Summary
Achieving efficient cotranslational folding of their complex proteomes poses a challenge for eukaryotic cells. Nascent polypeptides emerging vectorially from the ribosome often cannot fold stably and may be susceptible to misfolding and degradation. The extent to which nascent chains are subject to cotranslational quality control and degradation remains unclear. Here, we directly and quantitatively assess cotranslational ubiquitination and identify, at a systems level, the determinants and factors governing this process. Cotranslational ubiquitination occurs at very low levels, and is carried out by a complex network of E3 ubiquitin ligases. Ribosome-associated chaperones and cotranslational folding protect the majority of nascent chains from premature quality control. Nonetheless, a number of nascent chains whose intrinsic properties hinder efficient cotranslational folding remain susceptible for cotranslational ubiquitination. We find that quality control at the ribosome is achieved through a tiered system, where nascent polypeptides first have a chance to fold before becoming accessible to ubiquitination.
Introduction
The extent, specificity and significance of cotranslational quality control are long standing and important open questions in biology (Hartl et al., 2011). Polypeptides emerge vectorially from ribosomes and often are unable to fold stably until a complete domain has been synthesized (Hartl et al., 2011; Kramer et al., 2009; Preissler and Deuerling, 2012). This raises the question of whether nascent polypeptides are substrates of cellular quality control pathways, which target misfolded proteins for degradation by tagging through ubiquitination (Finley, 2009). Indeed, early studies indicated that a large fraction of nascent polypeptides (over 30%) were immediately degraded upon synthesis, leading to the so-called DRiPs (Defective Ribosomal Products) hypothesis (Schubert et al., 2000; Yewdell and Nicchitta, 2006). In contrast, subsequent studies pinpointed concerns regarding these initial studies, calling into question the significance of cotranslational ubiquitination (Vabulas and Hartl, 2005). The extent and significance of cotranslational quality control in vivo remains an important unanswered question.
A priori arguments could be presented both for and against robust cotranslational quality control at the ribosome, particularly in eukaryotic cells. On the one hand, the efficient biogenesis and maturation of functional proteins is critical for cell viability. Indeed, eukaryotic cells have evolved an elaborate machinery of ribosome-bound chaperones that interacts with and facilitates folding of nascent polypeptides (Hartl et al., 2011). Studies using model proteins have suggested that de novo folding is relatively efficient (Frydman et al., 1994; Vabulas and Hartl, 2005) although this may not be the case for all proteins (Sato et al., 1998). On the other hand, polypeptides emerge vectorially from the ribosome and often cannot complete folding until fully synthesized (Hartl et al., 2011; Kramer et al., 2009; Preissler and Deuerling, 2012). This places nascent polypeptides in a very precarious state, with a high potential for misfolding. Several studies observed cotranslational ubiquitination of the large membrane proteins CFTR (Sato et al., 1998) and Apolipoprotein B100 (Zhou et al., 1998) following in vitro translation. Furthermore, elegant studies engineering a synthetic N-end rule ubiquitination signal on the large enzyme β-galactosidase demonstrated that cotranslational ubiquitination and degradation can occur in vivo when the synthetic ubiquitination signal emerges at the N-terminus of a translating polypeptide (Turner and Varshavsky, 2000). Interestingly, placing the same signal on a shorter protein, Ura3, strongly reduced the extent of cotranslational ubiquitination. Interestingly, a ribosome-bound, folding-incompetent variant of actin was protected from ubiquitination and degradation until released from the ribosome (Frydman and Hartl, 1996).
Given the deleterious nature of misfolded species, it is clear that the cell must balance the need to maintain a properly folded proteome with the need to avoid premature degradation of nascent chains until they can complete their synthesis and folding. Surprisingly, until now this question has not been addressed directly (Figure 1A). Here, we use a direct and quantitative approach to assess the occurrence and extent of cotranslational ubiquitination in vivo in Saccharomyces cerevisiae and define the underlying principles governing quality control at the ribosome.
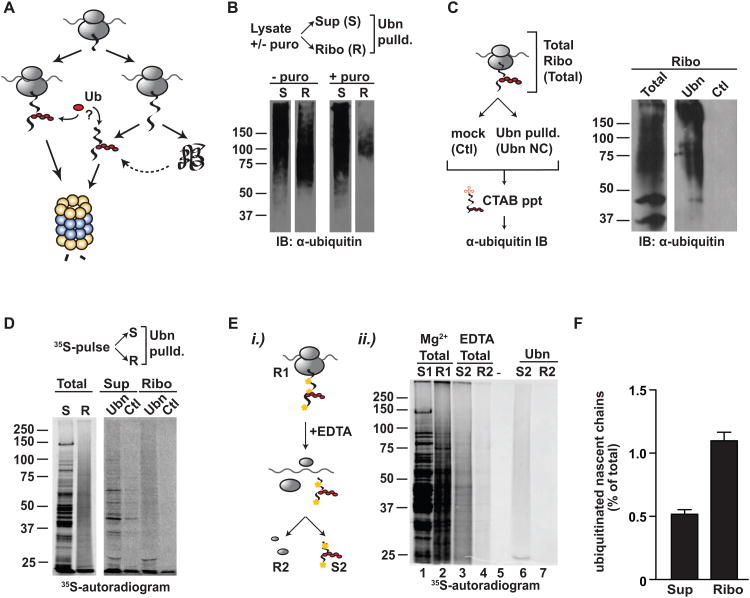
Figure 1. Cotranslational ubiquitination occurs at low levels in vivo.
(A) Nascent chains may be ubiquitinated as they emerge from the ribosome or post-translationally once the protein has been fully synthesized (B) Ribosome-nascent chain complexes (RNCs) contain ubiquitinated material that is released by puromycin. Ultrasedimentation with or without puromycin to release nascent chains separated the supernatant (S) from the ribosome-nascent chain complexes (R), which were subjected to polyUb-affinity isolation (Ubn). Totals and pulldowns were analyzed by SDS-PAGE and ubiquitin immunoblot. (C) Peptidyl-tRNA isolation through CTAB precipitation shows direct ubiquitination of nascent chains. Isolated RNCs were incubated with polyUb-affinity resin or GST-only and the nascent chain-tRNA complexes precipitated with CTAB. Samples run on SDS-PAGE were immunoblotted for ubiquitin. (D) 35S–pulse-labeled nascent chains are ubiquitinated while still ribosome-bound. Cells were pulse-labeled for 1 min with 35S-Met/Cys; fractionated into supernatant (S or Sup), containing newly made, full-length 35S-polypeptides and RNCs (R or Ribo), containing ribosome bound 35S-nascent chains and pulled-down either with GST (Ctl) or polyUb-affinity resin (Ubn). (E) 35S-labeled nascent chains are directly ubiquitinated and can be isolated independently of the ribosome. i) Cells were pulse-labeled and fractionated as before; S: supernatant; R: ribosomal pellet. EDTA was added to the RNCs (R1), which was resuspended and spun again to yield empty ribosomes (R2) and released nascent chains (S2). S2 and R2 were then subject to polyUb-affinity pulldown. Samples were analyzed by SDS-PAGE and autoradiography. ii.) Autoradiogram showing totals of S1/R1 and S2/R2, as well as the polyUb-affinity pulldown of S2 and R2 (Ubn). (F) A small fraction of nascent chains and newly made proteins is cotranslationally ubiquitinated; calculated as the ratio of 35S-labeled nascent chains isolated by Dsk2 over total 35S-labeled nascent chains in the ribosomal or in the supernatant fraction (mean ± SEM, n=59). See also Figure S1.
Results
A small fraction of nascent chains are ubiquitinated cotranslationally in vivo
To determine whether nascent chains are ubiquitinated as they emerge from the ribosome in vivo, we used a previously characterized polyubiquitin-affinity (polyUb-affinity) isolation approach that quantitatively captures polyubiquitinated proteins (Funakoshi et al., 2002; Mayor and Deshaies, 2005). Indeed, both ubiquitin immunoblot analyses and a Cy3-labeled tetra-ubiquitin tracer both demonstrated that this polyUb capture assay quantitatively depletes polyubiquitinated species from the cell extracts, leaving mono- and multi-ubiquitinated proteins in the supernatant (Figure S1A, B). We thus employed this approach to assess ubiquitination of nascent chains following ribosome-nascent chain complex (RNCs) isolation by sucrose cushion sedimentation from intact cells (Figure 1). Immunoblot analysis for ubiquitin indicated that both the ribosome-bound fraction (Figures 1 and S1C; Ribo or R) and the post-ribosomal supernatant (Sup or S) contained polyubiquitinated proteins (Figure 1B, left panel). If ubiquitinated nascent chains contribute to the polyubiquitin-signal in the ribosomal fraction, then releasing nascent polypeptides from ribosomes with either puromycin (Figure 1B, right panel) or EDTA (Figure S1D) prior to ribosome sedimentation should reduce the amount of ubiquitinated species in the ribosomal fraction. Either treatment dramatically decreased the presence of polyubiquitinated proteins in the ribosomal fraction, and concomitantly increased the polyubiquitin signal in the supernatant. The puromycin-reactive ribosome-nascent chain fraction contained a higher proportion of higher molecular weight ubiquitinimmunoreactive bands than low molecular weight, presumably mono-ubiquitinated, bands (Fig. 1B). Thus, RNCs contain polyubiquitinated polypeptides that shift from the ribosomal fraction to the post-ribosomal supernatant when nascent chains are released. A further direct test of nascent chains ubiquitination relied on the ability of the detergent cetyltrimethyl ammonium bromide (CTAB) to selectively precipitate peptidyl-tRNAs (Nakatogawa and Ito, 2001). Ribosome-nascent chain complexes were subjected to polyUb-affinity isolation as above; then, the peptidyl-tRNAs were released from ribosomes with EDTA, and the released tRNA-nascent chains were precipitated using CTAB. The subsequent ubiquitin immunoblot of the CTAB precipitates clearly indicated that nascent chains were directly ubiquitinated (Figures 1C and S1E). RNase treatment, which removes the tRNA moiety, leads to a reduction of the precipitated material, showing that CTAB is specific to the peptidyl-tRNA (Figure S1E). Together, these distinct lines of evidence demonstrate that nascent chains are directly ubiquitinated in vivo.
The extent of nascent chain ubiquitination was quantified using pulse-labeling analysis. Newly translated polypeptides were specifically labeled with a brief (1 minute) 35S-pulse, followed by isolation of ribosome-35S-nascent chain complexes. Since translation was not synchronized, a wide range of short and long nascent polypeptides are labeled during the pulse, including some that are close to completion. PolyUb-affinity isolation of both ribosome-bound and released polypeptides, followed by autoradiography, confirmed the presence of ubiquitinated ribosome-bound nascent chains (Figure 1D). Analysis of the post-ribosomal supernatant indicated that completed newly made proteins were also ubiquitinated, even after only 1 minute labeling, which is sufficient to synthesize approximately 250-300 amino acids (Figure 1D). We next examined whether all ubiquitinated nascent chains are captured by our polyUb affinity isolation. To this end, RNCs containing radiolabeled nascent chains were subjected to two sequential rounds of polyUb-affinity pulldown (Figure S1F). All ubiquitinated material was depleted after the first round of polyUb-affinity isolation (Figure S1F), demonstrating that the polyUb-affinity resin is not limiting in our experiments. Together with the experiments in Figure S1A and S1B, we conclude that the polyUb affinity approached used here quantitatively isolates poly-ubiquitinated polypeptides from the RNC fraction. The direct polyubiquitination of nascent chains was further confirmed in two additional experiments. First, 35S-nascent chains were released from purified RNCs and polyUb-affinity isolation was carried out after a further round of sucrose cushion sedimentation to separate vacant ribosomes from the released nascent chains (Figure 1E, lane 6, S2). This demonstrates that released nascent chains are directly captured by the polyUb-affinity resin. Secondly, we subjected isolated ribosome-35S-nascent chain complexes to a stringent salt wash (Fleischer et al., 2006) to remove associated factors prior to the ribosome dissociation step, and subsequent polyUb-affinity isolation confirmed that these salt-stripped nascent chains were ubiquitinated (Figure S1G, lanes 6 and 8, 10 and 12). Taken together, these experiments demonstrate that nascent chains are directly ubiquitinated while on the ribosome. Quantitative analysis showed that approximately 1.1 ± 0.07% (n=59) of the ribosome-bound nascent chains and 0.5 ± 0.04 % (n=59) of completed, newly made polypeptides were ubiquitinated in vivo (Figure 1F). We conclude that newly made proteins are ubiquitinated co- and posttranslationally during synthesis.
Ubiquitinated nascent chains are targeted to the proteasome
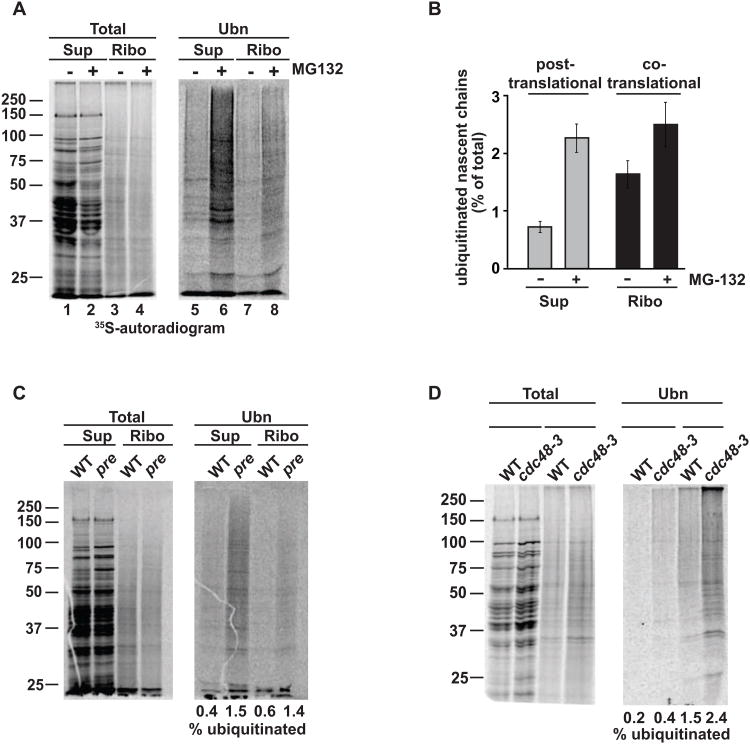
We next examined if ubiquitination of ribosome-bound and newly made polypeptides serves a quality control function, targeting them to the proteasome for degradation. Addition of the proteasome inhibitor MG132 shortly prior to 35S-labeling and analysis did not affect labeling efficiency or translation rates, but caused a marked accumulation of ubiquitinated ribosome-bound nascent chains, as well as ubiquitinated full-length polypeptides in the supernatant (Figures 2A and 2B). Similar results were obtained using another proteasome inhibitor, PS341 (Velcade; Figure S2A), as well as the proteasome defective strain pre1-1 2-1 (Figure 2C), indicating that ubiquitinated polypeptides are co-and posttranslationally targeted to the proteasome. In some quality control processes, proteasomal degradation requires the additional function of the AAA-ATPase Cdc48/p97 (Verma et al., 2011). We observed an increase of ubiquitinated nascent chains in the temperature-sensitive strain cdc48-3 (Figure 2D) as well as a mutation in its cofactor Npl4 (not shown). Therefore, Cdc48 appears to function in delivering ubiquitinated nascent chains to the proteasome. Thus, a small fraction of translating polypeptides is consistently ubiquitinated on the ribosome or soon after translation and is degraded by the proteasome with the help of Cdc48/p97. Consistent with this function, we find that the proteasome co-migrates with translating ribosomes on sucrose gradients and shifts towards lighter fractions upon EDTA dissociation of polysomes (Figure S2B), as reported (Sha et al., 2009). The quantitative nature of the polyUb-affinity isolation approach together with the sensitive and quantitative character of radiolabeling indicates that proteasome inhibition enhances the combined fraction of ubiquitinated nascent chains and newly made polypeptides from ∼1.5% to 5% (Figures 2A and 2B).
Figure 2. Co- and post-translationally ubiquitinated nascent chains are degraded by the proteasome.
(A) Ubiquitinated nascent chains and newly made proteins accumulate upon proteasome inhibition. Cells were pulse-labeled with 35S-Met/Cys with or without MG132. RNC isolation and polyUb-affinity isolation were performed as above and analyzed by SDS-PAGE and autoradiography. (B) Quantification of co- and post-translational ubiquitination in the presence and absence of proteasome inhibitors (mean ± SEM, n=3). (C) Genetic proteasome impairment in pre1-1 2-1 leads to accumulation of ubiquitinated nascent chains. Pulse-labeling and ubiquitin pulldown as described in (A). (D) Impairment of CDC48 increases levels of ubiquitinated nascent chains and newly made proteins. Ubiquitin-pulldown after radiolabeling in WT and cdc48-3 was carried out as in (A). See also Figure S2
To determine when, during or after synthesis, pulse-labeled proteins become ubiquitinated and degraded, we next performed pulse-chase experiments in the presence and absence of PS341. Without inhibitor, ubiquitinated nascent polypeptides are rapidly degraded and were not detectable after 5-10 min (Figures S2D and S2E). These ubiquitinated nascent polypeptides were degraded much more rapidly than most short-lived proteins, which have half-lives ranging from 30 min to 2 hours (Belle et al., 2006), and thus likely reflect co-and post-translational quality control. Addition of PS341 produced an accumulation of ubiquitinated newly made proteins, peaking at 2.5 minutes of chase time (Figure S2E). Thus, a small pool of newly made polypeptides is ubiquitinated and rapidly degraded during or soon after translation, presumably to serve a quality control function.
A network of ubiquitin ligases mediates quality control at the ribosome
The ubiquitination machinery mediating cotranslational ubiquitination was assessed using a panel of single and double deletion strains of ubiquitin-conjugating (E2) and ubiquitin ligase (E3) enzymes (Figures 3A and 3B and S3). Cotranslational ubiquitination was detected by pulse-labeling cells for 1 min with 35S-methionine, followed by RNC isolation and direct determination of the fraction of ubiquitinated nascent chains versus total nascent chains translated. This sensitive approach allowed us to reproducibly detect small differences in ubiquitination. No significant reduction in cotranslational ubiquitination was observed for single E2 deletions (data not shown), consistent with the known redundancy of E2 enzymes (Chen et al., 1993; Seufert and Jentsch, 1990). However, a strong reduction of ubiquitinated proteins was observed in a strain deleted for Ubc1/4, and less so in ubc4/5Δ and ubc4-7Δ (Figure S3), suggesting that these enzymes contribute to cotranslational ubiquitination of nascent polypeptides. Since these E2s also participate generally in quality control of short-lived, misfolded proteins, these results suggest an overlap between the machineries that carry out quality control on and off the ribosome.
Figure 3. A network of ubiquitin mediates cotranslational ubiquitination.
(A) WT and E3 ubiquitin ligase mutant strains were pulse-labeled with 35S-Met/Cys, and RNCs were isolated and subjected to polyUb-affinity isolation as before. Shown is a representative autoradiogram for pulldowns in different E3 ligase deletion strains. (B) Quantification indicates the strongest effect is observed for hel2Δ/rkr1Δ (red bar). * p < 0.05, (mean ± SEM, n=3). (C) Differential role of Rkr1, Hel2 and Not4 on the degradation of translation products of stalled or non-stop mRNAs. rkr1Δ abrogates degradation, while hel2Δ has a partial effect and not4Δ is without effect. (Left) Schematic of non-stop (NS) and stalled (K12) reporters (Bengtson and Joazeiro, 2010; Ito-Harashima et al., 2007). (Right) The indicated constructs were expressed in the presence or absence of MG132 and equal amounts of lysate analysed by SDS-PAGE and immunoblot anti-GFP. (D) Impairing mRNA quality control increases cotranslational ubiquitination. WT and indicated CCR4/NOT mutant strains were pulse-labeled with 35S-Met/Cys, RNCs isolated and subjected to polyUb-affinity isolation as described before. (E) Quantification: mean and SEM of 3 experiments. * p < 0.05. (F) not4Δ deletion is hypersensitive to AZC. Dilution series on YPD plates with or without 1 mg/ml AZC. Plates were imaged after 3 days at 30°C. See also Figure S3
We next examined which ubiquitin ligases play a role in ubiquitination of ribosome-bound chains. Previous studies identified a ribosome-bound ubiquitin ligase, Ltn1/Rkr1, dedicated to the degradation of non-stop mRNA-derived products (NS-mRNA) (Bengtson and Joazeiro, 2010). Notably, this ligase does not ubiquitinate misfolded quality control substrates posttranslationally (Bengtson and Joazeiro, 2010). Only a small reduction in cotranslational ubiquitination was observed in rkr1Δ cells (Figure 3B), even though these cells were blocked in degradation of NS-mRNA encoded polypeptides as well as in the degradation of stalled nascent chains, as shown using non-stop (NS) and poly-Lys (K12) reporters in the rkr1Δ deletion strain (Bengtson and Joazeiro, 2010; Ito-Harashima et al., 2007) (Figure 3C; NS and K12 respectively).
We also tested a panel of strains deleted in E3s previously implicated in quality control, as well as an additional uncharacterized ubiquitin ligase, Ydr266c/Hel2, previously identified as ribosome-bound through proteomics (Fleischer et al., 2006). No single E3 deletion abrogated cotranslational ubiquitination; however some ligase deletions caused small (5-10%) reductions in cotranslational ubiquitination, including the proteasome linked E3 hul5Δ (Fang et al., 2011) as well as doa10Δ and hrd1Δ (Figures 3A and 3B). The double deletion doa10/hrd1A afforded no further reduction in cotranslational ubiquitination (Figures 3A and 3B). No decrease in cotranslational ubiquitination was observed for single and double deletions of Ubr1 and Ubr2 (Figure 3B and data not shown). Interestingly, deletion of the ribosome-bound E3 Hel2 reduced cotranslational ubiquitination to a level comparable to rkr1Δ (Figure 3B). In addition, Hel2 deletion resulted in a slight stabilization of NS-mRNA products (Figure 3C, NS), as well as nascent chains carrying the K12 stalling signal (Figure 3F, K12). We next asked whether Rkr1 and Hel2 may have redundant functions in cotranslational ubiquitination. Indeed, a ∼25% reduction in ubiquitinated nascent chains was observed in the double deletion rkr1/hel2Δ (Figures 3A and 3B). Our results identify Hel2 as a novel E3 ligase involved in cotranslational ubiquitination and suggest that the ribosome bound ligases Rkr1 and Hel2 work in parallel with additional non-ribosomal ubiquitin ligases such as Doa10, Hrd1 and Hul5 to mediate cotranslational quality control.
Impairing mRNA quality control enhances cotranslational ubiquitination
Not4 is a component of the CCR4/Not complex, which associates with ribosomes but is also involved in mRNA maturation and quality control (Collart and Panasenko, 2012). Previous studies implicated Not4, the E3 ligase subunit of CCR4/NOT, in cotranslational quality control (Dimitrova et al., 2009). Surprisingly, deletion of Not4 dramatically enhanced the proportion of ubiquitinated nascent chains, in contrast to what is expected if Not4 mediates cotranslational ubiquitination (Figure 3E). Furthermore, Not4 was without effect on the degradation of NS-mRNA translation products or stalled mRNAs (Figure 3C). A plausible explanation for this paradoxical result is that Not4 deletion decreases mRNA quality control fidelity, resulting in an increase of aberrant nascent chains that are subject to enhanced ubiquitination. To test this idea, we examined a deletion of the deadenylase subunit of the CCR4-NOT complex, Ccr4. Similar to not4Δ, ccr4Δ also resulted in enhanced cotranslational ubiquitination (Figure 3E). These results indicate that a reduction in mRNA quality control induces nascent chain misfolding and ubiquitination. Interestingly, the tight interplay between mRNA and nascent chain quality control is highlighted by the fact that loss of Not4 renders cells hypersensitive to AZC-induced misfolding (Figure 3F).
Global identification of mRNAs encoding nascent chains susceptible to cotranslational ubiquitination
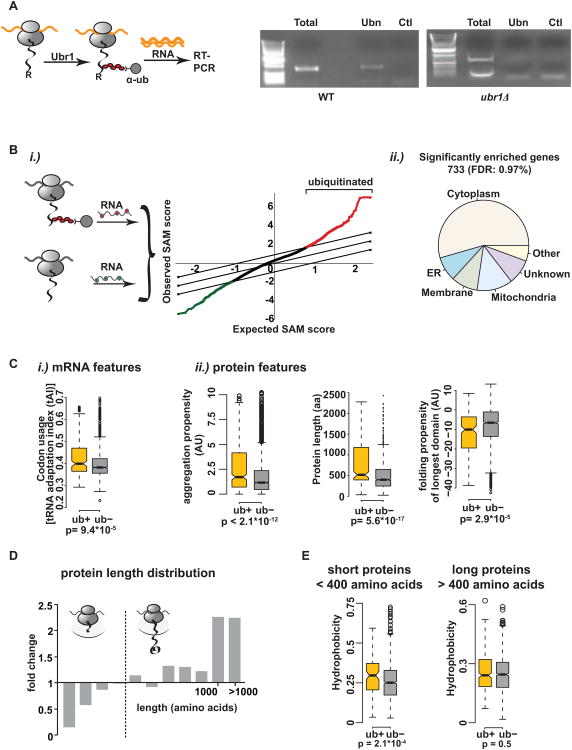
Are all nascent chains ubiquitinated to a small extent or are some proteins more susceptible to cotranslational ubiquitination? To define the principles of cotranslational ubiquitination, we identified at a systems-level which nascent polypeptides are ubiquitinated cotranslationally in vivo. Translating RNCs from actively growing cells were isolated and subjected to polyUb-affinity to purify RNCs bearing cotranslationally ubiquitinated nascent chains. The corresponding mRNAs encoding these ubiquitinated nascent chains were identified as previously described (del Alamo et al., 2011). The specificity of this approach was first established using the synthetic cotranslational N-end rule degrons of β-galactosidase, previously shown to be ubiquitinated and degraded cotranslationally (Turner and Varshavsky, 2000). Stable M-β-galactosidase or unstable R-β-galactosidase was expressed in vivo and translated with comparable efficiency (data not shown). For both stable and unstable variants, ribosomes were isolated and the RNC fraction was subject to polyUb-affinity isolation. The occurrence and extent of cotranslational ubiquitination of the β-galactosidase constructs was assessed by RT-PCR on the mRNA obtained in the polyUb-affinity pulldowns and compared to the total mRNAs present in the RNC complexes. The band corresponding to the β-galactosidase mRNA was observed following polyUb-affinity isolation of ribosomes from cells expressing the degron R-β-galactosidase, but not from those expressing M-β-galactosidase (data not shown), nor in the negative control reaction (Figure 4A, left, and S4A). Importantly, deletion of the corresponding E3 N-end rule ligase, Ubr1, abrogated the recovery of the β-galactosidase mRNA (Figure 4A, right, and S4A), indicating that ubiquitination is required for the polyUb-affinity isolation of the mRNA encoding this unstable nascent chain. Therefore the polyUb-affinity approach allows the identification of cotranslationally ubiquitinated nascent chains through the corresponding mRNAs.
Figure 4. Identification and characterization of ubiquitinated nascent chains.
(A) (Left) Experimental setup: R-β-galactosidase is ubiquitinated cotranslationally by Ubr1. After RNC isolation with polyUb-affinity resin, RNA is extracted and reverse transcribed. (Right) RT-PCR detects cotranslational ubiquitination of R-β-galactosidase in WT but not in ubr1Δ cells. T = total, Ubn = polyUb-affinity isolation, Ctl = GST-pulldown. (B)i.) Globally identification of ubiquitinated nascent chains in vivo. After RNC isolation and ubiquitin-pulldown, RNA was extracted. An aliquot of mRNAs from the initial RNCs serves as total translation reference. Both RNA samples are reverse transcribed, labeled with Cy3 and Cy5, respectively and labeled cDNAs hybridized to DNA microarrays. The graph shows a representative SAM plot for 10 WT replicates with a false discovery rate (FDR) of 1%. Red = significantly positively enriched genes; green = significantly negatively enriched genes in the data set. A set of 733 proteins is consistently ubiquitinated cotranslationally (ub+ set). ii.) Subcellular distribution of cotranslationally ubiquitinated proteins in WT yeast. (C) Features in the mRNA and polypeptide distinguishing the ub+ set from cytosolic proteins not in the dataset (ub-): i.) mRNA: tRNA adaptation index. ii.) protein features: aggregation propensity, protein length and folding propensity of the longest domain. (D) Cotranslational ubiquitination is disfavored in shorter proteins. Size analysis of the identified genes from WT experiments. Fold change of amino acid length compared to the genome. (E) Shorter ubiquitinated nascent chains (encoding proteins <400 amino acids) tend to be more hydrophobic. The cytosolic ub+ dataset was split into proteins shorter and longer than 400 amino acids and the overall hydrophobicity was compared separately to proteins of similar size distribution in the ub-data set. See also Figure S4 and Table S1.
To globally identify which polypeptides are ubiquitinated cotranslationally in vivo, we next combined the polyUb-affinity approach with a previously described sensitive and highly reproducible method to identify cotranslational interactions through the mRNA (Figure 4B) (del Alamo et al., 2011). The mRNAs encoding endogenous polypeptides subject to cotranslational ubiquitination were compared with the total translated mRNAs in the same cells, which served as reference. Notably, the analysis of 10 biological replicates with a stringent false discovery rate of 1%, reproducibly identified a subset of ∼ 700 genes encoding nascent chains that are ubiquitinated cotranslationally in actively dividing cells (Figure 4B; Table S1). Consistent with the detection of ribosomal subunits in the polyUb pulldown (Figure S1C), ribosomal rRNA was also observed in the polyUb pulldowns but not in the controls (Figure S4B), confirming that intact RNC complexes are isolated by this approach. The finding that not every polypeptide emerging from the ribosome is ubiquitinated cotranslationally, but rather a subset of nascent chains is more susceptible to cotranslational ubiquitination raised the question of which specific functional or sequence properties underlie their susceptibility to cotranslational ubiquitination.
Identifying the principles governing cotranslational ubiquitination
We examined possible functional or sequence-based underlying properties in both the polypeptide itself and the encoding mRNA that distinguish cotranslationally ubiquitinated polypeptides (herein ub+ set). Cotranslational ubiquitination occurred preferentially on nascent chains encoding cytoplasmic and nuclear proteins (Figures 4B and S4C). No significant differences were observed in the cellular function and activity of the ub+ set when compared to those that are less susceptible to cotranslational ubiquitination (herein ub- set). Similarly, there was no enrichment in proteins involved in cellular regulation, such as cyclins or other cell cycle components, indicating that cotranslational ubiquitination is not linked to cell cycle regulation (not shown). Of note, no link was found between cotranslational ubiquitination and the half-life of the mature protein in the cell, suggesting that cotranslational ubiquitination does not occur preferentially among short-lived proteins.
A comprehensive bioinformatics analysis was next used to identify common threads distinguishing the nascent chains that are ubiquitinated (ub+) from those that are not (ub−). No enrichment in lysines (Figure S4D), previously described consensus ubiquitination sites [not shown, (Radivojac et al., 2010)], or regions of intrinsic disorder (not shown) was observed in the ub+ subset, indicating that cotranslational ubiquitination does not occur preferentially on lysine-rich or disordered nascent chains. These data are in agreement with previously published results stating that ubiquitination sites are not preferentially located in regions of disorder and that disordered regions are not enriched in ubiquitinated proteins (Hagai et al., 2011). Importantly, our analysis did identify features in both the polypeptide and its corresponding mRNA distinguished the nascent chains in the ub+ and ub- datasets, indicating that cotranslational ubiquitination does depends on the properties of the nascent polypeptide as it synthesized by the ribosome. The ub+ nascent chains were encoded by mRNAs with a higher tRNA adaptation index and higher abundance than the ub-counterparts (Figures 4Ci and S4E). This suggests that more rapidly translated, highly expressed mRNAs are more susceptible to cotranslational ubiquitination. Of note, ribosomal proteins, which are among the most abundant and highly expressed proteins in the cell (Siwiak and Zielenkiewicz, 2010), are largely absent from the ub+ dataset. Thus, high mRNA abundance and rapid translation by themselves do not determine susceptibility to cotranslational ubiquitination, consistent with the fact that the ub+ and ub- sets also differed in their polypeptide features. At the protein level, the underlying theme distinguishing nascent polypeptides susceptible to cotranslational ubiquitination was their enrichment in properties that enhance aggregation and hinder folding, linking cotranslational quality control to inefficient folding. For instance, ubiquitinated proteins were significantly enriched in aggregation-prone sequence stretches (p<2.10−12, Figure 4C). Notably, analysis of the length distribution of cotranslationally ubiquitinated proteins versus the proteome showed that the ub+ dataset was highly enriched in nascent chains encoding longer polypeptides (Figure 4C). There was a marked depletion of smaller proteins, which constitute a large fraction of the proteins in the cell (Figures 4D and S4F). For those few shorter proteins that do get ubiquitinated cotranslationally, we find a significant enrichment in overall hydrophobicity, which was not observed for the longer ub+ nascent chains (Figure 4E). The enhanced hydrophobicity appears to promote recognition of these shorter nascent chains by the ubiquitination machinery. In addition to overall length, ubiquitinated nascent chains had on average longer domains with a lower propensity to fold compared to the ubiquitinated nascent chains than for the non-ubiquitinated proteins (p<10−5). Remarkably, protein length is the main determinant of protein folding kinetics (Ouyang and Liang, 2008) and thus long proteins are particularly challenged to fold cotranslationally. Conversely, slow translation rates and small domains, disfavored in the ub+ set, facilitate successful cotranslational folding (O'Brien et al., 2012). These data suggest that proteins that are rapidly translated, yet possess challenging folding properties, are selectively prone to cotranslational ubiquitination.
In summary, our systems level analysis indicates a link between global cotranslational ubiquitination and those mRNA and nascent chain properties that challenge efficient and rapid cotranslational folding, such as high translation rate, high hydrophobicity and aggregation propensity, and increased polypeptide length. The facts that most nascent polypeptides are not ubiquitinated cotranslationally and cotranslational ubiquitination is disfavored in shorter polypeptides, suggest the presence of mechanisms protecting polypeptides as they emerge from the ribosome. We next considered the roles of cotranslational folding and ribosome-bound chaperones in shielding nascent chains from ubiquitination at the ribosome.
Cotranslational folding protects nascent chains from ubiquitination
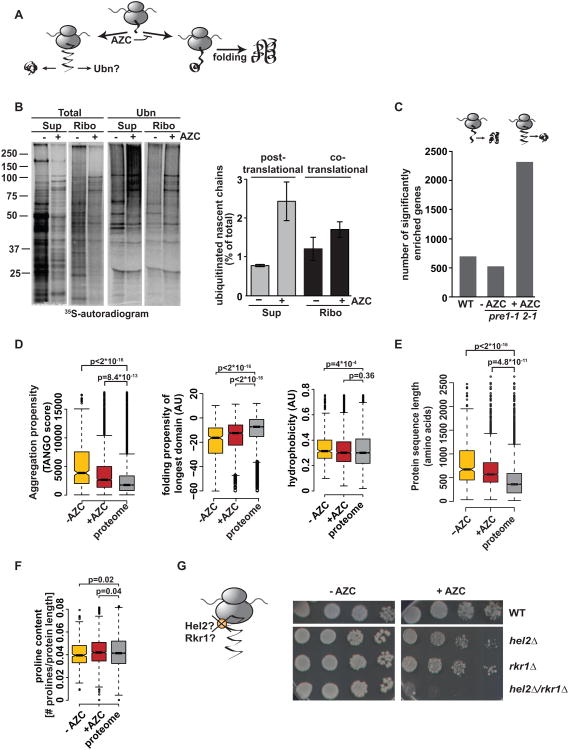
To test whether cotranslational folding antagonizes cotranslational ubiquitination, we specifically disrupted folding during translation with the proline analogue azetidine-2-carboxylic acid (AZC) which distorts the backbone when incorporated instead of proline (Trotter et al., 2002) (Figure 5A). Pulse-labeling experiments showed that brief treatment with AZC increased nascent chain ubiquitination, as well as post-translational ubiquitination (Figure 5B). Thus, disrupting the cotranslational folding process leads to enhanced misfolding and ubiquitination on- and off- the ribosome. To assess the global impact of impairing cotranslational folding, we defined the global repertoire of nascent chains that are ubiquitinated cotranslationally upon AZC treatment (Figure 5C) in pre1-1 2-1 cells. We observed a dramatic increase in cotranslational ubiquitination, whereby ∼ 40% of translated ORFs were represented in the ub+ set (Figure 5C). This reveals the far-reaching protection provided by cotranslational folding against cotranslational ubiquitination. A systems-level analysis of the properties of nascent chains ubiquitinated upon AZC treatment indicated that pharmacologic impairment of cotranslational folding by AZC treatment reduces the influence of the nascent chain sequence properties on ubiquitination susceptibility. Thus, the ub+ datasets in +AZC treated cells showed a relaxation of all the metrics associated with cotranslational ubiquitination in wildtype untreated cells, such as aggregation propensity, hydrophobicity and folding propensity, which now reflected more closely the proteomic distribution (Figure 5D). Similarly, although ubiquitination was still disfavored in short nascent chains, the size cutoff was also relaxed, so that the ub+ length distribution in the +AZC was more similar to the proteome (Figure 5E). Consistent with a preferential AZC-caused impairment of folding in proline-containing proteins, the proline content of proteins ubiquitinated in the presence of AZC was higher than in the untreated control (Figure 5F). Thus, AZC-induced deficiency of cotranslational folding leads to widespread ubiquitination of nascent polypeptides in the cell, in contrast to normal conditions where only a subset of polypeptides are prone to cotranslational ubiquitination. These results indicate that cotranslational ubiquitination serves a protein quality control function counteracted by efficient cotranslational folding.
Figure 5. Impairing cotranslational folding increases cotranslational ubiquitination.
(A) The proline analogue AZC impairs cotranslational folding. (B) AZC treatment leads to increased ubiquitination of 35S-labeled nascent chains and newly made proteins. (Left) Cells were pulse-labeled in the presence or absence of 1 mg/ml AZC. Supernatant (Sup) and RNC fractions (Ribo) were subject to polyUb-affinity isolation (Ubn), and analyzed by SDS-PAGE and autoradiography. (Right) Mean and SEM of 3 experiments. (C) Addition of AZC increases the number of mRNAs encoding cotranslationally ubiquitinated nascent chains. Number of identified genes in, pre1-1 2-1 − AZC and pre1-1 2-1 + AZC at 30°C. (D) Analysis of ub+ datasets in pre1-1 2-1 −/+ AZC identified as shown in (C). Comparison of aggregation propensity (TANGO score), folding propensity of the longest domain and hydrophobicity. (E) Size distribution of pre1-1 2-1 −/+ AZC datasets as in Figure 2. (F) AZC treatment increases the proline content of ubiquitinated nascent chains. (G) Role of Rkr1 and Hel2 in cotranslational quality control. hel2/rkr1Δ cells are hypersensitive to AZC. Plates were grown for 3 days at 30 °C.
The quality control function of cotranslational ubiquitination resonates with our finding that several ubiquitin ligases involved in general cellular quality control are also responsible for ubiquitination of nascent chains, including Hrd1, Doa10 and Hul5 (Figures 3D and 3E). However, we did not observe increased susceptibility of deletions in either Hrd1 or Doa10 to growth in AZC [data not shown, (Theodoraki et al., 2012)]. We next wondered if ribosome-bound ligases fulfill a quality control function for misfolded nascent chains, beyond a function linked to mRNA quality control (Bengtson and Joazeiro, 2010; Ito-Harashima et al., 2007). While rkr1Δ or hel2Δ cells are not sensitive to AZC (Figure 5G), hel2/rkr1Δ was extremely sensitive to AZC, suggesting a function for these ligases in nascent chain quality control (Figure 5G). The inability to clear misfolded proteins in the absence of these ligases likely leads to the enhanced toxicity of AZC and cell death.
Ribosome-associated chaperones counteract cotranslational ubiquitination
In eukaryotes, a chaperone network interacts with nascent chains upon their synthesis (Albanese et al., 2006; Hartl et al., 2011; Kramer et al., 2009; Preissler and Deuerling, 2012). To test whether ribosome-bound chaperones serve to protect nascent chains from ubiquitination (Figure 6A), we examined the role of the abundant ribosome-bound chaperone NAC (nascent polypeptide-associated complex). NAC is a ubiquitously conserved ATP-independent heterodimer with a well-defined nascent interactome (del Alamo et al., 2011); its isoforms bind most nascent chains and play a role in both sorting and trafficking of nascent chains (del Alamo et al., 2011; George et al., 1998). These features make NAC an ideal candidate for providing general protection to nascent chains. Pulse-labeling experiments in a strain deleted for all three NAC subunits, egd1/2Δbtt1Δ (herein nacΔ) indicated increased ubiquitination of nascent chains upon loss of NAC (Figures 6B and 6C). Global identification of the cotranslationally ubiquitinated nascent chains in nacΔ cells revealed a dramatic increase in the number of corresponding genes (Figure 6D). Interestingly, whereas in wildtype cells cotranslational ubiquitination targets preferentially cytoplasm-bound nascent chains, in nacΔ cells there was increased ubiquitination of nascent chains encoding membrane-bound and mitochondrial proteins (Figures 6E and S5C). This may reflect the previously reported roles of NAC in regulating trafficking to the secretory pathway (del Alamo et al., 2011) and mitochondria (George et al., 1998) (Figure 6E).
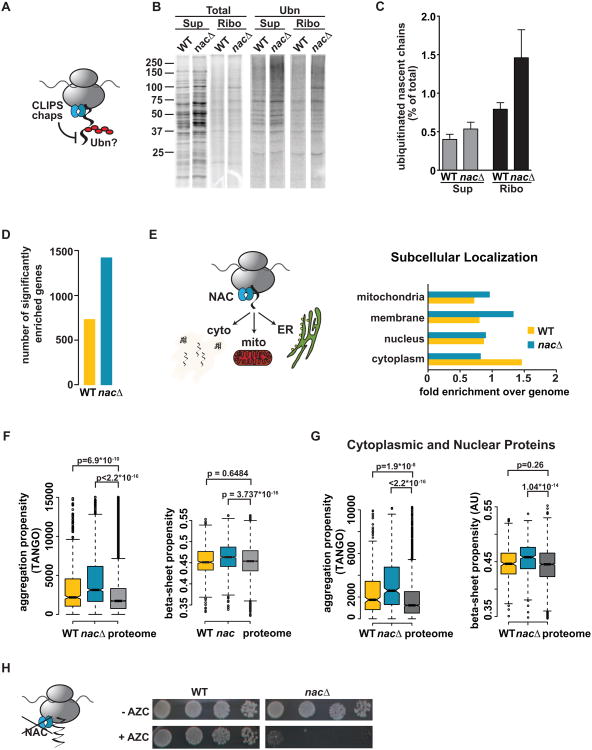
Figure 6. The ribosome-associated chaperone NAC protects nascent chains from cotranslational ubiquitination.
(A) Hypothesis: Ribosome-bound chaperones such as NAC protect nascent chains from ubiquitination (B) Pulse-labeling followed by polyUb-affinity isolation in egd1/2Δbtt1Δ (nacΔ) shows increased ubiquitination compared to WT. (C) Quantification: Mean ± SEM (n=4). (D) Deletion of NAC increases in the number of mRNAs encoding cotranslationally ubiquitinated nascent chains. (E) Deletion of NAC enhances ubiquitination of nascent chains trafficked to the secretory pathway and mitochondria. (Left) Role of NAC in targeting to the ER and mitochondria. (Right) Subcellular localization of ub+ proteins in WT and nacΔ datasets. (F) Deletion of NAC causes ubiquitination of nascent chains with very high aggregation- and beta-sheet propensity compared to the proteome. (G) As in (F) but analysis was restricted to cytoplasmic and nuclear proteins. (H) Synthetic effect of loss of NAC with impaired cotranslational folding: nacΔ cells are hypersensitive to AZC. Plates were grown for 3 days at 30 °C. See also Figure S5.
Further analysis of the nascent chains ubiquitinated in nacΔ cells suggested a rationale for the protective effect of NAC at the ribosome. Loss of NAC exacerbated the cotranslational ubiquitination of polypeptides with those folding-challenging features already enriched in the ub+ dataset. Perhaps most dramatic was the increased enrichment in aggregation propensity (Figure 6F), which suggests that extremely aggregation-prone proteins are normally protected by the action of NAC at the ribosome. Similarly, loss of NAC caused ubiquitination of nascent chains encoding longer proteins beyond what is observed in the wildtype dataset (Figures S5A and S5B). These proteins also possess a higher beta-sheet content, which is normally associated with slower folding and increased aggregation (Figure 6F). The enhancement in ubiquitination of aggregation-prone proteins was not biased by the increased levels of membrane proteins in the nacΔ dataset since similar results were obtained when comparing cytoplasmic and nuclear substrates of cotranslational ubiquitination in wildtype and nacΔ cells (Figure 6G). We hypothesize that NAC protects those nascent chains most susceptible to cotranslational quality control, giving them a chance to prioritize folding over degradation as they emerge from the ribosome. Many nascent chains protected by NAC are quite long, suggesting that NAC does not only function by direct binding at the ribosomal exit site, but perhaps also protects longer polypeptides.
To test whether NAC acts synergistically with cotranslational folding to protect nascent chains, we subjected a nacΔ strain to AZC treatment (Figure 6H). Indeed, AZC was extremely toxic to nacΔ cells, suggesting that these two processes, cotranslational folding and protection of the nascent chain by NAC, are together required for viability.
Discussion
The efficiency of protein biogenesis and the extent of cotranslational ubiquitination have remained long-standing open questions in biology. Our experiments resolve the longstanding paradox of how the cell balances the conflicting needs of promoting cotranslational protein folding while maintaining efficient quality control. We find that only a small fraction of nascent polypeptides is ubiquitinated cotranslationally. Specific nascent chain properties determine their susceptibility for cotranslational ubiquitination. However, the contribution of even this small fraction of nascent chains poses a significant load for the ubiquitin-proteasome pathway. Our studies begin to define the interplay between protein homeostasis components that promote folding and quality control at the ribosome (Figure 7).
Figure 7. Balancing protein folding and cotranslational ubiquitination at the ribosome.
Upon translation, most nascent chains are protected from degradation through cotranslational folding and interaction with NAC (shown here) and likely other ribosome-bound chaperones, which create a protected folding environment in the vicinity of the ribosomal exit site (shaded area). Damaged or non-stop mRNAs that escape mRNA quality control produce translation products eliminated by the ubiquitin ligase Rkr1. Most nascent chains fold quickly and efficiently with the assistance of chaperones or spontaneously. Nascent chains with features challenging efficient folding are susceptible to cotranslational ubiquitination by ribosome-bound and non-ribosome bound quality control ligases. See also Figure S6.
Balancing protein biogenesis and cotranslational quality control
Our work implies that protein biogenesis and cotranslational folding are largely efficient processes and suggests an environment that disfavors ubiquitination in the vicinity of the ribosome. Such protection cannot be attributed solely to steric hindrance by the ribosome, and likely involves additional factors. Cotranslational folding and association with ribosome-bound molecular chaperones contribute to protect nascent chains from premature ubiquitination. These observations resonate with previous studies indicating that in eukaryotes, nascent chains fold in a protected environment that is inaccessible to a bacterially-derived chaperone Trap (Frydman and Hartl, 1996; Thulasiraman et al., 1999). Our results thus suggest a tiered system at the ribosome, where nascent polypeptides first have a chance to fold and are then subjected to ubiquitination.
Despite the protective environment provided by the ribosome and associated chaperones, a subset of mostly cytoplasmic nascent polypeptides is nonetheless susceptible to cotranslational ubiquitination, mediated by a network of ribosome-bound and unanchored ubiquitin ligases. Degradation of these ubiquitinated nascent chains is carried out by the proteasome, likely in concert with Cdc48/p97 (Figure 7). The fact that no single ligase is solely responsible for cotranslational ubiquitination may reflect the range of properties and mechanisms by which nascent chains are targeted for ubiquitination. Our analysis of Rkr1 deletion strains suggests that only a relatively small fraction of ubiquitinated nascent chains arises from stalled or NS-mRNA derived products (Bengtson and Joazeiro, 2010). This likely reflects the robust nature of mRNA quality control (Doma and Parker, 2007), which rapidly eliminates aberrant mRNAs in an initial round of translation. Consistent with these findings, there are only 200 molecules of Rkr1 compared to 200,000 ribosomes per yeast cell (Ghaemmaghami et al., 2003; von der Haar, 2008). Of note, impairment of Listerin/Rkr1 in mice is linked to neurodegeneration (Chu et al., 2009), highlighting the importance of rigorous quality control for the cell. An additional ribosome-associated ubiquitin ligase, Hel2, is also involved in nascent chain quality control (Figure 3D,E). The synthetic effect of the double deletion rkr1/hel2Δ, which abrogates ∼25% of cotranslational ubiquitination, suggests that Rkr1 and Hel2 act on parallel, partially redundant pathways, consistent with the increased sensitivity of the double deletion rkr1/hel2Δ to nascent chain misfolding (Figure 5G). Of note, Hel2 is 10-fold more abundant than Rkr1 (Ghaemmaghami et al., 2003). It is tempting to speculate that the ribosome-associated ligases have more specialized roles sensing either local misfolding or stalling at the ribosome, whereas the bulk of de novo quality control is mediated by non-ribosome bound ligases, once nascent chains have been given a chance to fold.
Determinants of susceptibility to cotranslational ubiquitination
Susceptibility to cotranslational ubiquitination is linked to enrichment in biophysical properties that challenge efficient folding, such as higher hydrophobicity, larger domains and higher aggregation propensity; notably, it is also linked to a higher tRNA adaptation index. It thus appears that cotranslational quality control is intimately coupled to folding challenges which are exacerbated by high local translation rates (O'Brien et al., 2012; Pechmann and Frydman, 2013). Consistent with this idea, combining sequence properties with translation properties can be used to predict whether proteins are cotranslationally ubiquitinated (not shown).
It was noted that the complexity of the eukaryotic proteome, characterized by having longer multidomain proteins, creates challenges to cotranslational folding (Netzer and Hartl, 1998). Our work suggests that the vast majority of proteins are evolutionarily optimized to balance translation rate and cotranslational folding, thus avoiding aggregation and minimizing cotranslational ubiquitination and quality control. However, functional constrains may cause some of them to misfold to some extent and be ubiquitinated. For instance, we find that cotranslationally ubiquitinated proteins are more engaged in protein-protein interactions (Figure S6A), which may increase their hydrophobic interfaces, thereby decreasing the efficiency of folding. On the other hand, we find less essential proteins among cotranslationally ubiquitinated proteins, suggesting that essential proteins are optimized to reduce de novo misfolding (Figure S6B). Interestingly, when the chaperone NAC is deleted, the ubiquitinated nascent chains are even longer, more aggregation prone and containing higher beta-sheet propensity. It is thus tempting to speculate that the ribosome-bound chaperone network of eukaryotes evolved to protect the proteins with more complex folds and topologies from both misfolding (Willmund et al., 2013) and degradation (Figure 6).
In principle, some of the proteins in our dataset may use cotranslational ubiquitination to regulate protein levels during synthesis. Intriguingly many ubiquitin ligases themselves are ubiquitinated cotranslationally in our experiments, including San1, Ubr1, Ufd2 (Table S1). Since E3 ligases can autoubiquitinate in vivo and in vitro (Pickart, 2001), this may represent a mechanism to regulate and sense the need for a particular E3 ligase in the cell. Ubiquitinated nascent chains are enriched in PEST sequences, linked to regulated proteolysis (Rechsteiner and Rogers, 1996) (Figure S6C). Future studies exploiting our global analyses should determine whether cotranslational ubiquitination, largely disfavored for the vast majority of newly made proteins, has been co-opted by some proteins for rapid regulation upon synthesis.
Biological impact of cotranslational ubiquitination on cellular homeostasis
It is interesting to consider the impact of the low levels of cotranslational ubiquitination on overall cellular homeostasis. Ribosomes are present in ten-fold excess over proteasomes in both yeast and mammalian cells (Russell et al., 1999; von der Haar, 2008), and thus, a steady flow of 2-5% of ubiquitinated nascent chains represents a substantial burden for the proteasome pathway, even under non-stressed conditions, particularly since proteasomal degradation has similar processivity rates as translation (Henderson et al., 2011). This consideration may reconcile the seemingly contradictory observations that de novo folding is favored over degradation (Frydman and Hartl, 1996; Vabulas and Hartl, 2005) with findings that newly made proteins contribute significantly to proteasomal load (Kim et al., 2011) and to antigen presentation (Lelouard et al., 2004; Reits et al., 2000) Our analysis is also consistent with studies using model proteins and synthetic substrates which observed cotranslational ubiquitination on longer proteins, such as ApoB (Zhou et al., 1998), CFTR (Sato et al., 1998) and N-end rule β-galactosidase, but little to none on short nascent chains, such as those of actin (Frydman and Hartl, 1996) and even on the shorter synthetic N-end rule Ura3 (Turner et al., 2000). Thus, our analysis reconciles and explains the existing data while providing a coherent model for cotranslational quality control.
The idea that cotranslational quality control poses a significant load on the proteasome suggests that conditions challenging the delicate balance of cotranslational folding may have a significant impact on proteasomal function. These may include enhanced cotranslational misfolding, e.g., due to impaired RNA quality control, chromosomal abnormalities, aneuploidy or mutagens (Oromendia et al., 2012), as well as decreased chaperone function caused by stress, aggregation or aging (Balch et al., 2008; Taylor and Dillin, 2011). Consistent with this idea, recent studies observed that selected newly made polypeptides aggregate in cells expressing toxic amyloidogenic proteins (Olzscha et al., 2011). The lethality of combining AZC treatment with deletions in either ribosome-associated chaperones, mRNA quality control components, or quality control ligases underscores the idea that an imbalance in cotranslational quality control can lead to proteotoxicity and cell death. Indeed, the recently identified pathway to communicate stress at the ribosome to the heat shock transcription factor HSF highlights the central role of the ribosome in protein homeostasis (Brandman et al., 2012).
Experimental Procedures
GST and GST-UBA(Dsk2) were expressed and purified as described (Funakoshi et al., 2002) and coupled to glutathione sepharose beads (GE) or magnetic beads (Bioclone Inc.). Ubiquitin immunoblots used monoclonal antibody MMS-258R (Covance).
Pulse-labeling experiments, pulse-chase, cell lysis and ribosome isolation by sucrose cushion were as in (Yam et al., 2005). For AZC experiments, 1mg/ml AZC was added to the cells during the starvation phase, to give a brief (30 min) pulse of AZC. For MG132 experiments, a strain deleted for PDR5 was used and a control with DMSO included. MG132 was added during the 30 min starvation period. Sucrose gradient sedimentation was carried out as in (Albanese et al., 2006), and CTAB precipitation as in (Nakatogawa and Ito, 2001)
For RT-PCR experiments, cells expressing M/R-beta galactosidase were grown overnight in SC(Raf)-URA and shifted to SC(Gal)-URA. Cells were harvested, lysed and ribosomes were purified as above. An aliquot was taken for total RNA extraction and the remaining sample was split in half for pulldown with GST as negative control or with UBA(Dsk2) coupled to magnetic beads to retrieve ubiquitinated RNCs. RNA was extracted using RNeasy Mini kit according to the manufacturer. Reverse transcription of 100 ng RNA was performed with SuperScript II from Invitrogen. Controls included reverse transcription without enzyme as well as RNA from the GST pulldown. Plasmid DNA served as a positive control. The reactions were performed in WT and ubr1Δ cells.
For global analysis, cells were grown in 2L YPD, except for the AZC experiments, where cells were grown in 2L SC media and incubated with 0.5 mg/ml AZC for 1 hour before harvest. Cells were harvested and treated as in the RT-PCR experiments. After isolation and resuspension of the ribosomal pellet, an aliquot was taken as total RNA reference sample. The rest was split in half and pull-downs were performed with GST and GST-UBA(Dsk2) coupled to magnetic beads (Bioclone Inc.), respectively. RNA extraction was performed with the Qiagen RNeasy Mini Kit as described. Reverse transcription, microarray hybridization and microarray analysis were performed as described (del Alamo et al., 2011).
Supplementary Material
Highlights.
We assess determinants of cotranslational ubiquitination in vivo
Ubiquitination linked to nascent chain properties that challenge their folding
Cotranslational folding and chaperones protect nascent chains from ubiquitination
Ribosome-bound and cytosolic E3 ligases mediate cotranslational ubiquitination
Acknowledgments
We thank Drs. Felix Willmund and Marta Del Alamo for useful discussions and technical advice, Patrick Brown and Dan and Greg Hogan of the Brown lab for help with the microarray experiments and members of the Frydman lab for suggestions and critical reading of the manuscript. We thank M. Hochstrasser, T. Inada, H Kobayashi, A. Varshavsky, J. Warner, R. Deshaies, J. Brodsky and S. Michaelis for their kind gift of reagents and strains. This work was supported by grants from NIH to JF. SP was supported by an EMBO Long-term fellowship.
Footnotes
Author information: The authors declare no competing financial interest.
Supplemental Information: Supplemental Information includes 6 Supplemental Figures and can be found online at XXXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Science. Vol. 319. New York, NY: 2008. Adapting proteostasis for disease intervention; pp. 916–919. [DOI] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O'Shea E. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Chu J, Hong NA, Masuda CA, Jenkins BV, Nelms KA, Goodnow CC, Glynne RJ, Wu H, Masliah E, Joazeiro CA, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO. The Ccr4-Not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS biology. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. The Journal of biological chemistry. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nature cell biology. 2011;13:1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes & development. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Hartl FU. Science. Vol. 272. New York, NY: 1996. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms; pp. 1497–1502. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R, Beddoe T, Landl K, Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W, Bower K, Howson R, Belle A, Dephoure N, O'Shea E, Weissman J. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hagai T, Azia A, Toth-Petroczy Å, Levy Y. Intrinsic Disorder in Ubiquitination Substrates. Journal of molecular biology. 2011;412:319–324. doi: 10.1016/j.jmb.2011.07.024. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Henderson A, Erales J, Hoyt MA, Coffino P. Dependence of Proteasome Processing Rate on Substrate Unfolding. Journal of Biological Chemistry. 2011;286:17495–17502. doi: 10.1074/jbc.M110.212027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes & development. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Molecular Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nature structural & molecular biology. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- Lelouard H, Ferrand V, Marguet D, Bania J, Camosseto V, David A, Gatti E, Pierre P. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. The Journal of cell biology. 2004;164:667–675. doi: 10.1083/jcb.200312073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor T, Deshaies RJ. Two-step affinity purification of multiubiquitylated proteins from Saccharomyces cerevisiae. Methods in enzymology. 2005;399:385–392. doi: 10.1016/S0076-6879(05)99026-5. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. Secretion Monitor, SecM, Undergoes Self-Translation Arrest in the Cytosol. Molecular cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends in biochemical sciences. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- O'Brien EP, Vendruscolo M, Dobson CM. Prediction of variable translation rate effects on cotranslational protein folding. Nat Commun. 2012;3:868. doi: 10.1038/ncomms1850. [DOI] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes & development. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Liang J. Predicting protein folding rates from geometric contact and amino acid sequence. Protein Sci. 2008;17:1256–1263. doi: 10.1110/ps.034660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annual review of biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends in biochemical sciences. 2012;37:274–283. doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins: Structure, Function, and Bioinformatics. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends in biochemical sciences. 1996;21:267–271. [PubMed] [Google Scholar]
- Reits EAJ, Vos JC, Gromme M, Neefjes J. The major substrates for TAP invivo are derived from newly synthesized proteins. Nature. 2000;404:774–778. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Steger KA, Johnston SA. Subcellular Localization, Stoichiometry, and Protein Levels of 26 S Proteasome Subunits in Yeast. Journal of Biological Chemistry. 1999;274:21943–21952. doi: 10.1074/jbc.274.31.21943. [DOI] [PubMed] [Google Scholar]
- Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. The Journal of biological chemistry. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. The EMBO journal. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA. The eIF3 Interactome Reveals the Translasome, a Supercomplex Linking Protein Synthesis and Degradation Machineries. Molecular cell. 2009;36:141–152. doi: 10.1016/j.molcel.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwiak M, Zielenkiewicz P. A comprehensive, quantitative, and genome-wide model of translation. PLoS Comput Biol. 2010;6:e1000865. doi: 10.1371/journal.pcbi.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki MA, Nillegoda NB, Saini J, Caplan AJ. A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. The Journal of biological chemistry. 2012;287:23911–23922. doi: 10.1074/jbc.M112.341164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. The EMBO journal. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter EW, Kao CMF, Berenfeld L, Botstein D, Petsko GA, Gray JV. Misfolded Proteins Are Competent to Mediate a Subset of the Responses to Heat Shock in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2002;277:44817–44825. doi: 10.1074/jbc.M204686200. [DOI] [PubMed] [Google Scholar]
- Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- Turner GC, Varshavsky A. Science. Vol. 289. New York, NY: 2000. Detecting and measuring cotranslational protein degradation in vivo; pp. 2117–2120. [DOI] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU. Science. Vol. 310. New York, NY: 2005. Protein synthesis upon acute nutrient restriction relies on proteasome function; pp. 1960–1963. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 Mediates UV-Dependent Turnover of RNA Pol II. Molecular cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Systems Biology. 2008;2:87. doi: 10.1186/1752-0509-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F, Del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. The Cotranslational Function of Ribosome-Associated Hsp70 in Eukaryotic Protein Homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam AY, Albanese V, Lin HT, Frydman J. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. The Journal of biological chemistry. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Zhou M, Fisher EA, Ginsberg HN. Regulated Co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. The Journal of biological chemistry. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.