exchange saturation transfer
Certain protons that are loosely bound at specific chemical sites, such as amide protons, have a slightly different resonance frequency than water protons, from which the MRI signal is always collected. When these labile non-water protons are irradiated with a saturation pulse at their specific off-resonance frequency, the protons lose the ability to create an MRI signal. The saturated labile non-water protons exchange position with the water protons, which leads to a loss of MRI signal of the labelled cells on chemical exchange saturation transfer (CEST) images. The CEST irradiation can be turned on and off (see the figure).
Molecular probes containing 19F
The physical principles behind detection and image formation are the same for both 1H and 19F MRI. Unlike metal ion-based magnetic resonance contrast agents, which are detected through their indirect effects on the surrounding water protons, the 19F probe functions as a tracer agent in that 19F MRI directly detects the 19F nuclei that are associated with the labelled cells, with no background. The 19F signal is directly proportional to the number of fluorine atoms and the number of labelled cells that are present (see the figure). A composite 19F and 1H image is generally constructed, which shows the regions containing labelled cells within their anatomical context.
In this Innovation article, we describe emerging methods and applications of MRI-based tracking of immune cells. These methods use exogenous cell labels that are comprised of iron oxide nano-particles or perfluorocarbon (PFC) nano-emulsions (FIG. 1), or genetically-encoded MRI reporters (FIG. 2). The field of MRI-based cell tracking is rapidly evolving, and in the future we believe it will be possible to visualize not only the location and the number of cells in vivo but also more sophisticated biological processes, such as immune cell viability and activation status. These advances indicate a future in which MRI platforms will be used to elucidate a wide range of human cellular immunological processes in vivo.
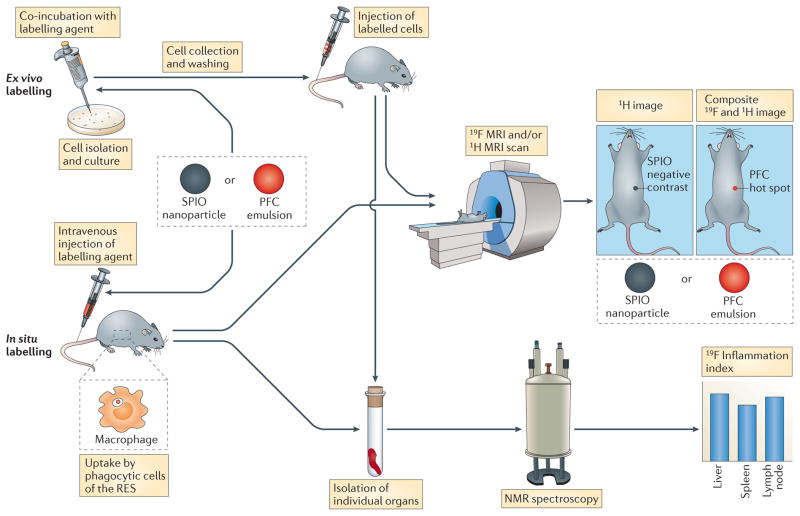
Figure 1. Schematic showing ex vivo and in situ labelling of cells with magnetic resonance nanoparticle contrast agents.
Cell labelling can be achieved ex vivo in pre-selected and cultured cells by adding the labelling reagent — superparamagnetic iron oxide (SPIO) nanoparticles or perfluorocarbon (PFC) emulsion — directly to the media followed by co-incubation. Often the labelling reagent is complexed with a cationic transfection agent before it is added to the cell culture to label non-phagocytic cells. Self-delivering formulation s of PFC emulsions have also been devised that do not require a transfection agent. Alternatively, electroporation can be used to label cells in culture. After collection and washing, labelled cells are then administered to the subject. In addition, the labelling agent can be intravenously injected; in this in situ labelling approach, the agent is intrinsically taken up by phagocytic cells of the reticuloendothelial (RES) system, particularly by monocytes and macrophages, which then accumulate at sites of inflammation. For both ex vivo and in situ labelling approaches an 1H magnetic resonance imaging (MRI) scan is then carried out to visualize the anatomy. The resulting images have a decreased signal in regions containing SPIO-labelled cells (known as negative contrast). In the case of PFC probes, a 19F MRI scan is also acquired in the same imaging session. A composite, pseudo-coloured 19F and 1H image is then constructed, in which labelled cells appear in the 19F colour channel (known as hot spot contrast). By quantifying the 19F MRI signal in individual organs using using nuclear magnetic resonance (NMR) spectroscopy an inflammation index can be calculated, which is a direct correlation between the 19F signal and the number of labelled macrophages.
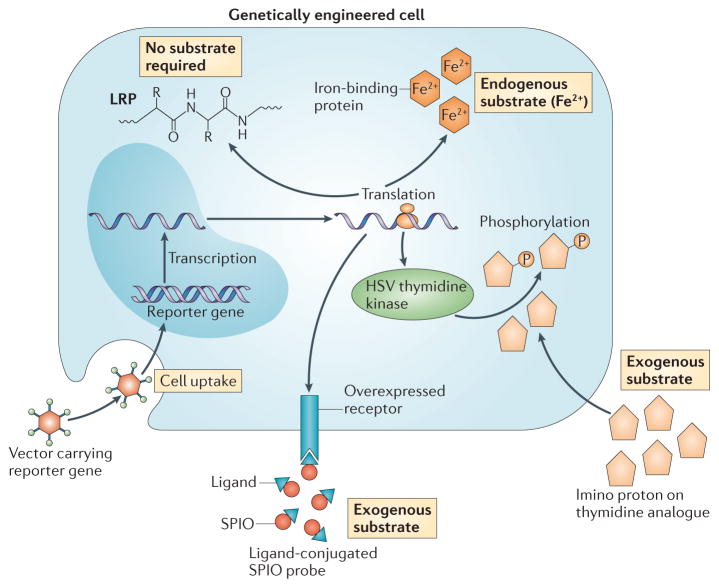
Figure 2. The development of MRI reporter genes.
Certain nucleic acid-based reporters encode cell surface receptors that bind to specific ligands that are conjugated to superparamagnetic iron oxide (SPIO) to render them magnetic resonance imaging (MRI)-detectable. They require delivery of the ligand–SPIO complex as an exogenous substrate. An example of this approach is an engineered transferrin receptor that binds transferrin-ligated SPIO. Another exogenous substrate-based approach involves a reporter gene encoding the thymidine kinase enzyme derived from herpes simplex virus (HSV). This enzyme phosphorylates thymidine analogues, which causes them to remain trapped in transduced cells. When thymidine analogues are used that are rich in specific (imino) protons, chemical exchange saturation transfer (CEST) MRI can be used to track the transduced cells. Other approaches use endogenous substrates, whereby the encoded reporter binds iron (Fe2+) that is naturally present in the body as the contrasting metal ion. Finally, a CEST reporter (such as lysine-rich protein (LRP)) can be used that does not require a substrate, as the protein itself contains multiple labile amide protons that can be saturated and detected.
Available in vivo cell tracking techniques
Much of our current knowledge of immune cell trafficking has come from static end points that have been obtained using conventional light microscopy and flow cytometry. In recent years, a wide range of imaging modalities have become available that can image immune cells in their native environment; however, each of these techniques has inherent advantages and limitations (TABLE 1). About a decade ago, the field saw a revolution with the introduction of intravital, two-photon microscopy, which enables precise determination of the movements of immune cells within lymph nodes and tumours1. However, because of limited tissue opacity, it is not possible to non-invasively look into tissues at a depth beyond ~1 mm using two-photon microscopy. Deeper tissue penetration and whole-body non-invasive tomography of small animals can be achieved by capturing the photons emitted by fluorescent or bioluminescent probes. Such bioluminescence imaging has become one of the most widely used techniques for non-invasive tracking of immune cells and inflammation in small animals, but it cannot be used in larger animals or clinically because of the physical limits of light penetration in deep tissues.
Table 1.
Comparison of non-invasive imaging modalities for in vivo cell tracking
| Imaging technique | Resolution* | Tissue penetration depth* | Sensitivity of cell detection* | Possibility of longitudinal studies* | Used generally in the clinic? | Used in clinical cell tracking? |
|---|---|---|---|---|---|---|
| MRI | +++ | +++ | +++ | ++ | Yes | Yes |
| SPECT | + | +++ | ++ | + | Yes | Yes |
| PET | + | +++ | ++ | +++ | Yes | Yes |
| CT or X-ray | +++ | +++ | + | + | Yes | No |
| Ultrasound imaging | ++ | ++ | ++ | + | Yes | No |
| BLI | + | + | ++ | +++ | No | No |
| Fluorescence imaging or NIR | + | + | ++ | ++ | No | No |
| 2PLSM | +++ | + | +++ | ++ | No | No |
| MPI84 | + | +++ | ++ | ++ | No | No |
2PLSM, two-photon laser scanning microscopy; BLI, bioluminescence imaging; CT, computed tomography; MPI, magnetic particle imaging; MRI, magnetic resonance imaging; NIR, near-infrared imaging; PET, positron emission tomography; SPECT, single-photon emission computed tomography.
Strengths and weaknesses are given using a relative scale in which + = poor, ++ = moderate and +++ = excellent.
Radionuclide labelling of cells is the oldest technique for tracking immune cells in larger animals and humans2; for example, 111In-oxine labelling of autologous white blood cells is used as a diagnostic agent for the imaging of occult inflammation and infection in humans and is currently the only cell tracking technique approved by the United States Food and Drug Administration Nuclear imaging is particularly useful for whole-body distribution studies, as there is no background signal.
Reporter genes became available in the field of nuclear medicine in the early 1990s following the development of the thymidine kinase enzyme that is derived from herpes simplex virus (HSV)3. After administration of positron-emitting substrates, such as 18FIAU (1-(2′-deoxy-2′-[18F]-β-D-arabinofuranosyl)-5-iodouracil) or 18FHBG (9-[4-[18F]fluoro-3-(hydroxymethyl)butyl]guanine), HSV thymidine kinase phosphorylates the radioactive probe and is responsible for its prolonged retention in transfected cells. In a. few studies, this approach has been used to monitor T cell trafficking4,5, including the use of positron emission tomography (PET) to visualize cytotoxic T cell homing to the tumour in a patient with glioma6.
However, there are cytotoxicity and patient safety concerns that limit the use of radionuclide-based methods for cell tracking. Moreover, radionuclide-based techniques are unable to provide anatomical imaging by themselves, and these scans must be combined, for example, with computed tomography (CT) or MRI, which adds to the methodological complexity and the cost. In addition, for the diagnostic imaging of inflammation using 111In-oxine labelling of white blood cells, there is a considerable cost associated with the invasive leukophoresis procedure and ex vivo labelling of the patient’s cells that is necessary before reinfusion. For applications in which the transferred cells, having been labelled ex vivo, are expected to persist in the body for extended periods of time (for example, in T cell and stem cell therapies), the effect of radio-cytotoxicity on cell viability is a concern. In addition, the finite half-life of the radioisotopes that are used precludes extended longitudinal studies. These limitations of radionuclide-based techniques have helped to stimulate the development of alternative imaging methods, particularly MRI-based methods (TABLE 2).
Table 2.
Overview of available in vivo MRI-based cell tracking techniques
| Probe | MRI technique | Type of contrast | Minimum number of detectable cells | Quantification of cell number? | Clinical trial approval? |
|---|---|---|---|---|---|
| Gd3+ or Mn2+ labelling | T1-weighted 1H MRI* | Positive | ~103 | No | No |
| SPIO labelling | T2-weighted 1H MRI‡ | Negative | 1 | No | Yes (in the Netherlands, China, Switzerland, Czech Republic, Israel, Poland and United Kingdom) |
| PFC labelling | 19F MRI | Hot spot (also known as tracer) | 103–105 | Yes | Yes (in USA) |
| Metal-binding reporter genes | T2-weighted 1H MRI‡ | Negative | 104 | No | No |
| CEST agents and reporter genes | 1H CEST MRI | Differential (can be colour encoded) | 104 | No | No |
CEST, chemical exchange saturation transfer; MRI, magnetic resonance imaging; PFC, perfluorocarbon; SPIO, superparamagnetic iron oxide.
T1 is the nuclear spin–lattice relaxation time.
T2 is the spin–spin relaxation time.
Metal ion-based MRI contrast agents
Positive and negative contrast agents
Intravascular MRI contrast agents that incorporate metal ions (such as Gd3+) are commonly used as positive agents to enhance the signal of lesions in clinical scans. The paramagnetic metal ions in these agents accelerate the relaxation of nearby water protons (1H), and this signal enhancement can be detected by 1H MRI (BOX 1) as a localized increase of image contrast. MRI contrast agents can also be used to label immune cells for cell tracking studies (TABLE 2). After cellular internalization by simple co-incubation, electroporation or conjugation to cell-labelling moieties, paramagnetic metal ions such as Mn2+ and Gd3+ can provide cells with positive contrast7. Positive contrast is generally preferred for image interpretation because the signal of the underlying anatomical structure is enhanced; the alternative, negative contrast (see below), decreases the signal, which makes the tissue invisible at times. However, labelling with paramagnetic metal ions only provides modest sensitivity in terms of the number of cells that can be detected (TABLE 2). Particulate formulations of metal ions, such as superparamagnetic iron oxide (SPIO) nanoparticles, create a very strong localized magnetic field disturbance in the MRI scanner because of the synergistic magnetic alignment of the individual iron ions. This localized magnetic perturbation affects nearby water protons, which causes a very strong negative contrast, in which the areas containing SPIO particles become dark on the images8,9. Immune cells can be labelled with SPIO nanoparticles using two main approaches (FIG. 1): sorted or mixed cell populations can be labelled ex vivo by incubation with SPIO nanoparticles in culture media; or non-selected phagocytic cell populations (for example, macrophages) can be labelled in situ following intravenous injection of SPIO nanoparticles. In many cases, SPIO particles have been conjugated to fluorochromes, which creates dual-mode agents that enable the validation of in vivo MRI cell tracking by post-mortem light microscopy10,11.
Labelling immune cells ex vivo
Inspired by clinical studies using radionuclide-based tracking of tumour-infiltrating lymphocytes, one of the first experimental applications of ex vivo SPIO labelling of cells was to label lymphocytes (FIG. 3a), first by labelling the cell membrane12, followed by labelling intra-cellular endosomes13,14. Membrane labelling can be achieved by targeting antibodies or peptides to specific cell surface epitopes, whereas intracellular labelling requires the use of cell-penetrating peptides or transfection agents if cells have a low level of phagocytosis or macropinocytosis activity. Intracellular labelling of lymphocytes has proven to be challenging, owing to the fairly small cytoplasmic volume of non-activated cells and the low phagocytic activity. The most widely used method for SPIO labelling of immune cells is to complex the negatively charged SPIO particles with various cationic transfection agents, such as poly-L-lysine or protamine sulphate, followed by co-incubation with cells in culture15–17. Receptor-mediated endocytosis using specific SPIO–antibody conjugates, such as conjugates that are specific for CD11c, has also been reported for the ex vivo labelling of both immature and mature dendritic cells (DCs)18. Electroporation of the imaging agent into immune cells is also feasible19.
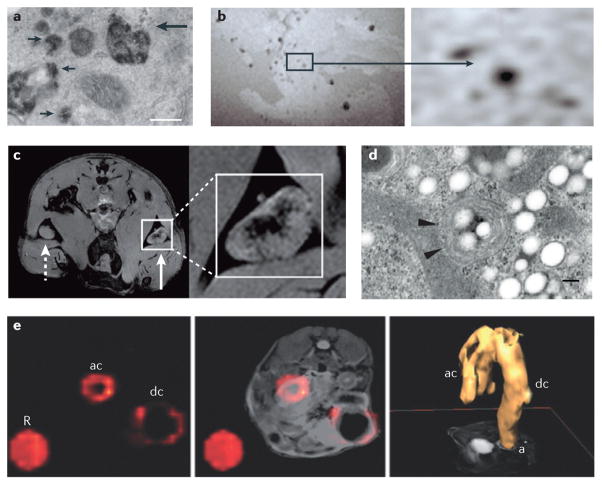
Figure 3. Tracking immune cells with MRI using SPIO nanoparticles and PFC emulsions.
a | Intracellular labelling of mononuclear cells with magnetoliposomes is shown13. The electron micrograph shows superparamagnetic iron oxide (SPIO) particles in secondary lysosomes (small arrows) and in primary lysosomes that are fusing with endosomes (large arrow). The scale bar represents 200 nm. b | Imaging of a non-obese diabetic (NOD) severe combined immunodeficient (SCID) mouse pancreas ex vivo is shown following the adoptive transfer of SPIO-labelled T cells22. Image hypointensities represent infiltrating T cells that are observed at 24 hours post transfer. c | Imaging of in vivo antigen capture and trafficking of dendritic cells (DCs) is shown26. Sentinel DCs were labelled in situ by intradermal injection of unlabelled (dashed arrow) or SPIO-labelled (solid arrow) irradiated cancer cells, which function as a vaccine. Following phagocytosis of both SPIO particles and tumour antigens in a process known as magnetovaccination, the hypointense DCs migrate into the medulla of the draining popliteal lymph node, as observed on day 8. d | An electron micrograph of a perfluorocarbon (PFC)-labelled DC is shown. Numerous bright spots (PFC droplets) are observed inside the cell. Particles appear as smooth spheroids45. Arrowheads indicate vesicles. The scale bar represents 200 nm. e | Inflammatory bowel disease (IBD) in an interleukin-10 (Il10)−/− mouse model was visualized using in situ PFC labelling and 19F magnetic resonance imaging (MRI) in vivo53. The 19F image (pseudo-colour) is shown on the far left, the composite 1H and 19F image is shown in the middle and a three-dimensional rendering of the in vivo 19F MRI data from the abdomen is shown on the far right. A reference tube that contains PFC emulsion (R) was placed alongside the torso of the mouse. The images through the abdomen show PFC accumulation (indicating inflammation) in the ascending colon (ac) and descending colon (dc), where (a) is the anus. Part a is reproduced, with permission, from REF. 13 © (1993) John Wiley and Sons. Part b is reproduced, with permission, from REF. 22 © (2002) John Wiley and Sons. Part c is reproduced, with permission, from REF. 26 © (2009) American Association for Cancer Research. Part d is reproduced, with permission, from REF. 45 © (2005) Macmillan Publishers Ltd. All rights reserved. Part e is reproduced, with permission, from REF. 52 © (2012) Macmillan Publishers Ltd. All rights reserved.
MRI-based immune cell tracking using SPIO particles has been applied to many types of preclinical study, ranging from the tumour homing of cytotoxic T cells20 and natural killer cells21 to the organ-specific homing of autoimmune T cells22,23 (FIG. 3b) and to studies investigating the migratory patterns of DCs used in cancer vaccines24–26. Iron oxide particles with a large, micrometre-sized diameter have been used to extracellularly label lymphocytes with greater sensitivity than SPIO nanoparticles27. As these iron oxide particles have a tenfold larger radius than SPIO nanoparticles, the labelled cells become much more magnetic, which enables the detection of single cells. However, the clinical translation potential of these fairly large particles remains uncertain. When dedicated gradient insert coils are used to enhance the performance of the MRI scanner, SPIO-labelled macrophages can also be imaged at the single-cell level28.
Labelling macrophages in vivo
Labelling of cells in vivo following systemic injection of SPIO particles (FIG. 1) is generally achieved by nanoparticles being taken up by the phagocytic cells of the reticuloendothelial system (RES), including circulating blood monocytes and tissue macrophages (and in much smaller numbers, neutrophils and DCs), which are frequently found at inflammatory sites. This approach has been broadly applied to image inflammatory events in both preclinical and clinical settings29–35. In a clinical context, this approach has been most widely used to image lymph nodes for tumour staging36 and to detect atherosclerotic plaques37. Immune cell labelling in situ can also be achieved by the direct injection of SPIO agents into a tissue. In a mouse study, SPIO labelling of DCs was accomplished following vaccination with irradiated labelled tumour cells from which antigens and SPIO were co-captured by the DCs. Labelled DCs could be visualized homing to the sentinel lymph node several days after this so-called ‘magnetovaccination’26 (FIG. 3c).
Concerns and limitations
It is important that any cell labelling protocol that is used for imaging does not substantially alter the immunological properties of the cells, and that it does not cause marked cytotoxicity or changes in the phenotype and the function of the cells. Changes in the phenotype of immune cells after labelling could reduce the efficacy of immunotherapeutic strategies. Overall, very few studies have noted an adverse effect of SPIO labelling on cell function or phenotype. In one study38, following SPIO labelling in culture, macrophages were found to have altered cytokine production, which was shifted towards an anti-inflammatory, less responsive phenotype. In other studies, the cytokine profile, surface expression of phenotypic markers, migratory capacity and ability to present antigens did not differ in SPIO-labelled DCs compared with in unlabelled DCs26,39,40. SPIO particles are biodegradable and, depending on the cell type, are broken down quickly; for example, when SPIO particles are taken up by Küpffer cells in the liver, the iron is metabolized and can be found in haemoglobin as soon as one week after administration41. Thus, SPIO-based cell labelling is best suited to short-term imaging studies; for example, MRI-guided injection, in which the infusion of cells and their initial engraftment can be imaged in real-time10, is viewed as a key early clinical use of SPIO-based cell tracking.
There are other limitations to SPIO-based cell tracking that are also common to essentially all nanoparticle-based and PFC nanoemulsion-based (see below) imaging reagents: in mitotic cells, cell division and subsequent dilution of the intracellular label can potentially limit long-term cell visualization; cell death can lead to dispersion of the reagent and loss of MRI detectability; the imaging reagent could potentially be transferred to resident phagocytes (such as macrophages); and if a large number of these labelled phagocytes remain in a region of interest, false positive contrast could result.
PFC cell labels and 19F MRI detection
In an effort to design next-generation cell tracking probes, there has recently been considerable interest in the use of PFC nano-emulsions. PFC emulsions can be used to track cells using ex vivo and in situ cell labelling approaches that are analogous to those used for SPIO nanoparticles (FIG. 1). PFC-based cell tracking enables high specificity cell detection and the quantification of cells. PFC-labelled cells are detected using 19F MRI (BOX 1), whereas metal ion-based contrast agents (such as SPIO particles) are detected using 1H MRI. The PFC functions as a tracer agent rather than as a contrast agent, as 19F MRI directly detects the 19F nuclei that are associated with the labelled cells. Importantly, because of the extremely low concentration of naturally occurring 19F in the body, 19F MRI has no background signal from the host’s tissues and therefore only labelled cells are observed. Thus, false positive cell detection is unlikely, which overcomes one of the major limitations of metal ion-based cell labelling approaches. Moreover, quantification of the 19F MRI signal is directly related to the apparent number of cells in the regions of interest or, alternatively, to the inflammation severity.
PFCs are among the most biologically inert organic molecules that have ever been produced42. They are water insoluble and immiscible in cell membranes and, for cellular use, they must be formulated into colloidal suspensions, such as nanoemulsions, ideally with a small droplet diameter (<200 nm)43. There are no known enzymes that metabolize PFCs in vivo and they are not degraded at typical lysosomal pH values42; thus, cell labelling with PFCs can potentially be long lasting.
Labelling immune cells ex vivo
The idea of using PFCs as tracer agents for 19F MRI followed shortly after the discovery of 1H MRI44, but their use in cell tracking is a relatively recent development45,46. Different nanoemulsion formulations have been designed for ex vivo (FIG. 3d) and in situ cell labelling43. For ex vivo labelling, an important innovation has been the formulation of nanoemulsions that intracellularly label cells in culture without the use of transfection methods47 — through the inclusion of charged moieties on the nanoemulsion droplet surface — thereby improving overall cell viability and the ease of use of PFCs. Numerous in vitro studies have investigated the effects of PFC labelling on cellular phenotype and function in primary immune cells, for example, using mouse DCs45 and T cells48. The most detailed study so far involved PFC-labelled primary human DCs49; cells were assayed for viability, maturation phenotype, cytokine production, T cell stimulatory capacity and chemotaxis, and no differences in these parameters were observed between labelled and unlabelled cells in vitro.
Tracking immune cells in vivo
Tracking cells in vivo using PFCs was first used to visualize DC migration in mice45. Several T cell studies have used PFC-based cell tracking to examine early pancreas inflammation in non-obese diabetic (NOD) mice50, which are an acute inflammation model48, and the biodistribution of mucin 1 (MUC1)-specific T cells in inflammatory bowel disease (IBD)51. A surprising finding of the IBD study was the localization of MUC1-specific T cells in the pancreatic duct, which indicates that pancreatitis, which is often diagnosed in patients with IBD, might be a true extra-intestinal manifestation of the disease. In addition, primary human DCs that are relevant to immunotherapeutic clinical trials have been labelled using PFCs and longitudinally tracked in vivo into draining lymph nodes in NOD severe combined immunodeficient (SCID) mice49.
Imaging of macrophage recruitment to inflammatory sites in vivo could help to elucidate the host inflammatory response, could provide considerable diagnostic value for a wide range of diseases and could be used as a surrogate marker and to monitor therapeutic interventions. By using PFC nanoemulsions that have been formulated for intravenous injection and that have a long blood half-life, nanoemulsion droplets enter into the RES and are intrinsically taken up in situ by monocytes and macrophages and, to a lesser extent, by neutrophils and DCs. As these in situ-labelled cells participate in inflammatory events in the body, the result is 19F accumulation at inflammatory sites. Importantly, the in vivo 19F MRI signal can be quantified in inflammatory sites, and this signal is linearly proportional to the macrophage burden52,53. 19F labelling in situ has been widely used to visualize inflammation in a large number of preclinical models of human disease, such as IBD53 (FIG. 3e), bacterial infection54, organ transplant rejection55,56, experimental allergic encephalomyelitis52,57, peripheral nerve inflammation58, pulmonary inflammation59 and cardiac and cerebral ischaemia60.
Extending the use of PFC emulsions
Similar to dual-mode SPIO agents, PFC nanoemulsions that can be detected by both 19F MRI and fluorescence have also been devised45,47; these reagents contain a bright fluorescent dye that is directly conjugated to the PFC molecule prior to nanoemulsion formulation, which thereby ensures that the originating 19F MRI and fluorescent signals are coincident in labelled cells. The fluorescent moiety enables the identification of the fate and the phenotype of labelled cells using flow cytometry, fluorescence microscopy or in vivo optical imaging; for example, dual-mode fluorescent PFC emulsions have been used to label primary murine CD4+ T cells that were then adoptively transferred into wild-type mice and imaged in multiple lymph nodes using MRI, followed by analysis using optical methods47. Near-infrared dyes have also been incorporated into PFC emulsions, which enables in vivo imaging at moderate tissue depths61. Moreover, fluorescent PFC emulsions have considerable potential as an optical-only cell labelling probe because of their brightness, low toxicity and long retention time in cells.
Conventional 19F nuclear magnetic resonance (NMR) spectroscopy can also be used to assay the biodistribution of PFC-labelled immune cells with high sensitivity in fixed, intact tissue panels. 19F NMR can measure the total cell count (or cell density) of labelled cells in tissue samples; for example, NMR cytometry has been used in T cell biodistribution studies in rodent IBD and diabetes models50,51,62. As NMR is non-destructive, the same tissues can then be processed for histology to further refine the tissue analysis. 19F-capable NMR spectrometers are ubiquitous in research laboratories and are commonly used for molecular structure determination, therefore the implementation of NMR cytometry is fairly straightforward. NMR instrumentation can also be used to rapidly and quantitatively assay inflammation in intact, excised tissue samples, which yields an inflammatory index52 that is proportional to the macrophage burden.
Future directions in immune cell tracking
Current MRI-based cell tracking technologies are poised to have a broad impact on basic and biomedical research, but there are still substantial efforts underway to devise next-generation cell imaging technologies.
MRI reporter genes
One important aim of these efforts is the development of robust MRI reporter genes. An ideal reporter gene would not be diluted by cell mitosis and would be cleared rapidly after cell death and could potentially elucidate cell viability, activation status and/or differentiation status. In recent years, researchers have been exploring schemes using exogenous (that is, injected) substrates, endogenous substrates or those that do not require a substrate (FIG. 2) in their quest to define optimal nucleic acid-based MRI reporters.
The first exogenous substrate approach involves genetically encoded, cell surface ligands that bind to MRI-detectable probes; for example, suitable ligands could include biotin63 or the transferrin receptor64, for use with a probe that consists of a SPIO nano-particle bound to streptavidin or transferrin, respectively. Alternatively, transfection of cells with the HSV thymidine kinase reporter gene used for PET imaging can be used to accumulate a thymidine analogue probe in these cells that is detectable by chemical exchange saturation transfer (CEST) MRI65 (BOX 1). One of the challenges of using ligand–probe-based reporter schemes is the efficient delivery of the probe to the tissues of interest.
The second class of MRI reporters use endogenous substrates (FIG. 2); several distinct technologies have been developed in this area. In one approach, transgenic or vector technologies are used to induce cells to express genes that encode engineered iron-binding proteins, for example, those in the ferritin family66–69. Following transgene expression in situ, the ferritin outer protein shell sequesters physiologically available iron, and the endogenous formation of an iron oxide crystal in the ferritin core renders the complex paramagnetic, which produces MRI contrast. Reporters that do not require a substrate have more recently been developed that involve the expression of amide-rich proteins that can be detected by CEST MRI70. So far, MRI reporter genes have not been used for immune cell tracking and their use has been mostly limited to tumour cells65,70 and to stem cells69.
Sensing the extracellular and intracellular environment
Another key area of innovation in the use of MRI to track cells has been the development of probes that can sense enzymatic activity and/or changes in the microenvironment, for example, changes in the local pH. Using arginine-filled liposomes as pH-sensitive contrast agents, changes in tissue pH that are associated with an immune response can potentially be detected by CEST MRI71,72. In addition, there are in vivo MRI probe technologies to detect enzymes that are overexpressed following cellular activation, such as protein kinase A (PKA). Following PKA-mediated phosphorylation of its peptide substrates, which can function as CEST contrast agents, a 50% change in the CEST MRI signal was observed73.
MRI measurements of intracellular oximetry have also been used to assay cell viability and metabolism74. Certain PFC molecules used for in vivo cytometry readily dissolve paramagnetic oxygen, which alters the MRI property (that is, the relaxation time) of the 19F signal in a manner that linearly increases with the absolute partial pressure of oxygen75. In a rodent brain glioma model, intracellular oximetry technology has recently been used to monitor the apoptosis dynamics of tumour cells as a result of their interaction with CD8+ T cells that are specific for glioma-associated antigens76.
Tracking cell–cell interactions
MRI also has the potential to visualize multiple cell populations in vivo to follow the spatial dynamics of cell–cell interactions, such as antigen presentation to B cells or T cells, and macrophage-mediated recruitment of lymphocytes; for example, ex vivo cell-labelling agents containing PFC molecules with different 19F NMR chemical shifts (that is, molecular signatures) can be used to label different immune cell subsets. Following injection of these labelled subsets, in vivo multicolour magnetic resonance images can be constructed, which show the distribution of each cell species according to its unique 19F spectral signature77 (a process known as multichannel in vivo cytometry). Alternatively, discrete immune cell populations could be labelled with two different modes of contrast, such as a positive contrast agent with a negative contrast agent (TABLE 2), a metal-based contrast agent with a CEST agent78, or multiple CEST agents of different off-resonance frequencies, which enables the generation of multiple colours79.
Prospects and challenges for translation
The use of MRI-based cell tracking in clinical trials is still in its infancy. Only a few small clinical pilot studies using SPIO-based cell tracking have been reported so far80. In the first clinical trial to use MRI-based cell tracking, an immunotherapeutic DC vaccine was labelled ex vivo with SPIO nanoparticles, and ultrasound imaging was used to guide the delivery of these cells into the inguinal lymph nodes in patients with melanoma40. Using MRI it was shown that the target lymph node was missed in four out of eight patients; this low success rate was not detected when 111In-oxine-labelled DCs were tracked using low-resolution radioscintigraphy40. In another study, peripheral blood mono-nuclear cells were labelled ex vivo with SPIO particles, injected systemically and visualized at sites of cutaneous inflammation81. As a setback for further clinical use, the SPIO agents that have been used for cell labelling — ferumoxides injectable solution (Feridex; Berlex laboratories in the United States (also known as Endorem in Europe)) and ferucarbotran (Resovist; Bayer Schering Parma AG) — are no longer being manufactured because of economic considerations. There are other experimental SPIO agents available but they would need large investments in order to be developed as a clinical MRI contrast agent.
Instead, there has recently been a technological push to use PFC nanoemulsions for clinical MRI-based cell tracking49. PFC labelling is clinically attractive because of the very low toxicity of the probes, the exceptionally high cell specificity in images and the cell quantification capacity in vivo. The sensitivity of PFC labelling to detect small cell numbers is potentially less than for SPIO agents (TABLE 2), but this disadvantage may be offset by the fact that there is no background signal in 19F images, cell detection is unambiguous (in other words, there are no false positives) and images can be readily quantitated to yield apparent cell numbers in regions of interest. The technical barriers that are associated with the implementation and the quantification of 19F MRI on a clinical scanner are surmountable; prior studies have shown the feasibility of efficient data collection schemes and the quantification of PFC signal intensity at clinical field strengths82.
Our view is that MRI-based cell tracking will routinely be used in the future for clinical trials to monitor therapeutic cell delivery and inflammation. We expect that MRI-based cell tracking will provide unexpected results regarding how cells behave in vivo following delivery and that this information will be crucial to obtain clinical success. Moreover, we foresee real-time SPIO-based MRI-guided immune cell delivery as a routine application of cell tracking, in conjunction with MRI-compatible injection catheters83. Finally, the development of cell tracking software for automated analysis of MRI scans may further facilitate image interpretation and intercomparison studies.
Conclusion
Overall, MRI-based cell tracking has great potential, not only in basic immunological research but also to guide the development of experimental immune cell-based therapies. SPIO-based cell tracking has already been used in the clinic and PFC cell labels, which are detected using 19F MRI, are currently being incorporated into clinical trials in the United States; these methods are likely to gain momentum in the future. In the meantime, emerging MRI-based cell tracking approaches could enable the visualization of cell survival, the activation status of immune cells and the identification of cell–cell interactions in vivo in ways that were previously impossible.
Acknowledgments
The authors are supported by US National Institutes of Health grants R01-CA134633, P41-EB0019772, R01-NS045062, R01-EB007825, R01-DA026299 and U54-CA151838, the Maryland Stem Cell Research Foundation and the California Institute for Regenerative Medicine.
Footnotes
Competing interests statement
The authors declare competing financial interests: see Web version for details.
Contributor Information
Eric T. Ahrens, Email: eta@ucsd.edu, Department of Radiology, University of California at San Diego, San Diego, California 92093–0695, USA
Jeff W. M. Bulte, Email: jwmbulte@mri.jhu.edu, Russell H. Morgan Department of Radiology and Radiological Science, Division of MR Research, Department of Oncology, Department of Biomedical Engineering, Department of Chemical & Biomolecular Engineering, and Cellular Imaging Section and Vascular Biology Program, Institute for Cell Engineering, The Johns Hopkins University School of Medicine, 217 Traylor Building, 720 Rutland Avenue, Baltimore, Maryland 21205, USA
References
- 1.Bousso P, Moreau HD. Functional immunoimaging: the revolution continues. Nature Rev Immunol. 2012;12:858–864. doi: 10.1038/nri3342. [DOI] [PubMed] [Google Scholar]
- 2.Thakur ML, Lavender JP, Arnot RN, Silvester DJ, Segal AW. Indium-111-labeled autologous leukocytes in man. J Nuclear Med. 1977;18:1014–1021. [PubMed] [Google Scholar]
- 3.Tjuvajev JG, et al. Imaging the expression of transfected genes in vivo. Cancer Res. 1995;55:6126–6132. [PubMed] [Google Scholar]
- 4.Koehne G, et al. Serial in vivo imaging of the targeted migration of human HSV-TK-transduced antigen-specific lymphocytes. Nature Biotech. 2003;21:405–413. doi: 10.1038/nbt805. [DOI] [PubMed] [Google Scholar]
- 5.Dubey P, et al. Quantitative imaging of the T cell antitumor response by positron-emission tomography. Proc Natl Acad Sci USA. 2003;100:1232–1237. doi: 10.1073/pnas.0337418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaghoubi SS, et al. Noninvasive detection of therapeutic cytolytic T cells with [18F]-FHBG PET in a patient with glioma. Nature Clin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki I, et al. Cell labeling for magnetic resonance imaging with the T-1 agent manganese chloride. NMR Biomed. 2006;19:50–59. doi: 10.1002/nbm.1000. [DOI] [PubMed] [Google Scholar]
- 8.Hawrylak N, et al. Nuclear-magnetic-resonance (NMR) imaging of iron oxide-labeled neural transplants. Exp Neurol. 1993;121:181–192. doi: 10.1006/exnr.1993.1085. [DOI] [PubMed] [Google Scholar]
- 9.Bulte JWM, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17:484–499. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 10.Gorelik M, et al. Use of MR cell tracking to evaluate targeting of glial precursor cells to inflammatory tissue by exploiting the very late antigen-4 docking receptor. Radiology. 2012;265:175–185. doi: 10.1148/radiol.12112212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cromer Berman SM, et al. Cell motility of neural stem cells is reduced after SPIO-labeling, which is mitigated after exocytosis. Magn Reson Med. 2013;69:255–262. doi: 10.1002/mrm.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulte JW, et al. Specific MR imaging of human lymphocytes by monoclonal antibody-guided dextran-magnetite particles. Magn Reson Med. 1992;25:148–157. doi: 10.1002/mrm.1910250115. [DOI] [PubMed] [Google Scholar]
- 13.Bulte JW, et al. Selective MR imaging of labeled human peripheral blood mononuclear cells by liposome mediated incorporation of dextran-magnetite particles. Magn Reson Med. 1993;29:32–37. doi: 10.1002/mrm.1910290108. [DOI] [PubMed] [Google Scholar]
- 14.Yeh TC, Zhang W, Ildstad ST, Ho C. Intracellular labeling of T-cells with superparamagnetic contrast agents. Magn Reson Med. 1993;30:617–625. doi: 10.1002/mrm.1910300513. [DOI] [PubMed] [Google Scholar]
- 15.Frank JA, et al. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228:480–487. doi: 10.1148/radiol.2281020638. [DOI] [PubMed] [Google Scholar]
- 16.Arbab AS, et al. Labeling of cells with ferumoxides-protamine sulfate complexes does not inhibit function or differentiation capacity of hematopoietic or mesenchymal stem cells. NMR Biomed. 2005;18:553–559. doi: 10.1002/nbm.991. [DOI] [PubMed] [Google Scholar]
- 17.Thu MS, et al. Self-assembling nanocomplexes by combining ferumoxytol, heparin and protamine for cell tracking by magnetic resonance imaging. Nature Med. 2012;18:463–467. doi: 10.1038/nm.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahrens ET, Feili-Hariri M, Xu H, Genove G, Morel PA. Receptor-mediated endocytosis of iron-oxide particles provides efficient labeling of dendritic cells for in vivo MR imaging. Magn Reson Med. 2003;49:1006–1013. doi: 10.1002/mrm.10465. [DOI] [PubMed] [Google Scholar]
- 19.Walczak P, et al. Magnetoelectroporation: improved labeling of neural stem cells and leukocytes for cellular magnetic resonance imaging using a single FDA-approved agent. Nanomedicine. 2006;2:89–94. doi: 10.1016/j.nano.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Kircher MF, et al. In vivo high resolution three-dimensional imaging of antigen-specific cytotoxic T-lymphocyte trafficking to tumors. Cancer Res. 2003;63:6838–6846. [PubMed] [Google Scholar]
- 21.Daldrup-Link HE, et al. In vivo tracking of genetically engineered, anti-HER2/neu directed natural killer cells to HER2/neu positive mammary tumors with magnetic resonance imaging. Eur Radiol. 2005;15:4–13. doi: 10.1007/s00330-004-2526-7. [DOI] [PubMed] [Google Scholar]
- 22.Moore A, et al. MRI of insulitis in autoimmune diabetes. Magn Reson Med. 2002;47:751–758. doi: 10.1002/mrm.10110. [DOI] [PubMed] [Google Scholar]
- 23.Anderson SA, et al. Magnetic resonance imaging of labeled T-cells in a mouse model of multiple sclerosis. Ann Neurol. 2004;55:654–659. doi: 10.1002/ana.20066. [DOI] [PubMed] [Google Scholar]
- 24.Baumjohann D, et al. In vivo magnetic resonance imaging of dendritic cell migration into the draining lymph nodes of mice. Eur J Immunol. 2006;36:2544–2555. doi: 10.1002/eji.200535742. [DOI] [PubMed] [Google Scholar]
- 25.Rohani R, et al. In vivo cellular MRI of dendritic cell migration using micrometer-sized iron oxide (MPIO) particles. Mol Imaging Biol. 2011;13:679–694. doi: 10.1007/s11307-010-0403-0. [DOI] [PubMed] [Google Scholar]
- 26.Long CM, van Laarhoven HW, Bulte JW, Levitsky HI. Magnetovaccination as a novel method to assess and quantify dendritic cell tumor antigen capture and delivery to lymph nodes. Cancer Res. 2009;69:3180–3187. doi: 10.1158/0008-5472.CAN-08-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro EM, Medford-Davis LN, Fahmy TM, Dunbar CE, Koretsky AP. Antibody-mediated cell labeling of peripheral T cells with micron-sized iron oxide particles (MPIOs) allows single cell detection by MRI. Contrast Media Mol Imaging. 2007;2:147–153. doi: 10.1002/cmmi.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heyn C, et al. In vivo magnetic resonance imaging of single cells in mouse brain with optical validation. Magn Reson Med. 2006;55:23–29. doi: 10.1002/mrm.20747. [DOI] [PubMed] [Google Scholar]
- 29.Chan TW, Eley C, Liberti P, So A, Kressel HY. Magnetic resonance imaging of abscesses using lipid-coated iron oxide particles. Investigative Radiol. 1992;27:443–449. doi: 10.1097/00004424-199206000-00009. [DOI] [PubMed] [Google Scholar]
- 30.Dousset V, et al. MR imaging of relapsing multiple sclerosis patients using ultra-small-particle iron oxide and compared with gadolinium. AJNR Am J Neuroradiol. 2006;27:1000–1005. [PMC free article] [PubMed] [Google Scholar]
- 31.Vellinga MM, et al. Pluriformity of inflammation in multiple sclerosis shown by ultra-small iron oxide particle enhancement. Brain. 2008;131:800–807. doi: 10.1093/brain/awn009. [DOI] [PubMed] [Google Scholar]
- 32.Sosnovik DE, Nahrendorf M. Cells and iron oxide nanoparticles on the move: magnetic resonance imaging of monocyte homing and myocardial inflammation in patients with ST-elevation myocardial infarction. Circ Cardiovasc Imaging. 2012;5:551–554. doi: 10.1161/CIRCIMAGING.112.978932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaglia JL, et al. Noninvasive imaging of pancreatic islet inflammation in type 1A diabetes patients. J Clin Invest. 2011;121:442–445. doi: 10.1172/JCI44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luciani A, et al. Adipose tissue macrophages: MR tracking to monitor obesity-associated inflammation. Radiology. 2012;263:786–793. doi: 10.1148/radiol.12111957. [DOI] [PubMed] [Google Scholar]
- 35.Kanno S, et al. Macrophage accumulation associated with rat cardiac allograft rejection detected by magnetic resonance imaging with ultrasmall superparamagnetic iron oxide particles. Circulation. 2001;104:934–938. doi: 10.1161/hc3401.093148. [DOI] [PubMed] [Google Scholar]
- 36.Harisinghani MG, et al. Noninvasive detection of clinically occult lymph-node metastases in prostate cancer. New Engl J Med. 2003;348:2491–2499. doi: 10.1056/NEJMoa022749. [DOI] [PubMed] [Google Scholar]
- 37.Kooi ME, et al. Accumulation of ultrasmall superparamagnetic particles of iron oxide in human atherosclerotic plaques can be detected by in vivo magnetic resonance imaging. Circulation. 2003;107:2453–2458. doi: 10.1161/01.CIR.0000068315.98705.CC. [DOI] [PubMed] [Google Scholar]
- 38.Siglienti I, Bendszus M, Kleinschnitz C, Stoll G. Cytokine profile of iron-laden macrophages: implications for cellular magnetic resonance imaging. J Neuroimmunol. 2006;173:166–173. doi: 10.1016/j.jneuroim.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Verdijk P, et al. Sensitivity of magnetic resonance imaging of dendritic cells for in vivo tracking of cellular cancer vaccines. Int J Cancer. 2007;120:978–984. doi: 10.1002/ijc.22385. [DOI] [PubMed] [Google Scholar]
- 40.de Vries IJM, et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nature Biotech. 2005;23:1407–1413. doi: 10.1038/nbt1154. [DOI] [PubMed] [Google Scholar]
- 41.Weissleder R, et al. Superparamagnetic iron oxide: pharmacokinetics and toxicity. AJR Am J Roentgenol. 1989;152:167–173. doi: 10.2214/ajr.152.1.167. [DOI] [PubMed] [Google Scholar]
- 42.Riess JG. Oxygen carriers (“blood substitutes”) — raison d’etre, chemistry, and some physiology. Chem Rev. 2001;101:2797–2919. doi: 10.1021/cr970143c. [DOI] [PubMed] [Google Scholar]
- 43.Janjic JM, Ahrens ET. Fluorine-containing nanoemulsions for MRI cell tracking. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1:492–501. doi: 10.1002/wnan.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holland GN, Bottomley PA, Hinshaw WS. F-19 magnetic-resonance imaging. J Magn Reson. 1977;28:133–136. [Google Scholar]
- 45.Ahrens ET, Flores R, Xu HY, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nature Biotech. 2005;23:983–987. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- 46.Bulte JWM. Hot spot MRI emerges from the background. Nature Biotech. 2005;23:945–946. doi: 10.1038/nbt0805-945. [DOI] [PubMed] [Google Scholar]
- 47.Janjic JM, Srinivas M, Kadayakkara DK, Ahrens ET. Self-delivering nanoemulsions for dual fluorine-19 MRI and fluorescence detection. J Am Chem Soc. 2008;130:2832–2841. doi: 10.1021/ja077388j. [DOI] [PubMed] [Google Scholar]
- 48.Srinivas M, et al. In vivo cytometry of antigen-specific t cells using 19F MRI. Magn Reson Med. 2009;62:747–753. doi: 10.1002/mrm.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helfer BM, et al. Functional assessment of human dendritic cells labeled for in vivo F-19 magnetic resonance imaging cell tracking. Cytotherapy. 2010;12:238–250. doi: 10.3109/14653240903446902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn Reson Med. 2007;58:725–734. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- 51.Kadayakkara DK, et al. Inflammation driven by overexpression of the hypoglycosylated abnormal mucin 1 (MUC1) links inflammatory bowel disease and pancreatitis. Pancreas. 2010;39:510–515. doi: 10.1097/MPA.0b013e3181bd6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahrens ET, Young WB, Xu H, Pusateri LK. Rapid quantification of inflammation in tissue samples using perfluorocarbon emulsion and fluorine-19 nuclear magnetic resonance. Biotechniques. 2011;50:229–234. doi: 10.2144/000113652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kadayakkara DK, Ranganathan S, Young WB, Ahrens ET. Assaying macrophage activity in a murine model of inflammatory bowel disease using fluorine-19 MRI. Lab Invest. 2012;92:636–645. doi: 10.1038/labinvest.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hertlein T, et al. Visualization of abscess formation in a murine thigh infection model of Staphylococcus aureus by F-19-magnetic resonance imaging (MRI) PLoS ONE. 2011;6:e18246. doi: 10.1371/journal.pone.0018246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hitchens TK, et al. 19F MRI detection of acute allograft rejection with in vivo perfluorocarbon labeling of immune cells. Magn Reson Med. 2011;65:1144–1153. doi: 10.1002/mrm.22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flogel U, et al. Noninvasive detection of graft rejection by in vivo 19F MRI in the early stage. Am J Transplant. 2011;11:235–244. doi: 10.1111/j.1600-6143.2010.03372.x. [DOI] [PubMed] [Google Scholar]
- 57.Noth U, et al. Perfluoro-15-crown-5-ether labelled macrophages in adoptive transfer experimental allergic encephalomyelitis. Artif Cells Blood Substit Immobil Biotechnol. 1997;25:243–254. doi: 10.3109/10731199709118914. [DOI] [PubMed] [Google Scholar]
- 58.Weise G, Basse-Luesebrink TC, Wessig C, Jakob PM, Stoll G. In vivo imaging of inflammation in the peripheral nervous system by 19F MRI. Exp Neurol. 2011;229:494–501. doi: 10.1016/j.expneurol.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 59.Ebner B, et al. Early assessment of pulmonary inflammation by 19F MRI in vivo. Circ Cardiovasc Imag. 2010;3:U202–U109. doi: 10.1161/CIRCIMAGING.109.902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flogel U, et al. In vivo monitoring of inflammation after cardiac and cerebral ischemia by fluorine magnetic resonance imaging. Circulation. 2008;118:140–148. doi: 10.1161/CIRCULATIONAHA.107.737890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Hanlon CE, Amede KG, O’Hear MR, Janjic JM. NIR-labeled perfluoropolyether nanoemulsions for drug delivery and imaging. J Fluor Chem. 2012;137:27–33. doi: 10.1016/j.jfluchem.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morel PA, et al. Gene expression analysis of dendritic cells that prevent diabetes in NOD mice: analysis of chemokines and costimulatory molecules. J Leukoc Biol. 2011;90:539–550. doi: 10.1189/jlb.0311126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tannous BA, et al. Metabolic biotinylation of cell surface receptors for in vivo imaging. Nature Methods. 2006;3:391–396. doi: 10.1038/nmeth875. [DOI] [PubMed] [Google Scholar]
- 64.Weissleder R, et al. In vivo magnetic resonance imaging of transgene expression. Nature Med. 2000;6:351–355. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 65.Bar-Shir A, et al. Transforming thymidine into a magnetic resonance imaging probe for monitoring gene expression. J Am Chem Soc. 2013;135:1617–1624. doi: 10.1021/ja312353e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahrens ET. Contrast agents for magnetic resonance imaging and methods related thereto. 10/384,496. US Patent. 2003
- 67.Genove G, DeMarco U, Xu HY, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nature Med. 2005;11:450–454. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 68.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iordanova B, Ahrens ET. In vivo magnetic resonance imaging of ferritin-based reporter visualizes native neuroblast migration. Neuroimage. 2012;59:1004–1012. doi: 10.1016/j.neuroimage.2011.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilad AA, et al. Artificial reporter gene providing MRI contrast based on proton exchange. Nature Biotech. 2007;25:217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 71.Sherry AD, Woods M. Chemical exchange saturation transfer contrast agents for magnetic resonance imaging. Annu Rev Biomed Eng. 2008;10:391–411. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chan KW, et al. MRI-detectable pH nanosensors incorporated into hydrogels for in vivo sensing of transplanted-cell viability. Nature Mater. 2013;12:268–275. doi: 10.1038/nmat3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Airan RD, et al. MRI biosensor for protein kinase A encoded by a single synthetic gene. Magn Reson Med. 2012;68:1919–1923. doi: 10.1002/mrm.24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kadayakkara DK, Janjic JM, Pusateri LK, Young WB, Ahrens ET. In vivo observation of intracellular oximetry in perfluorocarbon-labeled glioma cells and chemotherapeutic response in the CNS using fluorine-19 MRI. Magn Reson Med. 2010;64:1252–1259. doi: 10.1002/mrm.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sotak CH, et al. A new perfluorocarbon for use in fluorine-19 magnetic resonance imaging and spectroscopy. Magn Reson Med. 1993;29:188–195. doi: 10.1002/mrm.1910290206. [DOI] [PubMed] [Google Scholar]
- 76.Zhong J, Sakaki M, Okada H, Ahrens ET. In vivo intracellular oxygen dynamics in murine brain glioma and immunotherapeutic response of cytotoxic T cells observed by fluorine-19 magnetic resonance imaging. PLoS One. 2013;8:e59479. doi: 10.1371/journal.pone.0059479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Partlow KC, et al. F-19 magnetic resonance imaging for stem/progenitor cell tracking with multiple unique perfluorocarbon nanobeacons. FASEB J. 2007;21:1647–1654. doi: 10.1096/fj.06-6505com. [DOI] [PubMed] [Google Scholar]
- 78.Gilad AA, et al. Feasibility of concurrent dual contrast enhancement using CEST contrast agents and superparamagnetic iron oxide particles. Magn Reson Med. 2009;61:970–974. doi: 10.1002/mrm.21928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu G, et al. In vivo multicolor molecular MR imaging using diamagnetic chemical exchange saturation transfer liposomes. Magn Reson Med. 2012;67:1106–1113. doi: 10.1002/mrm.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bulte JW. In vivo MRI cell tracking: clinical studies. AJR Am J Roentgenol. 2009;193:314–325. doi: 10.2214/AJR.09.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Richards JMJ, et al. Clinical cell tracking of mononuclear cells using magnetic resonance imaging and superparamagnetic particles of iron oxide. Br J Surg. 2011;98:4–4. [Google Scholar]
- 82.Keupp J, et al. Simultaneous dual-nuclei imaging for motion corrected detection and quantification of 19F imaging agents. Magn Reson Med. 2011;66:1116–1122. doi: 10.1002/mrm.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aarntzen EH, et al. In vivo tracking techniques for cellular regeneration, replacement, and redirection. J Nucl Med. 2012;53:1825–1828. doi: 10.2967/jnumed.112.106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bulte JW, et al. MPI cell tracking: what can we learn from MRI? Proc Soc Photo-Opt Instrum Eng. 2011;7965:79650z. doi: 10.1117/12.879844. [DOI] [PMC free article] [PubMed] [Google Scholar]