Abstract
Enterotoxigenic Escherichia coli (ETEC), the leading bacterial pathogen of travelers' diarrhea, is routinely detected by an established DNA hybridization protocol that is neither sensitive nor quantitative. Quantitative real-time polymerase chain reaction (qPCR) assays that detect the ETEC toxin genes eltA, sta1, and sta2 in clinical stool samples were developed and tested using donor stool inoculated with known quantities of ETEC bacteria. The sensitivity of the qPCR assays is 89%, compared with 22% for the DNA hybridization assay, and the limits of detection are 10,000-fold lower than the DNA hybridization assays performed in parallel. Ninety-three clinical stool samples, previously characterized by DNA hybridization, were tested using the new ETEC qPCR assays. Discordant toxin profiles were observed for 22 samples, notably, four samples originally typed as ETEC negative were ETEC positive. The qPCR assays are unique in their sensitivity and ability to quantify the three toxin genes in clinical stool samples.
Introduction
Enterotoxigenic Escherichia coli (ETEC) is a Gram-negative pathogen and member of the Gammaproteobacteria. ETEC is endemic in developing countries and is a leading cause of travelers' diarrhea (TD) among persons who visit these regions and consume contaminated food or water.1 The organism encodes two toxins, heat labile enterotoxin (LT) and heat stable enterotoxin (ST), that initiate fluid secretion into the intestinal lumen resulting in watery diarrhea.2 In most cases, the genes that encode these toxins reside on plasmids.3 The ETEC strain H10407 is a human isolate collected during an acute diarrheal outbreak in Bangladesh.4 The strain has been extensively studied, and its chromosome and plasmids have been sequenced.5 H10407 carries two toxigenic plasmids, pETEC666, a 66.6 kb plasmid that carries the LT operon eltAB plus an allele of ST, designated sta1, and pETEC948, a 94.8 kb plasmid, that carries a second ST allele, sta2. Both of these plasmids belong to the IncFII incompatibility group.5 The strain also carries two smaller plasmids, pETEC58 and pETEC52 that are not believed to be associated with toxigenesis.5
Current clinical diagnostic methods for detection of ETEC in stool include polymerase chain reaction (PCR) and DNA hybridization assays. The PCR assays range from conventional multiplex assays to real-time quantitative PCR (qPCR) assays with limits of detection ranging from 1,000 to 107 ETEC colony-forming units (CFU)/mL. Detection assays rely on the amplification of eltA alone, eltA plus either sta1 or sta2, or all three toxin genes.6–16 The DNA hybridization assay is a culture-dependent method that requires transfer of cultured lactose-positive colonies from MacConkey agar to Whatman filter paper (Fisher Scientific, Waltham, MA) followed by cell lysis and probe hybridization. Probes targeting STP (sta1), STH (sta2), and LT (eltA) are then used for ETEC detection17,18; remarkably, many clinical manuscripts reporting the prevalence of ETEC use this less-sensitive DNA hybridization method or use PCR assays that target eltA plus only a single allele of ST. As a result, these studies are likely to have underestimated the prevalence of ETEC infections.
Our interest in both the specific toxin presence and toxin gene quantification in human clinical stool samples led us to develop independent qPCR assays for the ETEC toxin genes that are predominantly associated with human outbreaks: i.e., eltA, sta1, and sta2. We evaluated these assays in a control stool inoculation experiment, compared the assays to the established DNA hybridization method, and applied the assays using clinical samples from TD studies.
Materials and Methods
Bacterial strains and culture conditions.
The E. coli strains used were ETEC strain H10407 (ATCC 35401) and the non-toxigenic strains MG1655 (ATCC 700926) and NCTC 9001 (ATCC 11775). Strains were cultivated overnight at 37°C on sheep blood agar (Remel, Lenexa, KS), Luria Bertani (LB) agar, or MacConkey agar plus lactose (Becton, Dickinson and Company, Sparks, MD) as needed. For liquid culture, strains were cultivated in LB broth at 37°C with shaking overnight, and then pelleted for DNA extraction or diluted 1:100 in fresh medium and grown to mid-log phase (H10407 OD600 = 1.2, MG1655 OD600 = 1.6) at 37°C with shaking for cell enumeration and to serve as inocula for stool samples.
Preparation of stool samples inoculated with E. coli strains.
We prepared two sets of stool samples: one set was inoculated with serial dilutions of H10407 and a second control set was inoculated with serial dilutions of MG1655. Strains were grown to mid-log phase at 37°C, and then bacterial cell counts were determined using a Petroff-Hauser Counting Chamber (Electron Microscopy Sciences, Hatfield, PA). Serial dilutions of the cultures were prepared in phosphate buffered saline (PBS) and were plated on both LB and MacConkey agar plates. Plates were incubated overnight at 37°C and colonies were counted to determine CFU/mL.
A stool sample was collected from a healthy donor and was stored at −20°C.19 Aliquots of thawed stool (700 mg) were transferred to 1) MO BIO PowerSoil DNA Extraction PowerBead tubes (Carlsbad, CA) for DNA extraction; and 2) cryovials for DNA hybridization assays. Each aliquot was inoculated with a bacterial dilution of each strain, alone, to attain 10 to 109 bacterial cells per aliquot, and then the sample was briefly vortexed. Negative controls were prepared by adding 200 μL of PBS to a stool aliquot and leaving an aliquot uninoculated. Samples in PowerBead tubes were heated, as described below, before being stored at −80°C. Samples in cryovials were stored at −80°C.
Clinical stool samples.
Ninety-three stool samples from previous TD studies were selected for a comparative analysis of the DNA hybridization and qPCR assay methods. Samples were collected from adults participating in studies who had traveled from the United States to Mexico, India, or Guatemala between 2005 and 2007. Informed consent was obtained from all human adult participants and The University of Texas Health Science Center at Houston's institutional review board approved the studies. TD was defined as the passing of ≥ 3 unformed stools within a 24-hour period along with at least one additional enteric symptom including vomiting, abdominal cramps, fever, and nausea.20 Each stool sample was screened for enteric pathogens as described previously.20 ETEC was determined to be the causative agent in 70 samples. A pathogen was not identified in 11 samples—these were included as non-ETEC diarrheal controls. Twelve samples from healthy, diarrhea-free travelers were included as negative controls (DuPont HL, personal communication). 500–800 μL of each frozen stool sample was thawed and transferred to a MO BIO PowerSoil DNA Extraction PowerBead Tube, heated as described below, and stored at −80°C for DNA extraction.
DNA extraction from E. coli strains.
The DNA was extracted using the Omega Bio-tek E.Z.N.A Bacterial DNA Kit (Norcross, GA) with the following modifications. A cell pellet from 2 mL of an overnight bacterial culture was resuspended in 200 μL of TE buffer and incubated with 18 μL of 50 mg/mL lysozyme at 30°C for 30 minutes followed by centrifugation at 2,400 × g for 5 minutes. The pellet was resuspended in 200 μL of BTL solution, combined with 25 mg of glass beads, and then the cells were disrupted using an MP Biomedicals FastPrep-24 (Santa Ana, CA) for three 40 second beating cycles at 6.0 M/S followed by a 5-minute rest, and then a final bead beating for 40 seconds. The manufacturer's protocol was followed for the remaining DNA binding and wash steps.
DNA extraction from stool samples.
Stool aliquots in MO BIO PowerSoil DNA Extraction PowerBead Tubes were incubated at 65°C for 10 minutes followed by incubation at 95°C for 10 minutes and stored at −80°C before DNA extraction.19 The MO BIO PowerSoil DNA Isolation Kit was used, following the manufacturer's protocol. Samples were eluted in 100 μL of solution C6 and stored at −20°C.
Primer design.
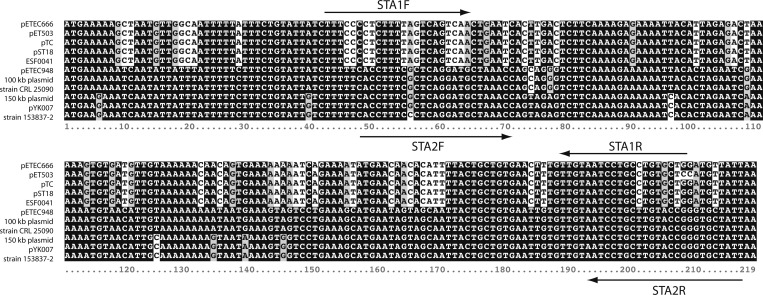
To design primer pairs for each qPCR assay, we collected independent nucleotide sequences of the A subunit of the LT gene, eltA (N = 23), and of the ST genes, sta (N = 11), available in GenBank as of July 2012 and used these to create multiple sequence alignments using Clustal X 2.1 to find consensus sequences.21 The high conservation of eltA permitted selection of a single set of primers directed to the 3′ end of the gene (Table 1); the eltA primers overlap with, but are not identical to, sequences selected for use in previous studies.11,16 Alignment of the sta sequences revealed a distinct clustering of variants that defined consensus regions for the sta1 and sta2 alleles (Figure 1). We designed optimal allele-specific primer pairs using Primer3 and Integrated DNA Technology's (IDT) PrimerQuest tools.22,23 We sought primer pairs that had similar melting temperatures (Tm) and were free of features such as high or low GC content, secondary structure, hetero- and homo dimer-formation, and off-target identities. For each primer pair, these features were analyzed using version 3.1 of IDT's OligoAnalyzer tool.23 Megablast against the NCBI nucleotide database was used to confirm target specificity. The sta primers designed for the assays described here overlap with, but are not identical to, primers described elsewhere.6,7,9,10,12–16,24,25 Primer sequences used in this work are listed in Table 1. Oligonucleotides were synthesized by Invitrogen (Carlsbad, CA).
Table 1.
Enterotoxigenic Escherichia coli quantitative real-time polymerase chain reaction primers developed for this study
| Gene | Primer sequence (5′-3′) | Sequence position* | Primer Tm (°C) | Amplicon size (bp) | Amplicon Tm† (°C) |
|---|---|---|---|---|---|
| eltA | ELTAF: ATTAGCAGGTTTCCCACCGGATCA | 543–566 | 60 | 138 | 78.4 (0.2) |
| ELTAR: TTGTGCTCAGATTCTGGGTCTCCT | 656–679 | 60 | |||
| sta1 | STA1F: TTTCCCCTCTTTTAGTCAGTCAA | 42–64 | 59 | 167 | 74.9 (0.3) |
| STA1R: CAGCACAGGCAGGATTACAA | 189–208 | 60 | |||
| sta2 | STA2F: ACCTTTCGCTCAGGATGCTAAACC | 48–71 | 59 | 171 | 75.6 (0.3) |
| STA2R: AATAGCACCCGGTACAAGCAGGAT | 194–217 | 60 |
Relative to the start codon of the open reading frame.
Mean (SD).
Figure 1.
Aligned enterotoxigenic Escherichia coli (ETEC) sta sequences. Sequences were aligned using Clustal X and then shaded using Boxshade 3.21 (www.ch.embnet.org). The sta1- and sta2-specific primers used for real-time quantitative polymerase chain reaction (qPCR) are indicated by arrows. GenBank accession nos. are as follows: pETEC666, GI 309705521; pTE503, GI 147877; pTC, GI 356598343; pST18, GI 145860; ESF0041, GI 43704; pETEC948, GI 309706192; 100 kb plasmid, GI 82697140; strain CRL 25090, GI 145862; 150 kb plasmid, GI 148029; pYK007, GI 147875; strain 153837-2, GI 146407.
Conventional PCR assay conditions.
Primer specificity was confirmed by conventional PCR using an Eppendorf vapo.protect Mastercycler (Hamburg, Germany). Each 20 μL reaction contained 2 μL of 10× Accuprime™ PCR Buffer I (Invitrogen), 0.15 μL of Accuprime™ Taq DNA Polymerase High Fidelity (Invitrogen), 1 μL of each forward and reverse primer at 5 μM, 2 μL of template DNA, and 13.85 μL of Sterile WFI-Quality, Cell Culture Grade Water (Mediatech, Inc. Manassas, VA). The H10407 DNA was used as a positive control template, MG1655 DNA as a negative control template, and water was used as a no template control. Cycling conditions were as follows: 95°C for 2 minutes, and then 30 cycles of 95°C for 30 seconds, 57°C for 1 minute, and 72°C for 2 minutes with a final extension at 72°C for 10 minutes. The optimal primer annealing temperature of 57°C for use in qPCR assays for the sta1 primers was determined using gradient PCR with the following annealing temperatures: 47.9, 48.2, 49.0, 50.2, 51.7, 53.4, 55.1, 56.8, 58.4, 59.7, and 60.6°C. The optimal primer annealing temperatures for the eltA and sta2 primer pairs were determined experimentally during development of the qPCR assays.
qPCR assay conditions.
The Applied Biosystems 7500 Fast Real-Time PCR System (Foster City, CA) was used with version 1.4 analysis software in Standard 7500 Mode. Optimal annealing temperatures for the eltA and sta2 primers and optimal primer concentrations for each primer pair were determined to be 57°C and 200 nM, respectively. Each 25 μL reaction was composed of 12.5 μL of Power SYBR© Green PCR Master Mix (Applied Biosystems), 1 μL of each forward and reverse primer at 5 μM, 2 μL of template DNA, and 8.5 μL of Sterile WFI-Quality, Cell Culture Grade Water (Mediatech, Inc.). Cycling conditions were as follows: Stage 1: 50°C for 2 minutes; Stage 2: 95°C for 10 minutes; Stage 3 (40 cycles): Step 1: 95°C for 15 seconds, Step 2: 57°C for 1 minute. A dissociation stage was added to each cycling profile to confirm specificity of amplified products. Data were collected following Stage 3, Step 2. Upon optimization, each assay plate included serial dilutions of H10407 DNA containing 10 to 107 copies of pETEC666 and pETEC948 as a standard curve, a stool sample inoculated with 106 H10407 cells as a positive stool control, an uninoculated stool sample as a non-diarrheal negative control, and a no template control.
Plasmid copy number determination.
We used qPCR to determine the copy number per chromosome equivalent of plasmids pETEC666 and pETEC948 by calculating the toxin gene copy number relative to the 16S rRNA genes in H10407. The relative 16S rRNA gene equivalent per mass of H10407 DNA was determined by performing qPCR on 10-fold dilutions of H10407 genomic DNA (1–100 pg) using the 16S rRNA gene primers: 340F: 5′ CGT ATT ACC GCG GCT GCT GG 3′; and 537R: 5′ TCC TAC GGG AGG CAG CAG T 3′.26 These primers are identical to their targets within each of the seven 16S rRNA genes in H10407. The optimal primer annealing temperature was determined to be 57°C using gradient PCR with the following annealing temperatures: 53.0, 53.2, 53.8, 54.7, 55.8, 57.1, 58.4, 59.7, 60.9, 61.9, and 62.6°C. For copy number determinations, all four qPCR assays (eltA, sta1, sta2, and 16S rRNA) were performed on a single assay plate, as described above, in triplicate. At least two technical replicates were performed. Toxin and 16S rRNA gene amplicons were quantified by product = (1 + E)Cq, where E is the reaction efficiency, and Cq is the cycle at which the fluorescent signal crosses an instrument-determined threshold. Plasmid copy number per chromosome equivalent was calculated as the ratio of toxin gene product to 16S rRNA gene product normalized to the seven copies of the gene.
DNA hybridization assay.
Control E. coli-inoculated stool samples and clinical stool samples were assayed for the presence of the LT and ST genes using the DNA hybridization method described previously.17 Briefly, the two sets of cryovials containing stool samples inoculated with dilutions of H10407 or MG1655, PBS, and the uninoculated samples were thawed and each streaked on MacConkey agar and grown at 37°C overnight to isolate single colonies. Ten lactose-positive colonies were transferred to a Whatman 541 filter paper and then lysed as described previously.27 Five prime 32P- end-labeled oligonucleotide probes: LT (eltA), 5′ GCG AGA GGA ACA CAA ACC GG 3′28; STP (sta1), 5′ GCT GTG AAC TTT GTT GTA ATC C 3′18; and STH (sta2), 5′ GCT GTG AAT TGT GTT GTA ATC C 3′18; were hybridized to the filters to detect the LT and ST genes, as described.27 The H10407 was used as a positive control and NCTC 9001 was used as a negative control.
Results
Performance of eltA, sta1, and sta2 assays.
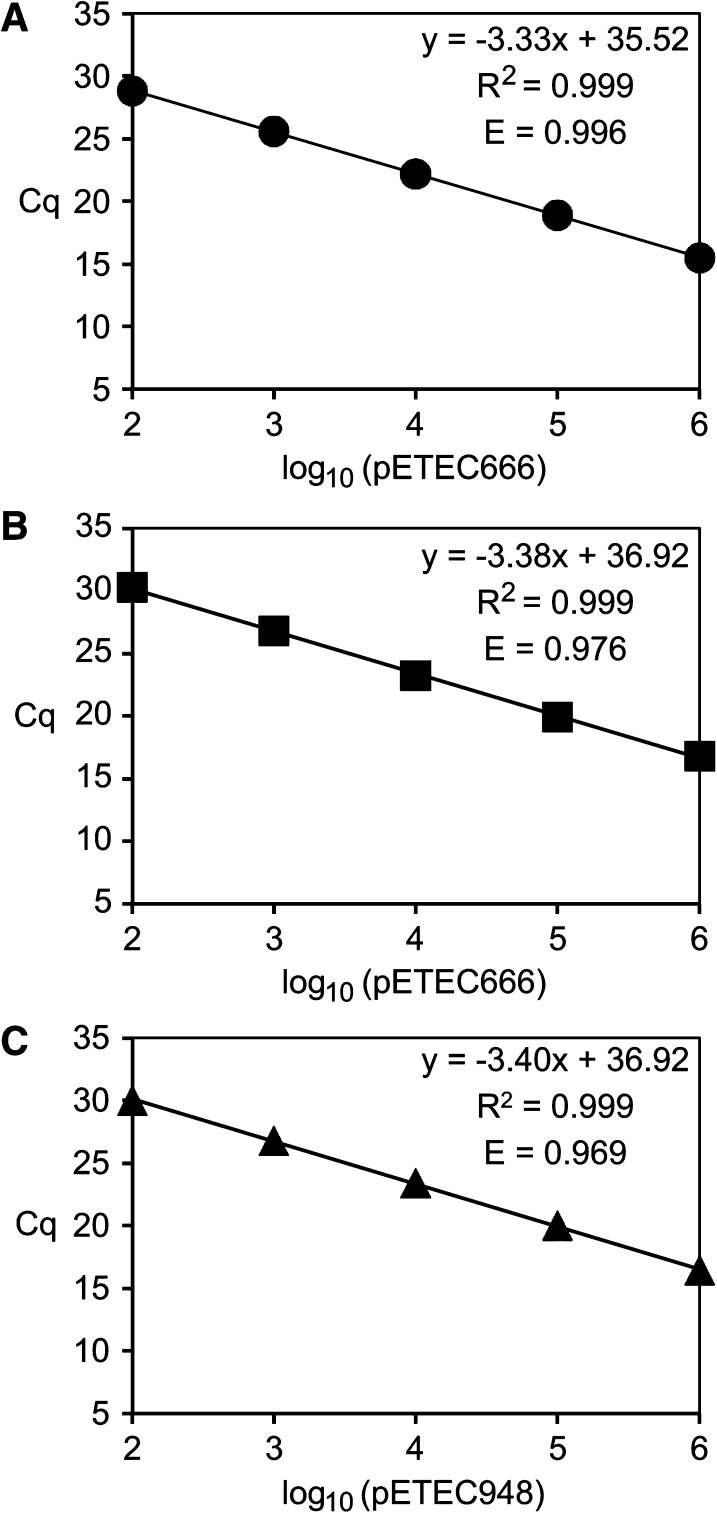
Primer specificity was first shown using conventional PCR. Each primer pair yielded a single product of the expected size from purified H10407 genomic DNA (plasmid plus chromosome), and no products were amplified using purified MG1655 DNA or water as template (data not shown). To determine the limit of toxin gene detection and quantification for each assay, standard curves were generated using serial dilutions of H10407 DNA containing 10 to 107 copies of pETEC666 and pETEC948 as template. A representative standard curve for each toxin qPCR assay is shown in Figure 2. The average efficiency of each assay is > 95%, and the average r2 values are 0.999. Specifically, average assay efficiencies (±SD) and correlation coefficients (± SD) for each are as follows: eltA (N = 7) E = 0.9969 ± 0.01, r2 = 0.999 ± 0.001; sta1 (N = 6), E = 0.9502 ± 0.02, r2 = 0.9996 ± 0.0004; sta2 (N = 5), E = 0.9825 ± 0.02, r2 = 0.9996 ± 0.0002. The limit of detection of each assay is 10 toxin gene copies per reaction. The lower limit of quantification for each assay is 100 toxin gene copies per reaction, and standard curves were linear to 106 toxin gene copies per reaction (Figure 2). Melt curve analysis showed that the products that amplified within the linear dynamic range had the correct Tm and template negative controls failed to generate toxin-specific melt curve amplicons (data not shown), also showing the specificity of each qPCR assay.
Figure 2.
Representative standard curves for each toxin-specific quantitative real-time polymerase chain reaction assay: (A) eltA; (B) sta1; (C) sta2. Cycle threshold (Cq) is plotted versus log10(plasmid molecules) per reaction.
pETEC666 and pETEC948 copy numbers.
To be able to estimate toxin gene copy number in stool samples inoculated with H10407, we first needed to determine the copy numbers of the toxigenic plasmids pETEC666 and pETEC948 in strain H10407. The plasmids carry single copies of eltA and sta1, and sta2, respectively. We used the toxin gene-specific qPCR assays coupled with a 16S rRNA-specific qPCR assay to calculate the ratio of toxin gene copies to chromosome copies. Based on eltA and sta1 qPCR assays, the copy number of pETEC666 is 16 ± 3 plasmid copies per chromosome equivalent, and the copy number of pETEC948 is 13 ± 4 based on sta2 assays (Table 2). Using these values, and assuming an average of one chromosome per E. coli cell,29 a 700 mg stool sample inoculated with 109 H10407 cells should contain ∼1010 copies of each plasmid. Assuming a 100% efficiency of microbial DNA extraction from a stool sample, 1 μL of DNA obtained from such a sample should theoretically contain 108 plasmid/toxin copies.
Table 2.
Plasmid copy number determinations
| Plasmid | Gene assayed | Input genomic DNA (pg) | Gene copy no.* | Average plasmid copy no. ± SD |
|---|---|---|---|---|
| pETEC666 | eltA | 100 | 19 | 16 ± 3 |
| 10 | 17 | |||
| 1 | 16 | |||
| sta1 | 100 | 16 | ||
| 10 | 16 | |||
| 1 | 14 | |||
| pETEC948 | sta2 | 100 | 14 | 13 ± 4 |
| 10 | 14 | |||
| 1 | 12 |
Average of 2–3 assays per DNA concentration.
Toxin gene detection and quantification in E. coli-inoculated stool samples.
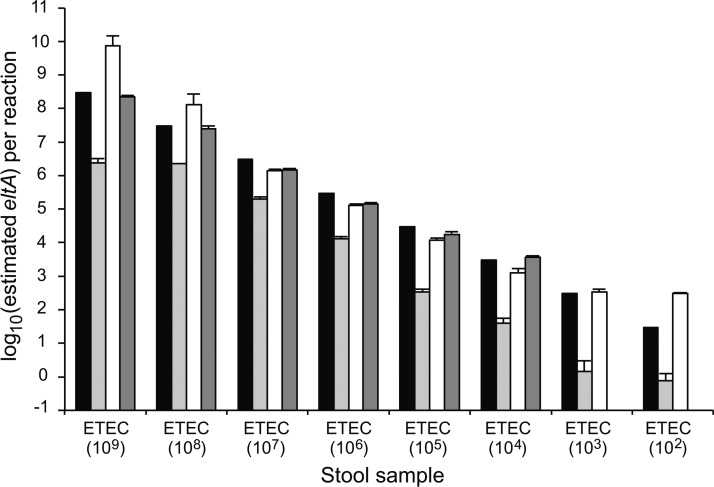
To establish the limits of detection and quantification of ETEC in a defined system, qPCR assays were performed using DNAs extracted from stool samples that were inoculated with serial dilutions of H10407 cells. In the eltA qPCR assay, we determined that 1:10 and 1:100 dilutions of stool DNA were ideal for both detection and quantification. Undiluted samples were non-quantitative regardless of the quantity of bacteria inoculated per stool aliquot (Figure 3). Ten copies of eltA could be detected in a reaction using a 1:10 dilution of stool DNA, which corresponds to a stool sample inoculated with 100 H10407 cells; however, quantification was most reliable between the ranges of 100 to 106 copies of eltA per reaction (1,000 to 107 H10407 cells per sample, respectively). A 1:100 dilution of H10407-inoculated stool DNA provided accurate quantification of 107 and 108 copies of eltA per reaction (108 and 109 H10407 cells per sample, respectively) with a limit of 1,000 copies per reaction (10,000 H10407 cells per sample) (Figure 3). In light of these results, all subsequent assays included both 1:10 and 1:100 dilutions of stool DNA to assure maximum quantification and accurate detection. For the sta1 and sta2 assays, both the limit of detection and quantification are 100 toxin gene copies per reaction (1,000 H10407 cells per sample, data not shown). In no case was toxin-specific amplification observed using DNA extracted from stool inoculated with MG1655 or PBS, the uninoculated stool sample, or in template negative reactions. This is a very important demonstration of the specificity of the assay even in complex mixtures such as stool.
Figure 3.
Effect of dilution of stool DNA on eltA quantitative real-time polymerase chain reaction accuracy for samples inoculated with serial dilutions of H10407 cells ranging from 109 to 102 cells/700 mg aliquot. Black bar: theoretical gene copy number per reaction. Assayed gene quantity using undiluted DNA (light gray bars), 1:10 stool DNA (white bars), and 1:100 stool DNA (dark gray bars) plotted for each stool sample, where the concentration of the H10407 inoculum (cells/sample) is indicated in parentheses. Each assay was performed at least twice; error bars represent the standard deviation.
Comparison of qPCR and DNA hybridization assays.
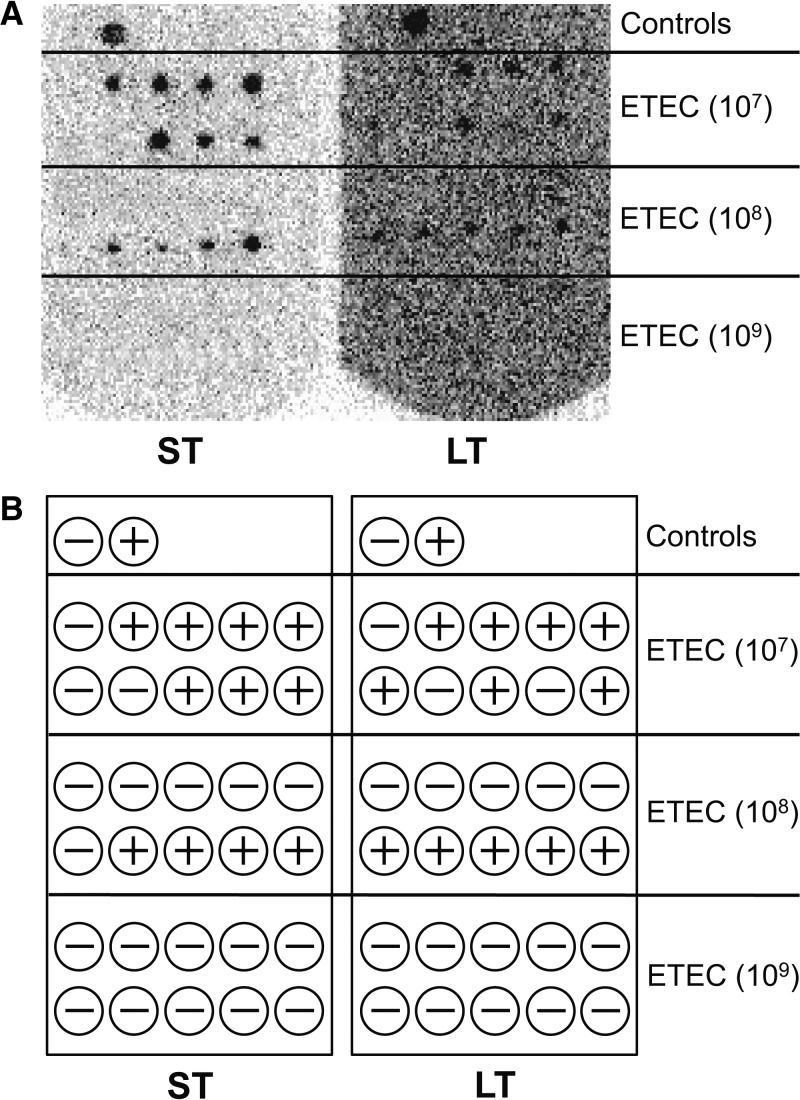
We used the H10407- and MG1655-inoculated stool samples to directly compare the sensitivities and specificities of our toxin gene qPCR assays with the established ETEC DNA hybridization method. From inoculated stool samples that had been stored in cryovials at −80°C, bacteria were cultivated on MacConkey agar, and then 10 presumptive lactose-positive E. coli colonies per sample were transferred to a filter for hybridization. These were directly compared with the samples where DNA was prepared as template for the qPCR assays. Using the hybridization method, H10407 was detected only in samples inoculated with 107 or 108 H10407 cells (108 and 109 toxin gene copies, respectively, Figure 4). Note that toxin was detected in 40–70% of the colonies tested. No toxin was detected in the samples inoculated with 109 H10407 cells (Figure 4) or in samples inoculated with fewer than 107 cells per sample (data not shown). The sensitivity of the DNA hybridization assay is 22% (2 of 9; 95% confidence interval [CI] = 5–41). Seven of the H10407-inoculated samples were not detected by DNA hybridization, therefore the negative predictive value is 65%. The lack of hybridization in samples with 109 H10407 cells per sample was surprising; the presence of the toxin genes in this sample was verified by PCR (data not shown). In the qPCR assays, the detection limit of eltA was 100 H10407 cells (or 1,000 gene copies), and 1,000 H10407 cells for sta1 and sta2 (or 10,000 gene copies) per stool sample. These limits of detection are four and five orders of magnitude lower than the detection limit of the DNA hybridization method performed in parallel. The sensitivity of the qPCR assays is 89% (8 of 9; 95% CI = 76–102). Only one of the H10407-inoculated samples was not detected by at least one of the qPCR assays, therefore the negative predictive value is 93%. Again, no toxin-specific amplification or probe hybridization was observed when assaying stool samples inoculated with MG1655, PBS, or the uninoculated control (data not shown), hence the specificities and positive predictive values for the assays are 100%. Differences were determined to be significant (p < 0.05) using a McNemar's Test with the Yate's Correction for Continuity.
Figure 4.
DNA hybridization filter showing results for 10 Escherichia coli colonies isolated from stool samples inoculated with 107, 108, or 109 H10407 cells and probed with end-labeled primers specific for heat stable enterotoxin (ST, left) and heat labile enterotoxin (LT, right) genes. (A) Hybridization membrane, hybridized first with the ST probes and then washed and reprobed with the LT probe; (B) key showing the location of individual spotted colonies, interpretation of results, and location of negative (E. coli NCTC 9001) and positive (E. coli H10407) control colonies. No positive signals were observed at dilutions below 107 H10407 cells per stool sample (filters not shown).
Toxin gene detection and quantification in clinical stool samples.
The ETEC qPCR assays were used to test 93 clinical stool samples collected from adults who traveled to Mexico, India, or Guatemala between 2005 and 2007. The samples were divided into five groups: eltA positive, sta positive, eltA and sta positive, no pathogen identified, and healthy travelers. Pathogen detection and toxin classifications were based on routine assays performed during the study,20 including LT and ST gene typing determined by the DNA hybridization method.17 Seventeen samples had matching toxin profiles determined by both DNA hybridization and qPCR (Table 3). Three of the 11 samples classified as “no pathogen identified” were toxin positive by qPCR: two samples contained eltA and sta1, with particularly high titers of eltA in sample 80028 and sta1 in sample 60025 (Table 4), and one contained eltA. Interestingly, eltA was detected by qPCR in one of the 12 healthy traveler samples. In 12 cases where samples were positive for either eltA or sta alone, based on DNA hybridization, the qPCR assays revealed that they carried alleles of both genes. We also observed that 35 of the 70 clinical samples typed as ETEC positive by DNA hybridization were ETEC negative by all three qPCR assays. Finally, in qPCR-positive samples, ETEC cells could be quantified, and quantities ranged from 103–108 bacteria per sample (104–109 toxin genes per sample) (Table 4).
Table 3.
Comparison of enterotoxigenic Escherichia coli detection in travelers' diarrhea samples by DNA hybridization and quantitative real-time polymerase chain reaction (qPCR) assays
| DNA hybridizationresults | LT+ ST+ 26 | LT+ ST− 21 | LT− ST+ 23 | LT− ST− 11* | LT−ST− 12† | |
|---|---|---|---|---|---|---|
| eltA+ sta1+ sta2+ | 3 | 3 | 4 | |||
| eltA+ sta1+ sta2- | 2 | 3 | 1 | 2 | ||
| qPCR | eltA+ sta1- sta2+ | 4 | 1 | |||
| Results | eltA+ sta1- sta2- | 2 | 3 | 1 | 1 | 1 |
| eltA- sta1+ sta2- | 2 | 3 | ||||
| eltA- sta1- sta2+ | 1 | 2 | ||||
| eltA- sta1- sta2- | 12 | 11 | 12 | 8 | 11 | |
| Total | 26 | 21 | 23 | 11 | 12 |
No pathogen identified as the diarrheal etiologic agent.
Samples collected from healthy travelers.
LT = heat labile enterotoxin; ST = heat stable enterotoxin.
Table 4.
Results of DNA hybridization and quantitative real-time polymerase chain reaction (qPCR) assays performed on travelers' diarrhea samples
| Sample ID | Year collected | Clinical presentation | DNA Hybridization | qPCR* | |||
|---|---|---|---|---|---|---|---|
| eltA | sta | eltA | sta1 | sta2 | |||
| 8 | 2007 | Diarrhea | + | − | − | − | − |
| 13 | 2007 | Diarrhea | + | − | + (107) | − | − |
| 59 | 2005 | Diarrhea | + | − | + (108) | + (108) | − |
| 231 | 2007 | Diarrhea | + | − | − | − | − |
| 367 | 2007 | Diarrhea | + | − | + (106) | + (106) | + (106) |
| 453 | 2007 | Diarrhea | + | − | − | − | − |
| 6128 | 2006 | Diarrhea | + | − | − | − | − |
| 6155 | 2006 | Diarrhea | + | − | + (105) | + (105) | − |
| 6165 | 2006 | Diarrhea | + | − | − | − | − |
| 6168 | 2006 | Diarrhea | + | − | − | − | − |
| 1574 | 2007 | Diarrhea | + | − | + (104) | + (103) | − |
| 1748 | 2007 | Diarrhea | + | − | + (105) | + (105) | + (105) |
| 1840 | 2007 | Diarrhea | + | − | + (103) | + (105) | + (103) |
| 50070 | 2005 | Diarrhea | + | − | − | − | − |
| 50076 | 2005 | Diarrhea | + | − | − | − | − |
| 50390 | 2005 | Diarrhea | + | − | + (106) | − | − |
| 50394 | 2005 | Diarrhea | + | − | + (104) | − | − |
| 50395 | 2005 | Diarrhea | + | − | − | − | − |
| TD-4 | 2005 | Diarrhea | + | − | + (103) | − | + (104) |
| TD-5 | 2005 | Diarrhea | + | − | − | − | − |
| TD-6 | 2005 | Diarrhea | + | − | − | − | − |
| 3 | 2007 | Diarrhea | − | + | − | − | − |
| 45 | 2007 | Diarrhea | − | + | − | − | − |
| 75 | 2007 | Diarrhea | − | + | − | − | − |
| 76 | 2005 | Diarrhea | − | + | − | − | + (103) |
| 79 | 2007 | Diarrhea | − | + | − | − | − |
| 101 | 2007 | Diarrhea | − | + | − | − | − |
| 103 | 2007 | Diarrhea | − | + | − | − | − |
| 137 | 2005 | Diarrhea | − | + | + (105) | + (105) | − |
| 147 | 2005 | Diarrhea | − | + | − | − | − |
| 201 | 2005 | Diarrhea | − | + | + (104) | + (105) | + (104) |
| 283 | 2007 | Diarrhea | − | + | − | + (106) | − |
| 403 | 2007 | Diarrhea | − | + | − | − | − |
| 421 | 2007 | Diarrhea | − | + | − | + (106) | − |
| 640 | 2007 | Diarrhea | − | + | − | + (107) | − |
| 6163 | 2006 | Diarrhea | − | + | − | − | − |
| 50091 | 2005 | Diarrhea | − | + | − | − | − |
| 50413 | 2005 | Diarrhea | − | + | + (103) | + (105) | + (105) |
| 50414 | 2005 | Diarrhea | − | + | − | − | − |
| PR124 | 2006 | Diarrhea | − | + | − | − | + (106) |
| RL124 | 2006 | Diarrhea | − | + | + (105) | + (107) | + (104) |
| TD-1 | 2005 | Diarrhea | − | + | + (106) | + (106) | + (107) |
| TD-2 | 2005 | Diarrhea | − | + | − | − | − |
| TD-3 | 2005 | Diarrhea | − | + | + (105) | − | − |
| 10 | 2007 | Diarrhea | + | + | − | − | − |
| 44 | 2007 | Diarrhea | + | + | + (104) | − | − |
| 78 | 2007 | Diarrhea | + | + | − | − | − |
| 93 | 2007 | Diarrhea | + | + | − | − | + (106) |
| 125 | 2007 | Diarrhea | + | + | + (105) | − | − |
| 127 | 2007 | Diarrhea | + | + | − | + (107) | − |
| 156A | 2007 | Diarrhea | + | + | − | − | − |
| 156B | 2007 | Diarrhea | + | + | + (106) | + (106) | + (103) |
| 160 | 2007 | Diarrhea | + | + | − | − | − |
| 171 | 2007 | Diarrhea | + | + | + (106) | + (106) | − |
| 173 | 2007 | Diarrhea | + | + | + (105) | + (105) | − |
| 195 | 2007 | Diarrhea | + | + | − | − | − |
| 207 | 2007 | Diarrhea | + | + | + (106) | − | + (106) |
| 400 | 2007 | Diarrhea | + | + | + (105) | − | + (105) |
| 407 | 2007 | Diarrhea | + | + | + (106) | − | + (106) |
| 1139 | 2007 | Diarrhea | + | + | + (105) | − | + (105) |
| 1510 | 2007 | Diarrhea | + | + | + (106) | + (104) | + (106) |
| 5024 | 2005 | Diarrhea | + | + | + (104) | + (105) | + (104) |
| 50005 | 2005 | Diarrhea | + | + | − | − | − |
| 50011 | 2005 | Diarrhea | + | + | − | − | − |
| 50012 | 2005 | Diarrhea | + | + | − | − | − |
| 50013 | 2005 | Diarrhea | + | + | − | − | − |
| 50027 | 2005 | Diarrhea | + | + | − | − | − |
| TD-7 | 2005 | Diarrhea | + | + | − | − | − |
| TD-8 | 2005 | Diarrhea | + | + | − | − | − |
| TD-9 | 2005 | Diarrhea | + | + | − | + (103) | − |
| 60025 | 2006 | Diarrhea† | − | − | + (103) | + (108) | − |
| 60104 | 2006 | Diarrhea† | − | − | − | − | − |
| 60108 | 2006 | Diarrhea† | − | − | − | − | − |
| 80028 | 2007 | Diarrhea† | − | − | + (106) | + (104) | − |
| 80045 | 2007 | Diarrhea† | − | − | − | − | − |
| 80077 | 2007 | Diarrhea† | − | − | − | − | − |
| 80129 | 2007 | Diarrhea† | − | − | − | − | − |
| 80134 | 2007 | Diarrhea† | − | − | − | − | − |
| 80142 | 2007 | Diarrhea† | − | − | − | − | − |
| 80144 | 2007 | Diarrhea† | − | − | + (104) | − | − |
| 80152 | 2007 | Diarrhea† | − | − | − | − | − |
| F1 | 2006 | Healthy Traveler | − | − | − | − | − |
| F2 | 2006 | Healthy Traveler | − | − | − | − | − |
| F3 | 2006 | Healthy Traveler | − | − | − | − | − |
| F4 | 2006 | Healthy Traveler | − | − | − | − | − |
| F5 | 2006 | Healthy Traveler | − | − | − | − | − |
| F6 | 2006 | Healthy Traveler | − | − | − | − | − |
| F7 | 2006 | Healthy Traveler | − | − | − | − | − |
| F8 | 2006 | Healthy Traveler | − | − | − | − | − |
| F9 | 2006 | Healthy Traveler | − | − | − | − | − |
| F10 | 2006 | Healthy Traveler | − | − | − | − | − |
| F11 | 2006 | Healthy Traveler | − | − | + (103) | − | − |
| F12 | 2006 | Healthy Traveler | − | − | − | − | − |
Gene presence (estimated quantity of enterotoxigenic Escherichia coli cells/stool sample).
No pathogen identified as the diarrheal etiologic agent.
Discussion
Detection of enteric pathogens has evolved with development of new methodologies. Microscopy is used for identification of parasites such as helminths and protozoa, and culture-based methods using selective media coupled with biochemical assays are used as clinical diagnostics for bacterial enteropathogens. Development of immunoassays allow for detection and quantification of pathogen-specific antigens. These methods are considered gold standard for pathogen identification, however the plus/minus nature of the results do not provide quantitative information for specific genes. Culture-independent PCR assays targeting specific virulence genes, such as adhesins, invasins, and toxins, or other pathogen-specific genes, for example, viral capsid or ribosomal genes, are rapid, sensitive, and generally do not require costly equipment and reagents. Real-time qPCR assays have increased specificity and sensitivity, can be performed in less time, and are quantitative. For detection of ETEC, DNA hybridization, endpoint PCR, and qPCR are routinely used. The DNA hybridization is culture-based, does not distinguish between the two variants of sta, and is not quantitative. Endpoint PCR assays are not quantitative. The PCR and qPCR assays developed for ETEC have varying limits of detection, not all assays target the three toxin genes, and some assays require specialized equipment and reagents (e.g., Luminex assays). Development of our qPCR assays focused on the need for specific, rapid, and inexpensive molecular assays for detection and quantification of eltA, sta1, and sta2 in stool samples. We considered use of a TaqMan type assay because of its characteristic specificity. Unfortunately, amplicon length constraints for the sta alleles led us instead to develop independent SYBR green-based qPCR assays for detection and quantification of the ETEC genes eltA, sta1, and sta2. The assays are specific, using either bacterial genomic DNA or stool DNA as template, have limits of detection that are four to five orders of magnitude lower than the traditional DNA hybridization method performed for ETEC toxin gene detection, and are quantitative using stool samples inoculated with H10407 cells.
Toxin typing of non-contemporary clinical travelers' diarrhea samples based on qPCR was, in many cases, discordant with results obtained by DNA hybridization. Since the DNA hybridization method using stool samples inoculated with H10407 was not particularly sensitive (22% CI = 5–41), we expected some clinical samples to be toxin-negative by DNA hybridization but toxin-positive by qPCR assay. This was the case in 15 of the clinical diarrheal samples. The detection of eltA and sta1 in two samples and eltA in one sample that had been classified as “no pathogen identified” (Table 3) is of important clinical and epidemiological significance. The co-occurrence of eltA and sta1 in samples, which occurred in eight clinical samples, is compelling because of their potential for linkage on a plasmid, as in H10407. Thirty-five of the 70 clinical samples that were ETEC positive by DNA hybridization were negative by all three qPCR assays. Since false positives were not detected in our DNA hybridization assays using H10407-inoculated stool, the negative qPCR results are most likely the result of sample degradation between the time of sample collection and DNA preparation. Indeed, we observed that samples collected in 2007 had a higher concordance than samples collected in 2005 or 2006 (Table 4), which highlights the importance of performing assays on clinical samples promptly after collection. Nevertheless, the large discrepancies between these two methods underscore the need for a rapid, culture-independent method for ETEC detection and toxin gene quantification that is both highly sensitive and specific.
Reliable quantification of ETEC toxin genes is an important feature of our qPCR assay. Although we observed similar quantification of toxin genes using 1:10 and 1:100 dilutions of stool DNA within the mid-range of H10407 inocula (Figure 3), the 1:100 dilution of stool DNA is critical for quantification of toxin genes ≥ 109 (≥ 108 H10407 cells) per stool sample, and a 1:10 dilution is required for quantifying lower concentrations caused by potential PCR inhibitory factors in stool. Clinical samples from subjects with acute ETEC-diarrhea are reported to contain 107–109 CFU per gram of stool, or 108–1010 toxin genes per gram.30,31 In the case of a diarrheal sample, however, a low ETEC toxin gene titer may indicate that ETEC is not necessarily the cause of disease. Subjects who remain asymptomatic upon rechallenge with ETEC have reported titers of 104 to 105 CFU per gram of stool,31 and ETEC has been isolated from asymptomatic subjects32 and was detected at low titer in one of our control healthy traveler samples (Tables 3 and 4). Twenty of the clinical samples we tested had ≤ 105 ETEC cells per stool sample (Table 4) suggesting that, although ETEC is present, it may not be the source of the diarrhea given the estimated quantities of toxin genes. Thus, use of a quantitative assay for toxin load could improve diagnosis and potentially prevent unnecessary treatment regimens.
We used the LT and ST genes as markers to determine the copy numbers of pETEC666 and pETEC948. Most eltA, sta1, and sta2 genes are plasmid borne3 although the sta1 allele, which is present on pETEC666, is usually associated with transposon Tn1681,33 therefore it may also be chromosome-associated in some strains. The copy numbers of pETEC666 and pETEC948 in H10407 are about 16 and 13 per chromosome equivalent, respectively. Based on these copy number calculations and assuming that a given eltA, sta1, and sta2 toxin gene could be present from about 2 (chromosomal) to 20 (plasmid) copies within an ETEC strain, we believe that our qPCR assays should be quantitative for most ETEC strains, within an order of magnitude.
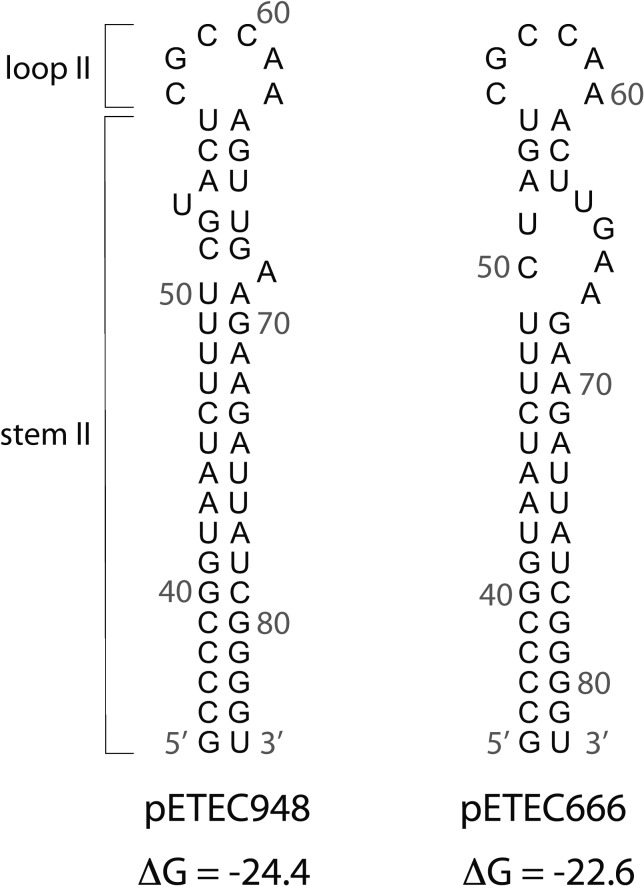
The copy numbers we obtained are 5- to 10-fold higher than we expected because both plasmids are members of the IncFII incompatibility group, which are generally maintained at 1–3 copies per cell.34 Replication of IncFII plasmids is negatively regulated, in part, by a ∼90 nucleotide (nt) antisense RNA, called CopA, that binds to an anti-sense target CopT-RNA, within the leader sequence of the mRNA that encodes the plasmid replication protein RepA.34 Binding of CopA-RNA inhibits translation of the RepA protein and thereby inhibits plasmid replication. Key base changes within the anti-sense regulator CopA-RNA can affect plasmid incompatibility and can also affect plasmid copy number.34,35 This led us to inspect the pETEC666 and pETEC948 CopA-RNA sequences and compare them with that of the well-studied large conjugative plasmid R1. The stem-loop II sequence of CopA encoded by pETEC948 is identical to that of R1 (Figure 5); the predicted structure contains a long stem (stem II) with a six nt loop (loop II) that, for R1, is the site of initial interaction between CopA and CopT.34 Although this same six-base loop is present in the predicted pETEC666 structure, the sequence has two single base deletions within the top of the stem II structure that cause a large bulge that is likely to destabilize loop II (Figure 5). We suggest that the differences in the sequences between the CopA-RNAs of pETEC666 and pETEC948 are responsible for their compatibility and coexistence within the same cell, as in H10407. CopA-RNA sequence inspection alone, however, cannot predict the mechanism by which the ETEC plasmid copy numbers are elevated relative to R1, because the ETEC plasmids also contain numerous sequence differences within their replication control regions.
Figure 5.
RNA structures of stem II and loop II for plasmids pETEC948 and pETEC666 predicted using mfold version 3.5.36 Base positions relative to the 5′ start of the complete RNA molecule are indicated, and the calculated free energies (kcal/mol) of the stem-loop structure alone are shown below the structure. The 5′ stem I and loop I sequences are not shown to simplify presentation.
The hybridization method of ETEC detection requires culturing stool samples on MacConkey agar to select for enteric pathogens and differentiates E. coli based on lactose fermentation. Although this method increases the likelihood of identifying E. coli isolates to assay, it does not differentiate commensal from pathogenic strains. Furthermore, the DNA hybridization assay targets both alleles of sta, but cannot differentiate between alleles. Most importantly, the DNA hybridization method is effective only when stools are heavily infected, and the assay is not quantitative. The qPCR assays described here provide a rapid and reliable means for detecting the ETEC eltA, sta1, and sta2 genes in clinical stool samples where sensitivity and quantification are important. These features may be especially useful for future epidemiological and investigational studies of ETEC-associated diarrhea.
ACKNOWLEDGMENTS
The American Committee on Clinical Tropical Medicine and Travelers' Health (ACCTMTH) assisted with publication expenses.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Financial support: BPY was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases by award no. F31DK094596.
Authors' addresses: Bonnie P. Youmans and Sarah K. Highlander, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX, E-mails: youmans@bcm.edu and sarahh@bcm.edu. Nadim J. Ajami and Joseph F. Petrosino, Alkek Center for Metagenomics and Microbiome Research, Baylor College of Medicine, Houston, TX, E-mails: Nadim.Ajami@bcm.edu and jpetrosi@bcm.edu. Zhi-Dong Jiang and Herbert L. DuPont, UT Health, School of Public Health, Houston, TX, E-mails: Zhi-Dong.Jiang@uth.tmc.edu and hdupont@StLukesHealth.org.
References
- 1.Hill DR, Beeching NJ. Travelers' diarrhea. Curr Opin Infect Dis. 2010;23:481–487. doi: 10.1097/QCO.0b013e32833dfca5. [DOI] [PubMed] [Google Scholar]
- 2.Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 2010;12:89–98. doi: 10.1016/j.micinf.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gyles C, So M, Falkow S. The enterotoxin plasmids of Escherichia coli. J Infect Dis. 1974;130:40–49. doi: 10.1093/infdis/130.1.40. [DOI] [PubMed] [Google Scholar]
- 4.Evans DJ, Jr, Evans DG. Three characteristics associated with enterotoxigenic Escherichia coli isolated from man. Infect Immun. 1973;8:322–328. doi: 10.1128/iai.8.3.322-328.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SG, Henderson IR. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol. 2010;192:5822–5831. doi: 10.1128/JB.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimata K, Shima T, Shimizu M, Tanaka D, Isobe J, Gyobu Y, Watahiki M, Nagai Y. Rapid categorization of pathogenic Escherichia coli by multiplex PCR. Microbiol Immunol. 2005;49:485–492. doi: 10.1111/j.1348-0421.2005.tb03752.x. [DOI] [PubMed] [Google Scholar]
- 7.Hegde A, Ballal M, Shenoy S. Detection of diarrheagenic Escherichia coli by multiplex PCR. Indian J Med Microbiol. 2012;30:279–284. doi: 10.4103/0255-0857.99485. [DOI] [PubMed] [Google Scholar]
- 8.Tobias J, Vutukuru SR. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli. Microbiol Res. 2012;167:564–570. doi: 10.1016/j.micres.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Fialho OB, de Souza EM, de Borba Dallagassa C, de Oliveira Pedrosa F, Klassen G, Irino K, Paludo KS, de Assis FE, Surek M, de Souza Santos Farah SM, Fadel-Picheth CM. Detection of diarrheagenic Escherichia coli using a two-system multiplex-PCR protocol. J Clin Lab Anal. 2013;27:155–161. doi: 10.1002/jcla.21578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujioka M, Otomo Y, Ahsan CR. A novel single-step multiplex polymerase chain reaction assay for the detection of diarrheagenic Escherichia coli. J Microbiol Methods. 2013;92:289–292. doi: 10.1016/j.mimet.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Gratz J, Amour C, Kibiki G, Becker S, Janaki L, Verweij JJ, Taniuchi M, Sobuz SU, Haque R, Haverstick DM, Houpt ER. A laboratory-developed TaqMan Array Card for simultaneous detection of 19 enteropathogens. J Clin Microbiol. 2013;51:472–480. doi: 10.1128/JCM.02658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reischl U, Youssef MT, Wolf H, Hyytia-Trees E, Strockbine NA. Real-time fluorescence PCR assays for detection and characterization of heat-labile I and heat-stable I enterotoxin genes from enterotoxigenic Escherichia coli. J Clin Microbiol. 2004;42:4092–4100. doi: 10.1128/JCM.42.9.4092-4100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bischoff C, Luthy J, Altwegg M, Baggi F. Rapid detection of diarrheagenic E. coli by real-time PCR. J Microbiol Methods. 2005;61:335–341. doi: 10.1016/j.mimet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Patel CB, Vajpayee P, Singh G, Upadhyay RS, Shanker R. Contamination of potable water by enterotoxigenic Escherichia coli: qPCR based culture-free detection and quantification. Ecotoxicol Environ Saf. 2011;74:2292–2298. doi: 10.1016/j.ecoenv.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Lothigius A, Janzon A, Begum Y, Sjoling A, Qadri F, Svennerholm AM, Bolin I. Enterotoxigenic Escherichia coli is detectable in water samples from an endemic area by real-time PCR. J Appl Microbiol. 2008;104:1128–1136. doi: 10.1111/j.1365-2672.2007.03628.x. [DOI] [PubMed] [Google Scholar]
- 16.Fukushima H, Tsunomori Y, Seki R. Duplex real-time SYBR green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J Clin Microbiol. 2003;41:5134–5146. doi: 10.1128/JCM.41.11.5134-5146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray BE, Mathewson JJ, DuPont HL, Hill WE. Utility of oligodeoxyribonucleotide probes for detecting enterotoxigenic Escherichia coli. J Infect Dis. 1987;155:809–811. doi: 10.1093/infdis/155.4.809. [DOI] [PubMed] [Google Scholar]
- 18.Hill WE, Payne WL, Zon G, Moseley SL. Synthetic oligodeoxyribonucleotide probes for detecting heat-stable enterotoxin-producing Escherichia coli by DNA colony hybridization. Appl Environ Microbiol. 1985;50:1187–1191. doi: 10.1128/aem.50.5.1187-1191.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, Patel S, Cutting M, Madden T, Hamilton H, Harris E, Gevers D, Simone G, McInnes P, Versalovic J. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27:1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang ZD, Lowe B, Verenkar MP, Ashley D, Steffen R, Tornieporth N, von Sonnenburg F, Waiyaki P, DuPont HL. Prevalence of enteric pathogens among international travelers with diarrhea acquired in Kenya (Mombasa), India (Goa), or Jamaica (Montego Bay) J Infect Dis. 2002;185:497–502. doi: 10.1086/338834. [DOI] [PubMed] [Google Scholar]
- 21.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO, Sailor CA, Dawson RB, Peek AS. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuwayama M, Shigemoto N, Oohara S, Tanizawa Y, Yamada H, Takeda Y, Matsuo T, Fukuda S. Simultaneous detection of virulence factors from a colony in diarrheagenic Escherichia coli by a multiplex PCR assay with Alexa Fluor-labeled primers. J Microbiol Methods. 2011;86:119–120. doi: 10.1016/j.mimet.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Duarte OG, Bai J, Newell E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;63:1–9. doi: 10.1016/j.diagmicrobio.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill WE, Wentz BA, Payne WL, Jagow JA, Zon G. DNA colony hybridization method using synthetic oligonucleotides to detect enterotoxigenic Escherichia coli: collaborative study. J Assoc Off Anal Chem. 1986;69:531–536. [PubMed] [Google Scholar]
- 28.Spicer EK, Kavanaugh WM, Dallas WS, Falkow S, Konigsberg WH, Schafer DE. Sequence homologies between A subunits of Escherichia coli and Vibrio cholerae enterotoxins. Proc Natl Acad Sci USA. 1981;78:50–54. doi: 10.1073/pnas.78.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pecoraro V, Zerulla K, Lange C, Soppa J. Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-)oligoploid and polyploid species. PLoS ONE. 2011;6:e16392. doi: 10.1371/journal.pone.0016392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DuPont HL, Formal SB, Hornick RB, Snyder MJ, Libonati JP, Sheahan DG, LaBrec EH, Kalas JP. Pathogenesis of Escherichia coli diarrhea. N Engl J Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- 31.Harro C, Chakraborty S, Feller A, DeNearing B, Cage A, Ram M, Lundgren A, Svennerholm AM, Bourgeois AL, Walker RI, Sack DA. Refinement of a human challenge model for evaluation of enterotoxigenic Escherichia coli vaccines. Clin Vaccine Immunol. 2011;18:1719–1727. doi: 10.1128/CVI.05194-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickering LK, DuPont HL, Evans DG, Evans DJ, Jr, Olarte J. Isolation of enteric pathogens from asymptomatic students from the United States and Latin America. J Infect Dis. 1977;135:1003–1005. doi: 10.1093/infdis/135.6.1003. [DOI] [PubMed] [Google Scholar]
- 33.So M, Heffron F, McCarthy BJ. The E. coli gene encoding heat stable toxin is a bacterial transposon flanked by inverted repeats of IS1. Nature. 1979;277:453–456. doi: 10.1038/277453a0. [DOI] [PubMed] [Google Scholar]
- 34.Nordstrom K. Plasmid R1–replication and its control. Plasmid. 2006;55:1–26. doi: 10.1016/j.plasmid.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Womble DD, Dong X, Luckow VA, Wu RP, Rownd RH. Analysis of the individual regulatory components of the IncFII plasmid replication control system. J Bacteriol. 1985;161:534–543. doi: 10.1128/jb.161.2.534-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]