Abstract
The Giardia and Cryptosporidium species are widespread and frequent diarrhea-related parasites affecting humans and other mammalian species. The prevalence of these parasites in Mongolia is currently unknown. Therefore, we performed molecular analyses of G. duodenalis and C. parvum in stool samples from 138 patients hospitalized with diarrhea in Mongolia using nested polymerase chain reaction (PCR). A total of 5 (3.62%) and 7 (5.07%) fecal samples were positive for G. duodenalis and C. parvum, respectively. Giardia duodenalis and C. parvum infections were prevalent in children < 9 years of age. The assemblage-specific fragment patterns for the β-giardin gene of G. duodenalis revealed that all five samples testing positive belonged to Assemblage A by the PCR-restriction fragment polymorphism method. For sequencing and phylogenetic analysis of the 18S rDNA and HSP70 genes of all seven patients testing positive the genes were further identified to be of the C. parvum bovine genotype. This study is the first to report the prevalence of G. duodenalis and C. parvum and its molecular characterization of fecal samples from individuals with diarrhea in Mongolia.
Introduction
Giardia and Cryptosporidium, genera of common protozoan parasites that infect domestic and wild animals and humans, generally cause diarrhea.1–3 The Giardia genus is composed of intestinal flagellates that infect a wide range of vertebrate hosts. The Giardia genus currently comprises six species that are distinguished on the basis of the morphology and ultrastructure of their trophozoites.4,5 Giardia duodenalis, Giardia intestinalis, and Giardia lamblia should be considered as a species complex, with little variation in morphology among them. Recently, genetic analyses using polymerase chain reaction (PCR) characterized isolates of Giardia directly from feces, allowing the identification of a comprehensive range of genotypes from humans and animals.6–8 The species G. duodenalis has assigned even assemblages from A to H. Assemblage A and B have been identified to infect humans and other mammalian hosts.9,10 Although “Assemblage C” infects only dogs, Assemblage F infects only cats, and Assemblage D infects both dogs and cats.11 Assemblage E infects cattle, sheep, and goats, and Assemblage G infects rats. Recently, Assemblage H infecting marine vertebrates has been reported.12 Regarding the Cryptosporidium species, 22 valid species have been identified on the basis of differences in oocyst morphology, the site of infection, vertebrate class specificity, and genetic differences.1 Among the Cryptosporidium species, Cryptosporidium parvum and Cryptosporidium hominis are known to infect cattle, humans, and other mammals.
The Giardia and Cryptosporidium are shed in feces as oocysts and cysts and can be directly transmitted by the fecal–oral route by contaminated water or food, especially raw vegetables.13 Clinical giardiasis and cryptosporidiosis accompanied by diarrhea are major public health concerns in developing nations.14,15 Approximately 200 million people currently have symptomatic giardiasis in Asia, Africa, and Latin America, and ∼500,000 new cases are reported each year16; alternatively, 300,000 persons in the United States are expected to be infected with Cryptosporidium species annually.17 In addition, the occurrence of Giardia and Cryptosporidium species has been reported in Russia and China.18,19
In Mongolia, which is located in central Asia and borders Russia to the north and China to the south; many people work in the livestock industry, such as pasturage of cattle, sheep, goats, and horses in steppes, and the agriculture industry. Therefore, individuals in Mongolia may be considered to have a naturally high risk of contact with zoonotic parasites. However, no studies to date have examined specific G. duodenalis and C. parvum infections among individuals who have diarrhea in Mongolia. The aim of this study was to perform molecular detection and phylogenetic characterization of G. duodenalis and C. parvum from diarrheal fecal samples of individuals in Mongolia.
Materials and Methods
Fecal sample collection and DNA isolation.
A total of 138 stool samples from 138 patients admitted to the intestinal ward of the National Center for Communicable Diseases located in Mongolia who had diarrhea were collected and transported to the Laboratory of Parasitology for diagnosis of parasitic diseases. Each fresh stool sample (5 g) was suspended in 15 mL of phosphate buffered saline and filtered using four layers of gauze to remove coarse material. The filtrate was then centrifuged at 3,000 rpm for 10 min. The supernatant was eliminated, and the sediment was mixed with 5 mL of phosphate buffered saline. The pellet underwent repeated boiling (100°C) and deep freezing (−70°C) 10 times to break the thick wall of the Cryptosporidium and Giardia cyst. Total genomic DNA was isolated from the pellet using DNAzol (MRC, Cincinnati, OH) and stored at −20°C until use.
PCR and characterization of G. duodenalis by PCR-restriction fragment length polymorphism (RFLP) assay.
The amplification of the β-giardin gene was performed using a nested PCR protocol. In the primary PCR reaction, a 753 basepair (bp) fragment was amplified using Accure PCR Master Mix (Bioneer, Daejeon, Korea) containing 1 μM of the forward primer Gia7 (5′-AAGCCCGACGACCTCACCCGCAGTGC-3′) and the reverse primer Gia759 (5′-GAGGCCGCCCTGGATCTTCGAGACGAC-3′), as previously described.20 In the nested PCR reaction, a 511 bp fragment was amplified using the forward primer (5′-GAACGAACGAGATCGAGGTCCG-3′) and the reverse primer (5′-CTCGACGAGCTT CGTGTT-3′).Thermal cycle reactions were set to an initial denaturing step (95°C for 5 min), 35 cycles of a denaturing step (95°C for 30 s), an annealing step (55°C for 30 s), an extension step (72°C for 60 s), and finally an extension step (72°C for 7 min). Amplification products were electrophoresed by an auto electrophoresis machine (QIAxcel, Hilden, Germany), as previously described.21 The PCR products were purified using the agarose gel extraction kit (Qiagen, Hilden, Germany) and digested using 10 U/μL of Hae III (Enzynomics, Daejeon, Korea) in a final volume of 20 μL for 4 h at 37°C for assemblage analysis, according to previous reports.22
Amplification of the 18S rDNA and heat-shock protein (HSP70) genes for C. parvum.
The primers used to amplify a 695 bp fragment from the 18S rDNA gene were 18SSF, forward primer (5′-AGTCATAGTCTTGTCTCAAAGATT-3′) and 18SR3B, reverse primer (5′-TTAACAAATCTAAGAATTTCACC-3′).23 Thermal cycle reactions were set to an initial denaturing step (96°C for 2 min), 35 cycles of a denaturing step (94°C for 30 s), an annealing step (55°C for 30 s), an extension step (72°C for 45 s), and finally an extension step (72°C for 10 min). A nested PCR protocol was used to amplify the HSP70 gene from genomic DNA of selected Cryptosporidium isolates for nucleotide sequencing.24 For the primary PCR reaction, a 448 bp fragment was amplified using the forward primer HSPF4 (5′-GGTGGTGGTACTTTTGATGTATC-3′) and reverse primer HSPR4 (5′-GCCTGAACCTTTGGAATACG-3′). Thermal cycle reactions were set to an initial denaturing step (94°C for 5 min), 40 cycles of a denaturing step (94°C for 30 s), an annealing step (56°C for 30 s), an extension step (72°C for 30 s), and finally an extension step (72°C for 10 min). For the secondary PCR, a 325 bp fragment was amplified using the primary PCR product and HSPF3 (5′-GCTGSTGATACTCACT TGGGTGG-3′) and HSPR3 (5′ CTCTTGTCCATACCAGCATCC-3′) primers. The condition for the secondary PCR was identical to the primary PCR. Secondary PCR products were sequenced directly in both directions.
Phylogenetic analysis of the 18S rDNA and HSP70 genes of C. parvum.
The PCR products were analyzed by electrophoresis, purified using an agarose gel DNA purification kit (Qiagen), and sequenced with an ABI PRISM 3730xl Analyzer (Applied Biosystems, Foster City, CA). A search of highly similar 18S rDNA gene fragment sequences was performed using nucleotide BLAST (National Center for Biotechnology Information, Bethesda, MD) to confirm the genotype. Cryptosporidium 18S rDNA sequences were obtained from GenBank. Sequence alignment was performed using CLUSTAL W (Multiple sequence alignment computer program, Histon, Cambridgeshire, UK). Phylogenetic trees were constructed using the neighbor-joining method25 with maximum composite likelihood distance correction in the Molecular Evolutionary Genetics Analysis (MEGA) program,26 with robustness of groupings assessed using 1,000 bootstrap replicates of the data.27
Results
Prevalence of G. duodenalis and C. parvum in human fecal samples in Mongolia.
The 138 patients comprised 85 children 1–15 years of age (mean age, 3.6 years) and 53 adults 16–74 years of age (mean age, 32.5 years). Of the 138 patients included, 5 (3.62%) and 7 (5.07%) tested positive for G. duodenalis and C. parvum, respectively. Four of the 5 patients with a G. duodenalis infection were < 4 years of age, and 3 of the 7 patients with a C. parvum infection were < 4 years of age and 3 patients were 5–9 years of age except 1 patient. Our results showed that the positive rate of G. duodenalis and C. parvum in children was higher than that in adults (Table 1).
Table 1.
Detection of Giardia duodenalis and Cryptosporidium parvum infections using polymerase chain reaction analysis in human fecal samples from Mongolia, by age and sex
| Variables | No. of sample | Positive no. of G. duodenalis (%) | Positive no. of C. parvum (%) |
|---|---|---|---|
| Age (year) | |||
| < 4 | 65 | 4 (6.15) | 3 (4.62) |
| 5–9 | 17 | – | 3 (17.65) |
| 10–14 | 3 | – | |
| 15–19 | 7 | – | – |
| 20–29 | 19 | 1 (5.26) | – |
| 30–39 | 15 | – | 1 (6.67) |
| 40–49 | 4 | – | – |
| 50–59 | 7 | – | – |
| 60+ | 1 | – | – |
| Sex | |||
| Female | 64 | 1 (1.56) | 2 (3.13) |
| Male | 74 | 4 (5.41) | 5 (6.76) |
| Total | 138 | 5 (3.62) | 7 (5.07) |
Identification of G. duodenalis assemblages by PCR-RFLP.
The five samples with G. duodenalis were confirmed by β-giardin gene amplification by nested PCR (Figure 1A). After digestion by Hae III, the assemblage-specific patterns were obtained, showing patterns of 201, 150, 110, and 50 bp (Figure 1B). All five samples with G. duodenalis belonged to Assemblage A.
Figure 1.
(A) Electrophoretic identification of Giardia β-giardin gene products (511 bp) by second polymerase chain reaction (PCR) using an auto electrophoresis machine. (B) Electrophoretic separation of Giardia β-giardin gene products after restriction with the Hae III enzyme using PCR-restriction fragment length polymorphism. Lane M, 2.5 kb molecular marker; lane P, positive for Giardia duodenalis; lanes 1–5, human fecal samples; lane N, negative.
Identification and phylogenetic analysis of C. parvum.
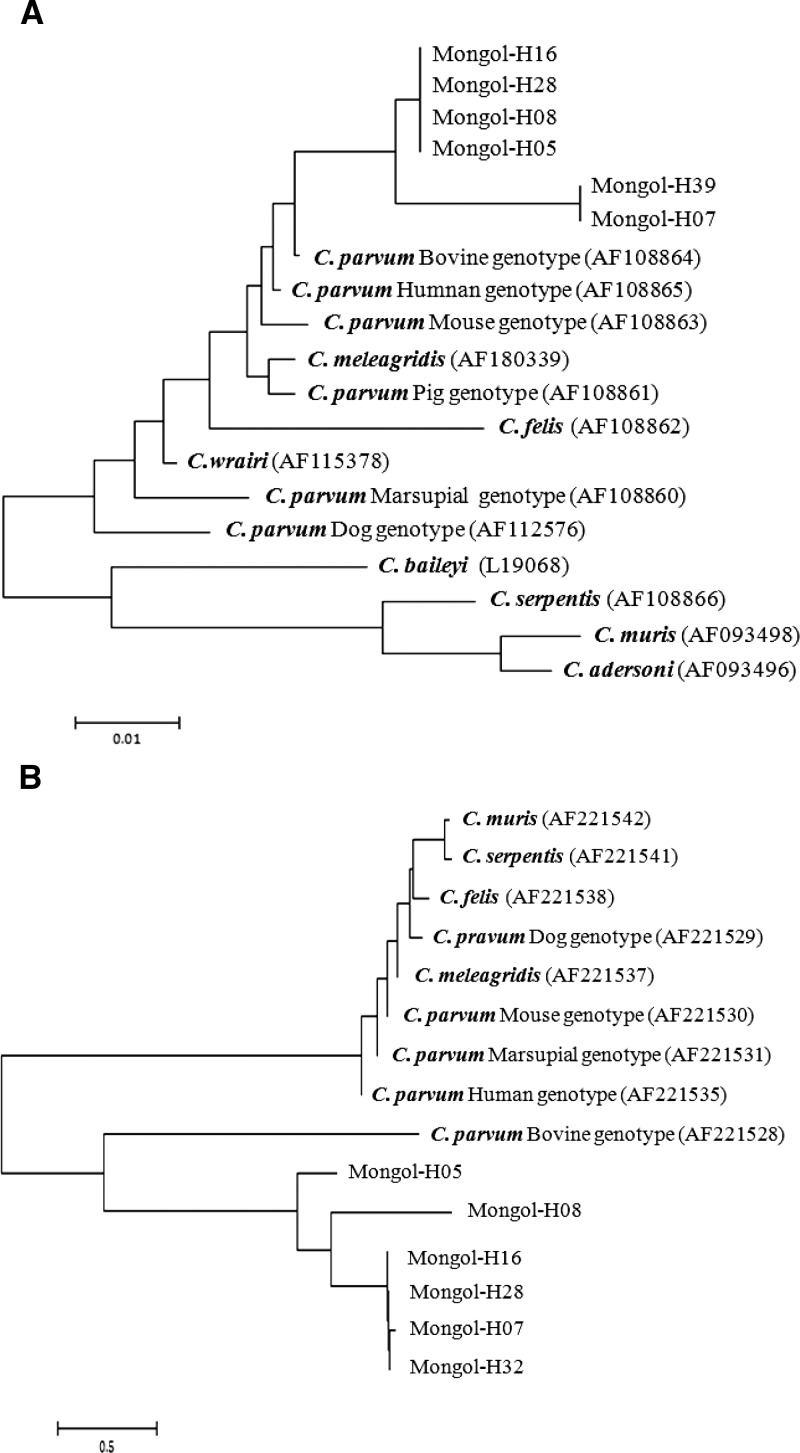
A total of seven fecal samples (sample numbers Mongol-H05, H07, H08, H16, H28, H32, and H39) tested positive for the 18S rDNA and HSP70 genes of C. parvum in the nested PCR. A sequence analysis of these seven samples suggested the presence of C. parvum in all patients, with homologies from 97% to 99% (Table 2). Phylogenetic analysis showed that the 18S rDNA gene fragments were of the C. parvum bovine genotype in all patients except Mongol-H32 (Figure 2A). An analysis of the HSP70 gene showed similar results (all patients except Mongol-H39) (Figure 2B).
Table 2.
Genotyping of the 18S rDNA and HSP70 genes for each human fecal sample of patients from Mongolia testing positive for Cryptosporidium parvum using nested polymerase chain reaction
| Specimen ID | Genotype (18S rDNA) | Genotype (HSP70) |
|---|---|---|
| Mongol-H05 | C. parvum | C. parvum |
| Mongol-H07 | C. parvum | C. parvum |
| Mongol-H08 | C. parvum | C. parvum |
| Mongol-H16 | C. parvum | C. parvum |
| Mongol-H28 | C. parvum | C. parvum |
| Mongol-H32 | ND | C. parvum |
| Mongol-H39 | C. parvum | ND |
ND = no detection.
Figure 2.
The phylogenetic relationships among Cryptosporidium species and genotypes according to the neighbor-joining analysis and the maximum composite likelihood distance correction (implemented using Molecular Evolutionary Genetics Analysis [MEGA]) of (A) a fragment from the partial 18S rDNA sequence and (B) HSP70 sequence. Sequences of other Cryptosporidium species and genotypes were obtained from GenBank.
Discussion
Giardia and Cryptosporidium are significant worldwide causes of diarrhea and nutritional disorders in humans. In Asia, among patients with diarrhea in a study from the Philippines, the prevalence rates for Giardia and Cryptosporidium species were 2.0% and 1.9%, respectively28; furthermore, the prevalence rates from a study in Malaysia were 0.7% for Giardia species and 0.3% for Cryptosporidium species.29 In our study, the percentage of patients with diarrhea infected with G. duodenalis and C. parvum in Mongolia was higher than the above rates from the Philippines and Malaysia. Most outbreaks of human giardiasis in developing countries have mainly been detected in children < 2 years of age.30 Furthermore, it has been reported that cryptosporidiosis generally affects children < 4 years of age.31 In our data, children were more frequently infected than adults, and it is a finding similar to the findings from studies in other countries. In previous reports, the reasons for high prevalence of giardiasis and cryptosporidiosis in young children may be caused by the lack of immunity, and because they are easily exposed to contaminated water through playing water games.31 In addition, Faubert reported that numerous factors contributed to infection with Giardia species, including the number of cysts ingested, the age of the host, the virulence of the Giardia strain, and the situation of the immune system at the time of infection.32 Interestingly, in the current study, there was only one case of an adult infected with G. duodenalis, whereas the rest were all children. The reason for the high prevalence in children was unclear because we did not acquire any information on the patients except that they had diarrhea. Almost all of the infected children were living in Gers or houses equipped with indoor latrines and without tap water located in a steppe. The poor hygiene conditions of a steppe, such as the low quality of water, poor cleanliness of containers for transporting water, and poor hand washing facilities, should be considered as contributing factors to infection with various pathogens, and these may be critical causes of infections. Further surveys for the detection of pathogens and the transmission through contamination of waters in poor environmental conditions should be performed. Additionally, in 3 of the 5 cases of C. parvum, Shigella flexneri (N = 2) and Salmonella enteritidis (N = 1), and in 2 of the 7 cases of G. duodenalis, S. flexneri were also detected (data not shown). These findings indicate that we detected the existence of mixed infections, both bacterial and parasitic, in patients with diarrhea in Mongolia. To understand the epidemiologic characteristics of the infections and to implement control measures, it is important to determine whether G. duodenalis and C. parvum can infect humans through a zoonotic route. Therefore, further epidemiologic studies examining the risk factors of infection among these protozoa in individuals with diarrhea should be carried out in the near future for improving public health.
Recently, molecular epidemiologic studies with Giardia DNA directly extracted from feces have been performed to amplify techniques, and several PCR assays have been developed.20,33 In this study, we successfully performed a molecular analysis of the β-giardin gene and pattern analysis using a PCR-RFLP assay with Hae III on Giardia DNA from fecal samples. An investigation of human isolates from stool samples in diverse geographic areas established that only G. duodenalis Assemblages A and B are related to almost all human infections.34 For example, the occurrence of Assemblage A and B of G. duodenalis have been reported in Thailand, China, and the Philippines.19,35,36 In this study population, only Assemblage A was identified, and this result is similar to the previous studies from Korea, Japan, Egypt, and Brazil.37–40
Cryptosporidium species are classified on the basis of different oocyst morphology, sites of infection, vertebrate class, and genetic differences; such classifications of Cryptosporidium species include C. parvum, (a parasite of humans, cattle, and other mammals), C. hominis (a parasite of humans), and Cryptosporidium felis (a parasite of cats).15,41 Particularly, Morgan and others42 reported that the C. parvum bovine genotype and C. hominis are responsible for the majority of human infections. Our results showed that the Cryptosporidium DNA isolated from diarrheal fecal samples belonged to the bovine genotype using phylogenetic analysis. Our results are meaningful because they reveal that zoonotic parasitic infection cycles from cattle to humans may be possible in Mongolia.
In this study, G. duodenalis and C. parvum genes from human diarrheal fecal samples in patients from Mongolia were identified by molecular analysis. In particular, G. duodenalis was classified as a zoonotic pathogen belonging to Assemblage A, and the C. parvum bovine genotype was discovered through phylogenetic analysis. From our results, we assume that C. parvum can possibly emerge important human pathogens with contact between humans and animals in Mongolia. Further epidemiological studies subject to humans and animals in different areas and/or a big population in Mongolia are needed to better characterize the transmission of Giardiasis and Cryptosporidiosis in humans. To our knowledge, this is the first study to report the prevalence and genetic identification of G. duodenalis and C. parvum, and it may contribute to the understanding of the epidemiologic characteristics and improve the preventive control of both parasites in Mongolia.
Footnotes
Financial support: This work is performed to collaborative research both Korea CDC and Mongolia NCCD and this is supported by funding (4847-302-210-13, 2012) from the Korea National Institute of Health, Korea Centers for Disease Control and Prevention.
Authors' addresses: Sung-Hee Hong, Young-Il Jeong, Shin-Hyeong Cho, Won-Ja Lee, and Sang-Eun Lee, Division of Malaria and Parasite Diseases, Korea National Institute of Health, Korea Centers for Disease Control and Prevention, Osong-up, Cheongwon-gun, Chungbuk 363-951, Korea, E-mails: h22h22h22@naver.com, tell74@naver.com, jo4u@cdc.go.kr, wonja@gmail.com, and ondalgl@korea.kr. Davaasuren Anu and Davaajav Abmed, Laboratory of Parasitology, National Center for Communicable Diseases, Ministry of Health, Ulaanbaator-210648, Mongolia, E-mails: davaasuren_a@ymail.com and abmed99@yahoo.com.
References
- 1.Xiao L, Fayer R. Molecular characterization of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol. 2008;38:1239–1255. doi: 10.1016/j.ijpara.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2000;30:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Thompson RC. Giardiasis as a re-emerging infectious disease and its zoonotic potential. Int J Parasitol. 2000;30:1259–1267. doi: 10.1016/s0020-7519(00)00127-2. [DOI] [PubMed] [Google Scholar]
- 4.Kulda J, Nohynkova E. In: Giardia in humans and other animals. Parasitic Protozoa. Kreier JP, editor. San Diego, CA: Academic Press; 1995. pp. 225–422. [Google Scholar]
- 5.Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amar CF, Dear PH, Pedraza-Díaz S, Looker N, Linnane E, McLauchlin J. Sensitive PCR-restriction fragment length polymorphism assay for detection and genotyping of Giardia duodenalis in human feces. J Clin Microbiol. 2002;40:446–452. doi: 10.1128/JCM.40.2.446-452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Read C, Walters J, Robertson ID, Thompson RC. Correlation between genotype of Giardia duodenalis and diarrhoea. Int J Parasitol. 2002;32:229–231. doi: 10.1016/s0020-7519(01)00340-x. [DOI] [PubMed] [Google Scholar]
- 8.Sprong H, Cacciò SM, van der Giessen JW. ZOOPNET network and partners: identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl Trop Dis. 2009;3:e558. doi: 10.1371/journal.pntd.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monis PT, Andrews RH, Mayrhofer G, Ey PL. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect Genet Evol. 2003;3:29–38. doi: 10.1016/s1567-1348(02)00149-1. [DOI] [PubMed] [Google Scholar]
- 10.Cacciò SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2010;40:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 11.McGlade TR, Robertson ID, Elliot AD, Thompson RC. High prevalence of Giardia detected in cats by PCR. Vet Parasitol. 2003;110:197–205. doi: 10.1016/s0304-4017(02)00322-9. [DOI] [PubMed] [Google Scholar]
- 12.Lasek-Nesselquist E, Welch DM, Sogin ML. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. Int J Parasitol. 2010;40:1063–1074. doi: 10.1016/j.ijpara.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karanis P, Kourenti C, Smith H. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J Water Health. 2007;5:1–38. doi: 10.2166/wh.2006.002. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, Xio L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fayer R, Morgan U, Upton SJ. Epidemiology of Cryptosporidium: transmission, detection and identification. Int J Parasitol. 2000;30:1305–1322. doi: 10.1016/s0020-7519(00)00135-1. [DOI] [PubMed] [Google Scholar]
- 16.WHO The World Health Report. 1996. http://www.who.int / whr/1996/en/index.html Available at. Accessed August 3, 2012.
- 17.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karanis P, Sotiriadou I, Kartashev V, Kourenti C, Tsvetkova N, Stojanova K. Occurrence of Giardia and Cryptosporidium in water supplies of Russia and Bulgaria. Environ Res. 2006;102:260–271. doi: 10.1016/j.envres.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Wang R, Zhang X, Zhu H, Zhang L, Feng Y, Jian F, Ning C, Qi M, Zhou Y, Fu K, Wang Y, Sun Y, Wang Q, Xiao L. Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp Parasitol. 2011;127:42–45. doi: 10.1016/j.exppara.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 20.Cacciò SM, De Giacomo M, Pozio E. Sequence analysis of the β-giardin gene and development of a polymerase chain reaction–restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human fecal samples. Int J Parasitol. 2002;32:1023–1030. doi: 10.1016/s0020-7519(02)00068-1. [DOI] [PubMed] [Google Scholar]
- 21.Talameh J, Misher A, Hoskins J. A capillary electrophoresis method for genotyping the 9-bp exon 1 insertion/deletion in BDKRB2. Pharmacogenomics. 2012;13:353–358. doi: 10.2217/pgs.11.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lalle M, Pozio E, Capelli G, Bruschi F, Crotti D, Cacciò SM. Genetic heterogeneity at the β-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int J Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Morgan UM, Monis PT, Xio L, Sulaiman I, Raidal S, O'Donoghue P, Gasser R, Murray A, Fayer R, Blagburn BL, Lal AA, Thompson RC. Molecular and phylogenetic characterization of Cryptosporidium from birds. Int J Parasitol. 2001;31:289–296. doi: 10.1016/s0020-7519(00)00164-8. [DOI] [PubMed] [Google Scholar]
- 24.Khramtsov NV, Tillet M, Blunt DS, Montelone BA, Upton SJ. Cloning and analysis of a Cryptosporidium parvum gene encoding a protein with homology to cytoplasmic form Hsp70. J Eukaryot Microbiol. 1995;42:416–422. doi: 10.1111/j.1550-7408.1995.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 28.Natividad FF, Buerano CC, Lago CB, Mapua CA, de Guzman BB, Seraspe EB, Samentar LP, Endo T. Prevalence rates of Giardia and Cryptosporidium among diarrheic patients in the Philippines. Southeast Asian J Trop Med Public Health. 2008;39:991–999. [PubMed] [Google Scholar]
- 29.Norhayati M, Fatmah MS, Yusof S, Edariah AB. Intestinal parasitic infections in man: a review. Med J Malaysia. 2003;58:296–305. quiz 306. [PubMed] [Google Scholar]
- 30.Jaros D, Zygner W, Jaros S, Wedrychowicz H. Detection of Giardia intestinalis assemblages A, B and D in domestic cats from Warsaw, Poland. Pol J Microbiol. 2011;60:259–263. [PubMed] [Google Scholar]
- 31.ANOFEL Cryptosporidium National Network Laboratory-based surveillance for Cryptosporidium in France, 2006–2009. Euro Surveill. 2010;15:19642. [PubMed] [Google Scholar]
- 32.Faubert G. Immune Response to Giardia duodenalis. Clin Microbiol Rev. 2000;13:35–54. doi: 10.1128/cmr.13.1.35-54.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss JB, van Keulen H, Nash TE. Classification of subgroups of Giardia lamblia based upon ribosomal RNA gene sequence using the polymerase chain reaction. Mol Biochem Parasitol. 1992;54:73–86. doi: 10.1016/0166-6851(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 34.Cacciò SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Siripattanapipong S, Leelayoova S, Mungthin M, Thompson RC, Boontanom P, Saksirisamphant W, Tan-Ariya P. Determination of discriminatory power of genetic markers used for genotyping Giardia duodenalis. Southeast Asian J Trop Med Public Health. 2011;42:764–771. [PubMed] [Google Scholar]
- 36.Yason JA, Rivera WL. Genotyping of Giardia duodenalis isolates among residents of slum area in Manila, Philippines. Parasitol Res. 2007;101:681–687. doi: 10.1007/s00436-007-0533-8. [DOI] [PubMed] [Google Scholar]
- 37.Yong TS, Park SJ, Hwang UW, Yang HW, Lee KW, Min DY, Rim HJ, Wang Y, Zheng F. Genotyping of Giardia lamblia isolates from humans in China and Korea using ribosomal DNA Sequences. J Parasitol. 2000;86:887–891. doi: 10.1645/0022-3395(2000)086[0887:GOGLIF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 38.Matsubayashi M, Kimate I, Abe N. Identification of genotypes of Giardia intestinalis isolates from a human and calf in Japan. J Vet Med Sci. 2005;67:337–340. doi: 10.1292/jvms.67.337. [DOI] [PubMed] [Google Scholar]
- 39.Abdel-Moneim SM, Sultan DM. Genetic characterization of Giardia lamblia isolates from Egyptian patients with relation to clinical giardiasis. J Egypt Soc Parasitol. 2008;38:547–560. [PubMed] [Google Scholar]
- 40.Volotão AC, Costa-Macedo LM, Haddad FS, Brandão A, Peralta JM, Fernandes O. Genotyping of Giardia duodenalis from human and animal samples from Brazil using β-giardin gene: a phylogenetic analysis. Acta Trop. 2007;102:10–19. doi: 10.1016/j.actatropica.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Pellitero P, Sitjà-Bobadilla A. Cryptosporidium molnari n. sp. (Apicomplexa: Cryptosporidiidae) infecting two marine fish species, Sparus aurata L. and Dicentrarchus labrax L. Int J Parasitol. 2002;32:1007–1021. doi: 10.1016/s0020-7519(02)00058-9. [DOI] [PubMed] [Google Scholar]
- 42.Morgan-Ryan UM, Fall A, Ward LA, Hijjawi N, Sulaiman I, Fayer R, Thompson RC, Olson M, Lal A, Xiao L. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J Eukaryot Microbiol. 2002;49:433–440. doi: 10.1111/j.1550-7408.2002.tb00224.x. [DOI] [PubMed] [Google Scholar]