Abstract
Rapid diagnostic tests are needed for typhoid fever (TF) diagnosis in febrile children in endemic areas. Five hundred children admitted to the hospital in Cambodia between 2009 and 2010 with documented fever (≥ 38°C) were investigated using blood cultures (BCs), Salmonella Typhi/Paratyphi A real-time polymerase chain reactions (PCRs), and a Typhoid immunoglobulin M flow assay (IgMFA). Test performance was determined by conventional methods and Bayesian latent class modeling. There were 32 cases of TF (10 BC- and PCR-positive cases, 14 BC-positive and PCR-negative cases, and 8 BC-negative and PCR-positive cases). IgMFA sensitivity was 59.4% (95% confidence interval = 41–76), and specificity was 97.8% (95% confidence interval = 96–99). The model estimate sensitivity for BC was 81.0% (95% credible interval = 54–99). The model estimate sensitivity for PCR was 37.8% (95% credible interval = 26–55), with a specificity of 98.2% (95% credible interval = 97–99). The model estimate sensitivity for IgMFA (≥ 2+) was 77.9% (95% credible interval = 58–90), with a specificity of 97.5% (95% credible interval = 95–100). The model estimates of IgMFA sensitivity and specificity were comparable with BCs and better than estimates using conventional analysis.
Introduction
Salmonella enterica serotype Typhi (S. Typhi) is the main causative organism of typhoid fever, although S. enterica Paratyphi A is becoming increasingly common in some areas, including north India, Nepal, and China.1 An estimated 21 million new cases of typhoid occur each year, resulting in approximately 216,000 deaths.2 The diagnosis of typhoid fever is challenging, because the clinical presentation can be confused with other infectious diseases, such as dengue, malaria, rickettsial infections, leptospirosis, and melioidosis; a secure diagnosis requires laboratory confirmation.3
Blood culture is the recommended diagnostic method, but it is reported to be positive in only 40–80% of cases.4 The sensitivity of blood culture varies according to the stage of illness, the volume of blood inoculated into the culture, and prior antimicrobial treatment.5 A low number of bacteria circulating in the blood is a crucial limitation.6 Culture of bone marrow is more sensitive than blood but not feasible in routine practice.7 S. Typhi and S. Paratyphi A DNA can be successfully detected in blood by nucleic acid amplification, but like with culture, sensitivity is limited by the low number of circulating organisms.8 Furthermore, few laboratories in endemic resource-limited countries have the capacity for bacterial culture or polymerase chain reaction (PCR). The Widal test is simple to perform and still widely used but limited by false-positive and -negative results.4 Enzyme-linked immunosorbent assays (ELISAs) and a number of rapid serological diagnostic tests have been evaluated with variable results.9–11
The Typhoid F immunoglobulin M flow assay (IgMFA) is a typhoid-specific rapid diagnostic test for use on human serum or whole-blood samples, which was developed by the Royal Tropical Institute (KIT) in Amsterdam, that detects S. Typhi lipopolysaccharide (LPS) -specific IgM antibodies using a one-step immunochromatographic lateral flow assay.12,13 Evaluations in Indonesia have suggested a sensitivity of 59% compared with blood culture, with a range from 41% to 90%, depending on the stage of illness, and a specificity of 98% based on results obtained for patients with clinical suspicion of typhoid fever when typhoid fever was later excluded.13
Recent studies have drawn attention to the importance of antimicrobial-resistant typhoid fever in Cambodia in Southeast Asia.14–17 At Angkor Hospital for Children (AHC), a pediatric hospital in Siem Reap in northwest Cambodia, S. Typhi is the most common isolate from the blood culture. Despite a capacity for blood culture confirmation at this hospital, many children with a negative blood culture are clinically diagnosed with typhoid fever. Alternative simple rapid diagnostic tests for typhoid are needed in such locations. Here, we have estimated the diagnostic accuracy of the KIT IgMFA test for the diagnosis of typhoid fever compared with blood culture, a real-time PCR assay, and clinical assessment in a group of children admitted to hospital with fever. Blood culture is an imperfect gold standard against which to compare new point of care tests; therefore, we have used a Bayesian latent class modeling approach to measure the sensitivity and specificity of all of the tests used.18–20
Materials and Methods
Study site.
AHC is a charitably funded hospital and one of two pediatric hospitals in the town of Siem Reap. It provides free medical care to children ages 0–15 years from the town, province, and surrounding provinces. The hospital has approximately 125,000 attendees and 4,000 admissions per year.
Patients.
Children consecutively admitted to AHC with a documented fever of ≥ 38°C within 48 hours of admission who were < 16 years of age were eligible for entry to the study. There were two periods of prospective study recruitment. The first period was between April and May of 2009 (N = 125), and it was the subject of a previous report.12 The second period of recruitment was between March and August of 2010 (N = 375), and it was part of a larger 1-year study of the causes of fever among children admitted to this hospital that is reported elsewhere.16 The two periods of recruitment were chosen to be during the season in which typhoid fever is most common in Cambodia. Blood was collected from the patients on admission and discharge where possible. Five hundred children were included in this study of typhoid rapid diagnostic tests if sufficient admission whole-blood and serum blood samples were available for analysis.
Ethical approval for the study was granted by the AHC Institutional Review Board, the Oxford Tropical Research Ethics Committee (Oxtrec 53-09), and the Research Ethics Committee of the Liverpool School of Tropical Medicine. Informed written or thumbprint consent was taken from the subjects' parents or caretakers.
Clinical procedures.
Demographic and clinical information was recorded on a study case report form at the time of admission. Blood was collected at the time of admission for complete blood count and blood culture; ethylenedinitrilo tetraacetic acid (EDTA) whole blood was collected for PCR, and serum was collected for the IgMFA. The serum samples were frozen at −80°C and tested later. A diagnosis or differential diagnosis was proposed and recorded by the junior clinician responsible for admitting the child. At the time of discharge, a judgment was made based on the blood culture results and assessment by an experienced pediatrician as to whether the clinical features, laboratory results, and inpatient progress of the patient were consistent with typhoid fever. These features included some (but not necessarily all) of the following features: a febrile illness of > 3 days duration, the presence of abdominal symptoms (abdominal pain, diarrhea, or constipation), a documented fever of ≥ 39°C, hepatomegaly and/or splenomegaly, a low or normal white cell count, elevation of liver enzymes (serum glutamic oxaloacetic transaminase [SGOT] and serum glutamic pyruvic transaminase [SGPT]) two to three times above the normal range, a slow defervescence with ceftriaxone treatment (the standard antibiotic used for hospital-admitted febrile children), and no alternative confirmed diagnosis established.
Blood culture.
A volume of 1–2 mL blood, dependent on age, was inoculated into 20 mL tryptic soy broth with sodium polyanethole sulfonate (SPS). All blood bottles were examined daily and subcultured at 24 hours, 48 hours, and 7 days after incubation or if turbid, on examination.16 Subcultures were onto 5% sheep blood agar and chocolate agar incubated in a candle jar and MacConkey agar incubated in air for 48 hours (all media; Oxoid, Basingstoke, United Kingdom; blood bottles, stoppers and caps supplied by PN Laboratories, Bangkok, Thailand). Bacterial isolates were identified by standard methods, including biochemical test using API test strips (bioMérieux, Marcy l'Etoile, France) and agglutination with specific antisera (Biorad, Hertfordshire, United Kingdom). All media and tests are subject to regular internal quality assessment.
Real-time PCR.
DNA extraction was performed from whole blood stored in EDTA using a QIAmp DNA Mini Kit (Qiagen, United Kingdom). The real-time PCR method previously published by Nga and others8 was followed using 25-μL reactions containing 5 μL extracted DNA for the detection of S. Typhi and S. Paratyphi A.
IgMFA.
The IgMFA was tested on 500 admission serum samples according to the manufacturer's instructions. For the first 125 children, the tests were performed at the time of admission. For the remaining 375 children, the tests were not performed at the time of admission but 1 year later; the tests were on serum samples taken on admission that had been stored at −80°C. In all cases, the testers were blind to the clinical details and laboratory results. The result for each sample was graded as recommended using the band intensity from 0 to 4+.21 For the first 125 samples, the test was performed and read by a single observer. For the remaining samples, each test was performed by one scientist, and the results were read, after a short period of training, independently by three technicians blinded to study number. The assay card was relabeled, and each card was read again independently by the same three blinded technicians. Cards were read within 30 minutes of preparation. For each case, the average of all readings was taken as the final result.
Analysis.
Analysis was performed using Stata version 10.0 (College Station, TX). The κ-statistic22 was used to measure interrater and intrarater agreement of the IgMFA-positive band intensity in 375 assays. The κ-statistic ranges between zero and one, with one representing perfect agreement and zero representing no more agreement than would be expected to occur on the basis of chance alone. By convention, κ-values of ≤ 0.20 are considered to reflect poor agreement, κ-values from > 0.20 to ≤ 0.40 are considered to reflect fair agreement, κ-values from > 0.40 to ≤ 0.60 are considered to reflect moderate agreement, κ-values from < 0.60 to ≤ 0.80 are considered to reflect good agreement, and κ-values of > 0.80 are considered to reflect very good agreement. Sensitivity, specificity, and predictive values with 95% confidence intervals were calculated using laboratory-confirmed tests (blood culture- and/or PCR-positive) and all cases (blood culture- and/or PCR-positive and/or clinical diagnosis of typhoid fever) as comparators. The Standards for Reporting of Diagnostic Accuracy (STARD) reporting guidelines were followed.23 The Bayesian latent class model (LCM) as described by Dendukuri and Joseph24 was used to approximate the prevalence, sensitivities, and specificities of all tests.18 The Bayesian LCM does not assume that any test is perfect but considers that each test could be imperfect in diagnosing the true disease status. The true disease status of the patient population is then defined on the basis of overall prevalence (the probability that a patient with suspected typhoid fever is truly infected with S. Typhi). LCMs estimate the prevalence and accuracy of each test based on the observed frequency of the possible combinations of test results. To estimate the accuracy of different diagnostic tests, the model that we used assumed (1) some correlation between the blood culture and PCR results, (2) that blood culture specificity was fixed at 100%, and (3) that there was a random effect variable representing different amounts of bacteria in each infected subject. The model assumed that no other prior information (non-informative priors) about the unknown parameters (prevalence, sensitivities, and specificities) was available. The median values of all the parameters were reported together with the 95% credible intervals.25 The Bayesian LCM was run using WinBUGS 1.4 (Medical Research Council and Imperial College London, United Kingdom).
Results
Patients.
In total, 500 children with fever were studied. The median (interquartile range; range) age of the children was 2.7 years (1.1–8.0 years; 1 month to 15 years), and 264 (53%) children were male. The median (interquartile range; range) duration of illness before hospital admission, which was reported for 489 (98%) patients, was 3 days (2–6 days; 0–30 days). Children were not systematically tested for human immunodeficiency virus (HIV), although 20 (4%) children were known to be positive; 9 of 398 children tested had microscopy-positive malaria: 2 children with Plasmodium falciparum, 3 children with P. vivax, and 4 children with mixed P. falciparum and P. vivax. There were 19 (3.8%) in-hospital deaths, none of which were attributed to typhoid fever.
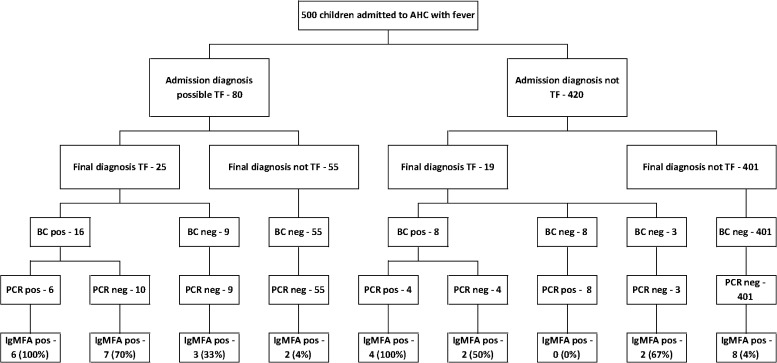
Typhoid fever was considered in the differential diagnosis by the admitting clinician in 80 (16.0%) children (Figure 1). At the time of discharge, 25 (31.3%) of these children were diagnosed with typhoid fever. Typhoid was not suspected at the time of admission in 420 (84.0%) children, and in 19 (4.5%) of these children, the final diagnosis was considered to be typhoid. In total, 44 (8.8%) children were diagnosed with typhoid fever: 24 (54.5%) cases were confirmed by blood culture, including 10 cases that were also positive by PCR, 8 (18.2%) cases were confirmed with a negative blood culture but positive PCR, and 12 (25.0%) cases were confirmed with negative blood culture and PCR as well as based on clinical assessment, laboratory results, and in-patient progress. Common diagnoses in the remaining 456 patients without typhoid included lower respiratory tract infection (154), diarrhea or dysentery (82), dengue (38), upper respiratory tract infection (27), and central nervous system infection (22); 15 patients had a significant positive blood culture containing Streptococcus pneumoniae (5), Escherichia coli (3), Haemophilus influenzae (2), Acinetobacter baumanii (1), Burkholderia cepacia (1), B. pseudomallei (1), Pseudomonas aeruginosa (1), or Staphylococcus aureus (1). An unidentified Gram-negative bacillus was isolated in five other cases, which was inconsistent with the clinical picture and thought to be an environmental contaminant.
Figure 1.
Flowchart showing enrolment of 500 children admitted to the study. BC = blood culture; pos = positive.
The clinical and laboratory features of each group of patients are shown in Table 1. Previous consumption of an antimicrobial was documented in 112 (22.4%) patients, including an antimicrobial potentially active against S. Typhi in 83 (16.6%) patients. In another 156 (31.2%) cases, the medication given was unknown. The median (interquartile range; range) volume of blood taken for culture for all the children was 2.0 mL (1.6–2.5 mL; 0.1–13.2 mL). In the blood culture-positive group, the median (interquartile range) volume was 2.0 mL (1.83–2.3 mL) compared with 1.9 mL (1.6–2.8 mL) in the blood culture-negative group (P = 0.791; Mann–Whitey U test).
Table 1.
Demographic, clinical, and laboratory features of studied children
| BC- and PCR-positive | BC-positive and PCR-negative | BC-negative and PCR-positive | BC- and PCR-negative (clinical typhoid) | BC- and PCR-negative (not typhoid) | |
|---|---|---|---|---|---|
| Number | 10 | 14 | 8 | 12 | 456 |
| Age (years) | 5.8 (3.3–12.5) | 9.3 (4.6–12.9) | 0.9 (0.7–2.7) | 8.1 (4.8–11.2) | 2.4 (1.0–7.5) |
| Sex (male) | 5 (50) | 5 (36) | 4 (50) | 8 (68) | 242 (53) |
| Duration of illness | 5.0 (5.0–7.0) | 6.0 (4.0–10.0) | 2.0 (2.0–3.0) | 6.5 (3.5–10.0) | 3.0 (2.0–5.0) |
| Previous antibiotics | 7 (70) | 6 (43) | 2 (25) | 5 (42) | 69 (15) |
| Headache | 5 (50) | 8 (57) | 0 (0) | 8 (67) | 94 (21) |
| Abdominal pain | 8 (80) | 9 (64) | 1 (13) | 9 (75) | 111 (24) |
| Diarrhea | 2 (20) | 5 (36) | 7 (88) | 5 (42) | 154 (32) |
| Constipation | 2 (20) | 1 (7) | 0 (0) | 1 (8) | 24 (5) |
| Vomiting | 5 (50) | 8 (57) | 4 (50) | 6 (50) | 210 (46) |
| Cough | 1 (10) | 4 (29) | 7 (88) | 4 (33) | 245 (54) |
| Shortness of breath | 0 (0) | 3 (21) | 2 (25) | 0 (0) | 191 (42) |
| Rigors | 3 (30) | 5 (36) | 0 (0) | 3 (25) | 28 (6) |
| Temperature ≥ 39°C | 6 (60) | 6 (43) | 1 (13) | 5 (42) | 133 (29) |
| Jaundice | 0 (0) | 0 (0) | 1 (13) | 1 (8) | 14 (3) |
| Hepatomegaly | 6 (60) | 5 (36) | 1 (13) | 2 (17) | 117 (26) |
| Splenomegaly | 1 (10) | 1 (7) | 0 (0) | 0 (0) | 22 (5) |
| Altered consciousness | 0 (0) | 2 (14) | 0 (0) | 0 (0) | 61 (13) |
| Hemoglobin (g/dL) | 8.0 (6.6–9.3) | 9.5 (7.3–10.3) | 10.1 (9.0–11.9) | 8.8 (7.1–11.0) | 9.9 (8.6–11.2) |
| White cell count (×109/L) | 4.4 (4.0–7.0) | 6.0 (5.1–9.7) | 10.7 (9.6–14.4) | 8.6 (6.3–11.2) | 12.4 (7.7–17.5) |
| Neutrophils (%) | 3.4 (2.1–4.1) | 4.2 (2.9–6.2) | 7.1 (4.8–8.8) | 5.9 (4.3–6.6) | 7.1 (4.3–12.1) |
| Lymphocytes (×109/L) | 1.9 (1.0–2.6) | 1.5 (0.9–2.2) | 3.8 (2.7–6) | 2.2 (1.1–4.0) | 2.8 (1.6–5) |
| Monocytes (×109/L)* | 0.3 (0.1–0.6) | 0.4 (0.3–0.6) | 0.7 (0.1–1.1) | 0.5 (0.4–0.7) | 0.6 (0.3–1.0) |
| Eosinophils (×109/L)* | 0 | 0 | 0 (0–0.2) | 0 | 0 |
| Platelet count (×106/L) | 162 (128–213) | 292 (186–328) | 356 (300–482.5) | 313 (241–486.5) | 328 (222–466) |
| Volume of blood taken for culture (mL) | 2.0 (1.8–2.3) | 2.1 (1.9–2.2) | 2.1 (1.9–2.9) | 1.7 (1.5–2.6) | 2.0 (1.6–2.5) |
Proportions are expressed as number (%), and continuous variables are expressed as median (interquartile range). BC = blood culture.
These results were from 491 samples available for analysis.
IgMFA results.
In total, 106 patients had a positive IgMFA test. In 72 patients, the reading was 1+. In 10 patients, it was 2+. In 14 patients, it was 3+, and in 10 patients, it was 4+. For 375 cases in which three separate raters scored the result, the κ-statistic for the interobserver variability for all IgMFA results was 0.84 (range = 0.56–0.92), suggesting very good agreement. The intraobserver variability for all IgMFA results for the three raters was 0.86, 0.83, and 0.87, also suggesting very good agreement. For scoring the result positive or negative based on a cutoff of ≥ 2+, the interobserver κ-statistic was 0.92, and the intraobserver κ-values were 0.90, 0.90, and 0.89.
The sensitivity, specificity, and positive and negative predictive values of the different intensities of the IgMFA result are shown in Table 2. The values were calculated for children with a laboratory-confirmed diagnosis of typhoid as well as those children with a laboratory and/or clinical diagnosis compared with the children without a diagnosis of typhoid. For both comparisons, the IgMFA sensitivity gradually decreased as increasing cutoff intensities were used. There was stepwise increase in specificity between the reading of ≥ 1+ at 83% and ≥ 2+ at 98%.
Table 2.
Conventional sensitivity and specificity results for each laboratory method (blood culture, PCR, and IgMFA with differing cutoff values of 1+, 2+, 3+, and 4+) compared with laboratory-confirmed typhoid cases and all typhoid cases (laboratory-confirmed plus clinically suspected cases)
| Test | Test positive in typhoid cases | Test positive in non-typhoid cases | Sensitivity (95% credible interval) | Specificity (95% credible interval) | PPV (95% credible interval) | NPV (95% credible interval) |
|---|---|---|---|---|---|---|
| Laboratory-confirmed typhoid (N = 32) compared with non-typhoid cases | ||||||
| Blood culture | 24/32 | 0/456 | 75 (56.6–88.5) | 100 (99.2–100) | 100 (85.8–100) | 98.3 (96.6–99.3) |
| PCR | 18/32 | 0/456 | 56 (37.7–73.6) | 100 (99.2–100) | 100 (81.5–100) | 97 (95.1–98.4) |
| ≥ IgMFA 1+ | 22/32 | 78/456 | 68.8 (50.0–83.9) | 82.9 (79.1–86.2) | 22 (14.3–31.4) | 97.4 (95.3–98.8) |
| ≥ IgMFA 2+ | 19/32 | 10/456 | 59.4 (40.6–76.3) | 97.8 (96.0–98.9) | 65.5 (45.7–82.1) | 97.2 (95.2–98.5) |
| ≥ IgMFA 3+ | 16/32 | 4/456 | 50.0 (31.9–68.1) | 99.1 (97.8–99.8) | 80 (56.3–94.3) | 96.6 (94.5–98.0) |
| ≥ IgMFA 4+ | 8/32 | 1/456 | 25.0 (11.5–43.4) | 99.8 (98.8–100) | 88.9 (51.8–99.7) | 95.0 (92.6–96.8) |
| Laboratory-confirmed clinically suspected typhoid (N = 44) compared with non-typhoid cases | ||||||
| Blood culture | 24/44 | 0/456 | 54.5 (38.8–69.6) | 100 (99.2–100) | 100 (85.8–100) | 95.8 (93.6–97.4) |
| PCR | 18/44 | 0/456 | 40.9 (26.3–56.8) | 100 (99.2–100) | 100 (81.5–100) | 94.6 (92.2–96.4) |
| ≥ IgMFA 1+ | 28/44 | 78/456 | 63.6 (47.8–77.6) | 82.9 (79.1–86.2) | 26.4 (18.3–35.9) | 95.9 (93.5–97.7) |
| ≥ IgMFA 2+ | 24/44 | 10/456 | 54.5 (38.8–69.6) | 97.8 (95.9–98.9) | 70.6 (52.5–84.9) | 95.6 (93.3–97.3) |
| ≥ IgMFA 3+ | 20/44 | 4/456 | 45.5 (30.4–61.2) | 99.1 (97.8–99.8) | 83.3 (62.6–95.3) | 95.0 (92.6–96.7) |
| ≥ IgMFA 4+ | 9/44 | 1/456 | 20.5 (9.8–35.3) | 99.8 (98.8–100) | 90.0 (55.5–99.7) | 92.9 (90.2–95.0) |
NPV = negative predictive value; PPV = positive predictive value.
The IgMFA was positive at ≥ 2+ in 10 children who were considered not to have typhoid. Two children, one with juvenile idiopathic arthritis and one with acute glomerulonephritis, may potentially have had cross-reactive antibodies. The others were diagnosed with pneumonia (four), dysentery (two), bowel obstruction (one), and dental abscess (one).
Evaluation using a Bayesian LCM.
We applied LCMs to explore the sensitivity and specificity of blood culture, real-time PCR, and IgMFA in typhoid fever. Sensitivities of each possible cutoff for the intensity of IgMFA reading (≥ 1+, ≥ 2+, ≥ 3+, and 4+) were assessed from the model. The specificity of blood culture was assumed to be 100%, and the blood culture and PCR were assumed to be correlated. The sensitivity of blood culture was 81.0% (95% credible interval = 54–99) (Table 3). The sensitivity of the real-time PCR was 37.8%, with a specificity of 98.2%. The IgMFA sensitivity ranged with the different cutoff intensities from ≥ 1+ (90%; 95% credible interval = 79–94%) to 4+ (29.6%; 95% credible interval = 22–35%). The reverse was observed for specificity with the highest at 4+ (99.6%; 95% credible interval = 99–100%) and lowest at 1+ (82.9%; 95% credible interval = 82–85%). The calculated prevalence from the model was 6% (95% credible interval = 4–9%), which is consistent with the prevalence of 8.8% based on laboratory-confirmed and clinically diagnosed cases.
Table 3.
Bayesian model sensitivity and specificity results for the different IgMFA cutoffs
| Model | Sensitivity (95% credible interval) | Specificity (95% credible interval) |
|---|---|---|
| Culture | 81.0 (54.1–99.1) | 100 |
| PCR | 37.8 (25.9–55.1) | 98.2 (96.7–99.2) |
| IgMFA | ||
| ≥ 1+ | 90.0 (78.6–93.8) | 84.7 (82.9–84.7) |
| ≥ 2+ | 77.0 (58.2–90.4) | 97.5 (95.4–99.5) |
| ≥ 3+ | 62.5 (51.3–69.2) | 99.8 (98.7–99.8) |
| ≥ 4+ | 29.6 (22.2–34.6) | 99.6 (99.6–100) |
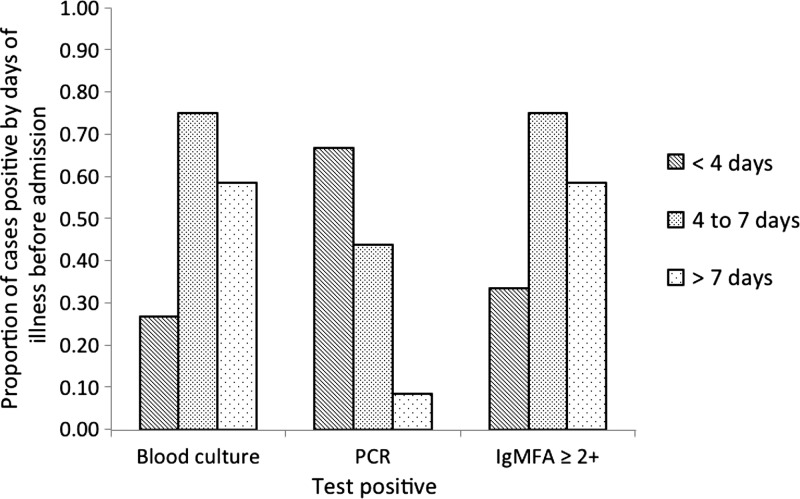
Using the IgMFA cutoff of ≥ 2+, the proportion of IgMFA-positive results for each category of patient is in Figure 1. In the patients with a clinical suspicion of typhoid at the time of admission, an IgMFA of ≥ 2+ had a positive predictive value of 16 out of 18 (89%) for laboratory-confirmed and clinically diagnosed typhoid and a negative predictive value of 85%. The corresponding values in the patients not thought to have typhoid on admission were 50% and 95%. The proportion of positive samples for each test at different duration of illness is shown in Figure 2.
Figure 2.
Duration of illness before admission to hospital in patients with a positive laboratory test (blood culture-, PCR-, or IgMFA-positive at two or higher). Each bar denotes the duration of illness (< 4, 4–7, and > 7 days).
Discussion
Evaluations of rapid diagnostic tests for typhoid fever have been hampered by the lack of a suitable gold standard because of low sensitivity of blood and bone marrow culture.5,26–29 We have used an S. Typhi and Paratyphi A real-time PCR assay on blood to increase the number of laboratory-confirmed cases.8 In addition, we have used a Bayesian LCM to provide an unbiased statistical approach to this issue without the need for a gold standard. This approach is being increasingly used in the evaluation of diagnostic tests for infectious diseases.18,20 This use is the first use of this type of analysis to evaluate typhoid diagnostic tests.
Using a conventional approach, the sensitivity and specificity of the typhoid IgMFA compared with laboratory-confirmed typhoid cases ranged from 25% to 69% and 83% to 99% respectively. The results were dependent on the intensity of the band within the IgMFA window (ranging from 1+ to 4+). The Bayesian LCM model gave generally higher values for the IgMFA sensitivity at each intensity cutoff but similar specificities. The results suggest that the IgMFA may be more sensitive than previously shown.13 A result of ≥ 2+ in the IgMFA seemed to be the optimum cutoff, with a sensitivity of 77.0% (95% credible interval = 58–90%) and a specificity of 98% (95% credible interval = 95–100%).
The sensitivity of blood culture in the Bayesian LCM model was 81.0% (95% credible interval = 54–99%). Previous published sensitivities, using the combined cultures of blood, bone marrow, and other samples as a reference standard, have varied between 40% and 80%.5 The median volume of blood taken for culture in this study was quite low at 2.0 mL but reflects the young age of the children studied. These results suggest that blood culture may be more sensitive than is generally appreciated. The negative blood culture in the group with a clinical diagnosis of typhoid cannot be explained by smaller volumes of blood taken for culture or a history of taking an antimicrobial potentially active against typhoid.
The real-time PCR sensitivity of 38% (95% credible interval = 26–55%), which was evaluated in the Bayesian LCM, was low, although the specificity was high at 98%. More than one-half of the blood culture patients were PCR-negative. This counterintuitive result may relate to the difference in the volumes of blood in the culture (median = 2.0 mL) compared with smaller volumes used to extract the DNA for PCR. The children who were blood culture-negative but PCR-positive represent an interesting group. They were younger than the other patients with blood culture-confirmed typhoid (with a median age of 0.9 years [interquartile range = 0.7–2.7 years]), with a short duration of illness before admission and clinical features that did not suggest typhoid. They may represent a previously described group of young children who have a non-specific illness not recognized as typical typhoid but with S. Typhi isolated from their blood culture.2,3
The IgMFA test is simple, can be used at the bedside using whole blood, and requires very little training to perform and read the results. The test is rapid, with a result available within 30 minutes, and it contains stabilizing compounds for use in hot countries. The interrater and intrarater κ-values for reading the intensity of the bands suggested very good agreement, showing that a cutoff of ≥ 2+ could be used as a reliable end point. The use of the IgMFA varied according to the duration of illness before testing. The test was more sensitive when the duration of illness was longer (> 5 days), presumably because of an increasing IgM antibody response, which is consistent with previous observations.13
The positive predictive value of the IgMFA test was better in patients in whom there was a clinical suggestion of typhoid fever at the time of admission, and the negative predictive value was better in those patients in whom typhoid was not suspected. Such a test could potentially be used to rule in typhoid in those patients with clinically suspected disease and rule it out in those patients in whom typhoid is not suspected. A clinical diagnostic rule to help standardize the selection of patients with a higher or lower probability of typhoid could be helpful if that approach was used, but as of now, a clinical diagnostic rule has not been developed for children, which has been proposed for adults.30,31
The study is limited, like in many typhoid diagnostic studies, by the low number of laboratory-confirmed typhoid cases. We did not include bone marrow culture as part of our study, and the lack of a satisfactory gold standard led to the diagnosis of clinically suspected cases without laboratory confirmation. The use of Bayesian LCM should help to address this issue. An additional limitation was that we were unable to test any cross-reactions from the test in patients with typhoid fever caused by S. Paratyphi A, because it is a rare cause of typhoid fever in our setting. There was a limited number of children with bacteremia caused by other organisms. The only potential cross-reactivity with other causes of bacteremia noted for the IgMFA test occurred in two patients who were blood culture-positive for E. coli and S. pneumoniae. In both instances, the result was positive at 1+ and would have been scored negative with a 2+ cutoff. The inclusion of consecutive febrile children in this endemic setting is a strength of the study and makes the findings more generalizable.
The IgMFA test with a cutoff of 2+ seems to be a promising rapid diagnostic test. Larger evaluations in other settings and a wide range of ages are needed to confirm and extend these findings. The contribution of real-time PCR to the laboratory confirmation of typhoid and the value of the Bayesian LCM in the evaluation of new typhoid diagnostic tests require additional study.
ACKNOWLEDGMENTS
The authors thank Dr. Bill Housworth and Dr. Ngoun Chanpheaktra at the Angkor Hospital for Children for their support of this work and the staff of the Microbiology Laboratory, particularly Sar Poda and Suy Kuong, and the Hospital Records Department for their help with the conduct of this study. The authors thank Dr. Direk Limmathurotsakul for providing technical advice with the Bayesian latent class models and Mr. James I. Campbell for his microbiological assistance. The authors also thank Mr. Rob Pastoor and Dr. Henk Smits of Royal Tropical Institute (KIT) Biomedical Research, KIT, Amsterdam, The Netherlands for providing the Typhoid IgMFA kits. This study was presented in part at the 15th International Congress on Infectious Disease, Bangkok, Thailand, held June 13–16, 2012.
Disclaimer: The study sponsors had no role in the study design; collection, analysis, or interpretation of the data; writing of the report; or decision to submit the paper for publication. All authors had full access to all the data in the study and approved the final version of the paper.
Footnotes
Financial support: The work was supported by the University of Oxford Li Ka Shing Global Health Foundation and the Wellcome Trust of Great Britain. S.B. is supported by a Sir Henry Dale Fellowship jointly funded by Wellcome Trust and Royal Society Grant 100087/Z/12/Z.
Authors' addresses: Catrin E. Moore and Kate Emary, Angkor Hospital for Children, Siem Reap, Cambodia, and Mahidol-Oxford Tropical Medicine Research Unit (MORU), Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mails: catrin.moore@ndm.ox.ac.uk and kate.emary@googlemail.com. Wirichada Pan-Ngum, Lisa White, and Nicholas P. J. Day, Department of Tropical Hygiene and Mahidol-Oxford Tropical Medicine Research Unit (MORU), Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand, E-mails: pan@tropmedres.ac, lisa@tropmedres.ac, and nickd@tropmedres.ac. Lalith P. M. Wijedoru, Centre of Clinical Vaccinology and Tropical Medicine, Churchill Hospital, Oxford, United Kingdom, E-mail: lwijedoru@hotmail.com. Soeng Sona, Microbiology Department, Angkor Hospital for Children, Siem Reap, Cambodia, E-mail: na_sona@yahoo.com. Tran Vu Thieu Nga, Pham Thanh Duy, Phat Voong Vinh, and Stephen Baker, Hospital for Tropical Diseases, Wellcome Trust Major Overseas Programme, Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam, E-mails: ngatvt@oucru.org, duypt@oucru.org, phatvv@oucru.org, and sbaker@oucru.org. Kheng Chheng and Varun Kumar, Medical Department, Angkor Hospital for Children, Siem Reap, Cambodia, E-mails: kh_chheng@yahoo.com and varun@angkorhospital.org. Michael Carter, Institute of Child Health, University College London, London, United Kingdom, E-mail: michael.james.carter@googlemail.com. Christopher M. Parry, Angkor Hospital for Children, Siem Reap, Cambodia; Mahidol-Oxford Tropical Medicine Research Unit (MORU), Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand; and Liverpool School of Tropical Medicine, Liverpool, United Kingdom, E-mail: chrisp@tropmedres.ac.
Reprint requests: Catrin E. Moore, Pneumococcal Vaccine Group, Microbiology, Level 7, John Radcliffe Hospital, Headington, Oxford, United Kingdom, E-mail: catrin.moore@ndm.ox.ac.uk.
References
- 1.Karkey A, Aryjal A, Basnyat B, Baker S. Kathmandu, Nepal: still an enteric fever capital of the world. J Infect Dev Ctries. 2008;2:461–465. doi: 10.3855/jidc.162. [DOI] [PubMed] [Google Scholar]
- 2.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 3.Petit PL, Wamola IA. Typhoid fever: a review of its impact and diagnostic problems. East Afr Med J. 1994;71:183–188. [PubMed] [Google Scholar]
- 4.Bhutta ZA. Current concepts in the diagnosis and treatment of typhoid fever. BMJ. 2006;333:78–82. doi: 10.1136/bmj.333.7558.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dance D, Richens JE, Ho M, Acharya G, Pokhrel B, Tuladhar NR. Blood and bone marrow cultures in enteric fever. J Clin Pathol. 1991;44:1038. doi: 10.1136/jcp.44.12.1038-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wain J, Diep TS, Ho VA, Walsh AM, Nguyen TT, Parry CM, White NJ. Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. J Clin Microbiol. 1998;36:1683–1687. doi: 10.1128/jcm.36.6.1683-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wain J, Pham VB, Ha V, Nguyen NM, To SD, Walsh AL, Parry CM, Hasserjian RP, HoHo VA, Tran TH, Farrar J, White NJ, Day NP. Quantitation of bacteria in bone marrow from patients with typhoid fever: relationship between counts and clinical features. J Clin Microbiol. 2001;39:1571–1576. doi: 10.1128/JCM.39.4.1571-1576.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nga TV, Karkey A, Dongol S, Thuy HN, Dunstan S, Holt K, Tu le TP, Campbell JI, Chau TT, Chau NV, Arjyal A, Koirala S, Basnyat B, Dolecek C, Farrar J, Baker S. The sensitivity of real-time PCR amplification targeting invasive Salmonella serovars in biological specimens. BMC Infect Dis. 2010;10:125. doi: 10.1186/1471-2334-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parry CM, Wijedoru L, Arjyal A, Baker S. The utility of diagnostic tests for enteric fever in endemic locations. Expert Rev Anti Infect Ther. 2011;9:711–725. doi: 10.1586/eri.11.47. [DOI] [PubMed] [Google Scholar]
- 10.Dutta S, Sur D, Manna B, Sen B, Deb AK, Deen JL, Wain J, Von Seidlein L, Ochiai L, Clemens JD, Kumar Bhattacharya S. Evaluation of new-generation serologic tests for the diagnosis of typhoid fever: data from a community-based surveillance in Calcutta, India. Diagn Microbiol Infect Dis. 2006;56:359–365. doi: 10.1016/j.diagmicrobio.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 11.Keddy KH, Sooka A, Letsoalo ME, Hoyland G, Chaignat CL, Morrissey AB, Crump JA. Sensitivity and specificity of typhoid fever rapid antibody tests for laboratory diagnosis at two sub-Saharan African sites. Bull World Health Organ. 2011;89:640–647. doi: 10.2471/BLT.11.087627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijedoru LP, Kumar V, Chanpheaktra N, Chheng K, Smits HL, Pastoor R, Nga TV, Baker S, Wuthiekanun V, Peacock SJ, Putchhat H, Parry CM. Typhoid fever among hospitalized febrile children in Siem Reap, Cambodia. J Trop Pediatr. 2012;58:68–70. doi: 10.1093/tropej/fmr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastoor R, Hatta M, Abdoel T, Smits H. Simple, rapid, and affordable point-of-care test for the serodiagnosis of typhoid fever. Diagn Microbiol Infect Dis. 2008;61:129–134. doi: 10.1016/j.diagmicrobio.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Kasper MR, Sokhal B, Blair PJ, Wierzba TF, Putnam SD. Emergence of multidrug-resistant Salmonella enterica serovar Typhi with reduced susceptibility to fluoroquinolones in Cambodia. Diagn Microbiol Infect Dis. 2010;66:207–209. doi: 10.1016/j.diagmicrobio.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Emary K, Moore CE, Chanpheaktra N, An KP, Chheng K, Sona S, Duy PT, Nga TV, Wuthiekanun V, Amornchai P, Kumar V, Wijedoru L, Stoesser NE, Carter MJ, Baker S, Day NP, Parry CM. Enteric fever in Cambodian children is dominated by multidrug-resistant H58 Salmonella enterica serovar Typhi with intermediate susceptibility to ciprofloxacin. Trans R Soc Trop Med Hyg. 2012;106:718–724. doi: 10.1016/j.trstmh.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Chheng K, Carter MJ, Emary K, Chanpheaktra N, Moore CE, Stoesser N, Putchhat H, Sona S, Reaksmey S, Kitsutani P, Sar B, van Doorn HR, Uyen NH, Van Tan L, Paris D, Blacksell SD, Amornchai P, Wuthiekanun V, Parry CM, Day NP, Kumar V. A prospective study of the causes of febrile illness requiring hospitalization in children in Cambodia. PLoS One. 2013;8:e60634. doi: 10.1371/journal.pone.0060634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlieghe ER, Phe T, De Smet B, Veng HC, Kham C, Lim K, Koole O, Lynen L, Peetermans WE, Jacobs JA. Bloodstream infection among adults in Phnom Penh, Cambodia: key pathogens and resistance patterns. PLoS One. 2013;8:e59775. doi: 10.1371/journal.pone.0059775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limmathurotsakul D, Jamsen K, Arayawichanont A, Simpson JA, White LJ, Lee SJ, Wuthiekanun V, Chantratita N, Cheng A, Day NP, Verzilli C, Peacock SJ. Defining the true sensitivity of culture for the diagnosis of melioidosis using Bayesian latent class models. PLoS One. 2010;5:e12485. doi: 10.1371/journal.pone.0012485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limmathurotsakul D, Turner EL, Wuthiekanun V, Thaipadungpanit J, Suputtamongkol Y, Chierakul W, Smythe LD, Day NP, Cooper B, Peacock SJ. Fool's gold: why imperfect reference tests are undermining the evaluation of novel diagnostics: a reevaluation of 5 diagnostic tests for leptospirosis. Clin Infect Dis. 2012;55:322–331. doi: 10.1093/cid/cis403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan-ngum W, Blacksell SD, Lubell Y, Pukrittayakamee S, Bailey MS, de Silva HJ, Lalloo DG, Day NP, White LJ, Limmathurotsakul D. Estimating the true accuracy of diagnostic tests for dengue infection using bayesian latent class models. PLoS One. 2013;8:e50765. doi: 10.1371/journal.pone.0050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Royal Tropical Institute . Typhoid F IgM Flow Assay. Amsterdam, The Netherlands: Royal Tropical Institute Koninklijk Instituut voor de Tropen (KIT); 2007. [Google Scholar]
- 22.Altman DG. Practical Statistics for Medical Research. London, UK: Chapman and Hall; 1991. [Google Scholar]
- 23.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, Lijmer JG, Moher D, Rennie D, de Vet HC. Standards for Reporting of Diagnostic Accuracy Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD Initiative. Radiology. 2003;226:24–28. doi: 10.1148/radiol.2261021292. [DOI] [PubMed] [Google Scholar]
- 24.Dendukuri N, Joseph L. Bayesian approaches to modeling the conditional dependence between multiple diagnostic tests. Biometrics. 2001;57:158–167. doi: 10.1111/j.0006-341x.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- 25.Lunn D, Spiegelhalter D, Thomas A, Best N. The BUGS project: evolution, critique and future directions. Stat Med. 2009;28:3049–3067. doi: 10.1002/sim.3680. [DOI] [PubMed] [Google Scholar]
- 26.Gasem MH, Dolmans WM, Isbandrio BB, Wahyono H, Keuter M, Djokomoeljanto R. Culture of Salmonella typhi and Salmonella paratyphi from blood and bone marrow in suspected typhoid fever. Trop Geogr Med. 1995;47:164–167. [PubMed] [Google Scholar]
- 27.Akoh JA. Relative sensitivity of blood and bone marrow cultures in typhoid fever. Trop Doct. 1991;21:174–176. doi: 10.1177/004947559102100415. [DOI] [PubMed] [Google Scholar]
- 28.Farooqui BJ, Khurshid M, Ashfaq MK, Khan MA. Comparative yield of Salmonella typhi from blood and bone marrow cultures in patients with fever of unknown origin. J Clin Pathol. 1991;44:258–259. doi: 10.1136/jcp.44.3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilman RH, Terminel M, Levine MM, Hernandez-Mendoza P, Hornick RB. Relative efficacy of blood, urine, rectal swab, bone-marrow, and rose-spot cultures for recovery of Salmonella typhi in typhoid fever. Lancet. 1975;1:1211–1213. doi: 10.1016/s0140-6736(75)92194-7. [DOI] [PubMed] [Google Scholar]
- 30.Ross IN, Abraham T. Predicting enteric fever without bacteriological culture results. Trans R Soc Trop Med Hyg. 1987;81:374–377. doi: 10.1016/0035-9203(87)90139-8. [DOI] [PubMed] [Google Scholar]
- 31.Hosoglu S, Geyik MF, Akalin S, Ayaz C, Kokoglu OF, Loeb M. A simple validated prediction rule to diagnose typhoid fever in Turkey. Trans R Soc Trop Med Hyg. 2006;100:1068–1074. doi: 10.1016/j.trstmh.2005.12.007. [DOI] [PubMed] [Google Scholar]