Abstract
We determined the prevalence of rifampin-monoresistant tuberculosis (RMR-TB) in Iran. Because development of RMR-TB is not common, we also identified the major risk factors associated with RMR-TB reported from different provinces of Iran. Data for 3,020 TB patients who remained or became smear positive after two, four, six, and nine months of standard first-line chemotherapy were retrospectively analyzed. Of 3,020 patients, 1,242 patients (41.1%) were culture and DNA positive for Mycobacterium tuberculosis. Of these patients, 73 (7.4%) patients had monoresistant isolates to rifampin, which was significantly higher than that for multidrug-resistant TB (5.8%). The average rate of RMR-TB in the studied population ranged from 5% to 10%. Classical investigation showed that 33.6% of patients had either a previous or family history of TB. Molecular epidemiology methods (i.e., spoligotyping and Mycobacterium intespersed repetitive unit–variable number tandem repeat), defined transmission link in three clusters (13%). These results outline the urgent need for a comprehensive plan for detection and treatment of RMR-TB cases.
Introduction
Based on recommendation from National Tuberculosis Control Program (NTP) in Iran, all new sputum smear-positive tuberculosis (TB) cases are treated with four first-line drugs: isoniazid (INH), rifampin (RMP), pyrazinamide, and ethambutol.1 Among these drugs, RMP is the most effective agent and the its absence from treatment regimens has considerable implications for the individual patient and the for NTP.2,3 Generally, patients infected rifampin-monoresistant (RMR) bacilli need a longer treatment duration (up to 18 months) and more expensive drugs. It is important to include INH in this regimen, and the risk of misclassification of RMR as multidrug-resistant (MDR) TB is common. In addition, these patients continued to expectorate smear-positive sputum longer than patients with drug-sensitive bacilli, increasing the probability of spread of infection.4,5 Therefore, resistance to RMP represents a major risk factor for treatment failure with recommended standard TB therapy.
With advances in molecular biology, the basis of resistance to RMP is well characterized.6,7 Resistance to RMP is caused by point mutations in the beta subunit of the DNA-dependent RNA polymerase rpo gene. These mutations can be detected by using single polymerase chain reaction (PCR), multiplex PCR, or DNA sequencing.8,9 In Iran, prevalence data for drug-resistant strains for entire country are not available.10 During 1998–2000, Masjedi and others reported information about drug-resistant TB, which was comparable with international data.11 Subsequent investigations reported emergence of highly drug-resistant strains of Mycobacterium tuberculosis in Iran.11,12 Likewise, a few other studies documented drug-resistant TB in different provinces of Iran.13,14 However, the true extent of drug-resistant TB is not clear. Considering the impact of drug-resistant TB, we retrospectively analyzed susceptibility patterns of strains that were collected in previous years.15 We reviewed laboratory and clinical information for smear-positive TB cases.
The study included 85% of all diagnostic health centers that used the directly observed therapy, short course (DOTS) strategy in Iran. Results showed high levels of MDR and RMR strains. In Iran, the DOTS strategy was initiated in 1995, and treatment protocols are uniformly implemented by NTP for registered pulmonary TB patients. Because RMP is never prescribed as a single drug for treatment of active TB, analyzed the clinical and/or epidemiologic features that promote RMR-TB. To our knowledge, this is the first report that describes and highlights the high prevalence of RMR in Iran.
Materials and Methods
Data and specimen collection.
This study was conducted during February 2010–February 2011. The study included TB patients who remained or became sputum smear positive at the after two, four, six, and nine months of standard therapy. These patients were referred to DOTS health units throughout Iran. A total of 5,362 pulmonary specimens from 3,020 patients were collected. Samples with questionnaires were transported to National Reference TB Laboratory of Iran for culture, direct susceptibility testing, and genotyping. The required questionnaires were completed and signed by DOTS medical officers. The questionnaire included information on sex, date and country of birth, close contact (> 16 hours contact), family or previous history, previous TB therapy, human immunodeficiency virus (HIV) infection, previous laboratory results (if any), and the category (I and/or II) treatment, accordingly. The Scientific Board of the National Research Institute of Tuberculosis and Lung Disease of Iran approved the study.
Genomic DNA preparation.
Mycobacterial genomic DNA was extracted directly from sputum smear-positive specimens. In brief, the digested and decontaminated suspension was inactivated by heating at 95°C for 30 minutes in a water bath. The DNA was extracted from heat-inactivated sputum suspension by using the QIAGEN DNA Extraction Kit (QIAGEN, Hilden, Germany). DNA was further quantified by using a Microliter Spectrophotometer (Picodrop Ltd., Cambridge United Kingdom).
Genotyping of isolates.
Isolates were characterized by using spoligotyping and mycobacterial interspersed repetitive unit–variable number tandem repeat (MIRU-VNTR) genotyping for 15 loci. Spoligotyping was performed as described.16 The DR region was amplified by PCR using primers DRa (5′-biotin-CCG AGA GGG GAC GGA AAC-3′) and DRb (5′- GGT TTT GGG TCT GAC GAC-3′), 20–50 ng of DNA, and 0.5 units of Taq DNA polymerase. The PCR conditions were 35 cycles for 1 minute at 95°C, 1 minute at 55°C, and 30 seconds at 72°C. The initial denaturation and final extension steps were each 10 minutes.
Amplified DNA was hybridized to 43 immobilized oligonucleotides derived from the spacer sequences of M. tuberculosis H37Rv and M. bovis BCG P3 by reverse line blotting. Hybridized DNA was detected by enhanced chemiluminesence (ECL; Amersham, Little Chalfont, United Kingdom) and by exposing ECL Hyper Film (Amersham) to the membrane for 10 minutes. Results were entered in binary formats into Excel (Microsoft, Redmond, WA) spreadsheet and compared with the SpolDB4 database.17 The MIRU-VNTR genotyping was performed by PCR amplification of a panel of 15 MTB MIRU loci using primers described in the MIRU-VNTR standard protocol.16 The PCR products were subjected to electrophoresis on 1.5% agarose gels, and the number of MIRU-VNTR copies for each isolate was determined. The MIRU-VNTR analysis was performed by using MIRU-VNTR plus database.18 Discrimination power was evaluated by using the Hunter-Gaston Index with the equation
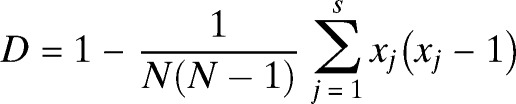 |
where N is the total number of isolates in the sample and S is the number of different MIRU-VNTR types described.19
PCR-based method to detect mutations.
RMP resistance-related mutations in the rpoB gene (codons 531, 526, 516, 513, 518, 511).
Multiplex PCR was performed to detect mutations in rpoB gene for RMP resistance.20 The PCR was conducted in 25-μL mixtures containing 10 pmol of specific primers (Table 1), 5.5 mM MgCl2, 0.2 mM dNTP, 1× PCR buffer, 1.25 units of Hot Start Taq DNA polymerase, and 50 ng of DNA. The PCR conditions were an initial denaturation step at 95°C for 5 minutes; 35 cycles of denaturation at 95°C for 30 seconds, annealing at 70°C for 30 seconds, and extension at 72°C for 30 seconds; and a final extension at 72°C for 7 minutes. The PCR products were separated by electrophoresis on 8% polyacrylamide gels. Fragments were visualized by staining the gels with ethidium bromide staining and viewing under ultraviolet light.
Table 1.
rpoB, katG, and inhA primer sequences*
| Multiplex allele-specific PCR (MAS-PCR) | Primer name | Primer oligo sequence, 5′ → 3′ |
|---|---|---|
| Multiplex-PCR (rpoB 516, 526, 531) | rpoB 516 (forward primer) | CAGCTGAGCCAATTCATGGA |
| rpoB 526 (forward primer) | CTGTCGGGGTTGACCCA | |
| rpoB 531 (forward primer) | CACAAGCGCCGACTGTC | |
| Multiplex-PCR (rpoB 507, 518, 533) | rpoB 507 (forward primer) | GCGATCAAGGAGTTCTTCGG |
| rpoB 518 (forward primer) | TGAGCCAATTCATGGACCAGA | |
| rpoB 533 (forward primer) | CGCCGACTGTCGGCGCT | |
| Multiplex-PCR (rpoB 511, 513, 522) | rpoB 511 (forward primer) | TTCGGCACCAGCCAGCT |
| rpoB 513 (forward primer) | CACCAGCCAGCTGAGCC | |
| rpoB 522 (forward primer) | GACCAGAACAACCCGCTGT | |
| RIRm (reverse primer) | TTGACCCGCGCGTACAC | |
| PCR (katG 315) | katG (forward primer) | GCAGATGGGGCTGATCTACG |
| PCR (katG 315) | katG5 (reverse primer) | ATACGACCTCGATGCCGC |
| PCR (inhA) | inhA-P15 (forward primer) | GCGCGGTCAGTTCCACA |
| PCR (inhA) | inhA-PF2 (reverse primer) | CACCCCGACAACCTATCG |
PCR = polymerase chain reaction.
INH resistance-related mutations in katG 315 and the inhA promoter.
Two codons of katG 315, and inhAP-15 were used to detect INH-resistant strains according to the protocol of Kazumi and Mitara with minor modifications.21 The PCRs were conducted in 25-μL mixtures containing 10 pmol of specific primers (Table 1), 5.5 mM MgCl2, 0.2 mM dNTP, 1× PCR buffer, and 1.25 units of Hot Start Taq DNA polymerase, 2% dimethylsulfoxide, and 50 ng of DNA. The PCR conditions were an initial denaturation step at 95°C for 5 minutes; 40 cycles of denaturation at 95°C for 30 seconds, annealing at 62°C (inhA) and 66°C (katG) for 30 seconds, extension at 72°C for 30seconds; and a final extension at 72°C for 7 minutes.
Statistical analyses
Statistical analyses for the studied parameters were performed by using SPSS version 20 (SPSS Inc., Chicago, IL). The chi-square and Fisher's exact test were used. Variables with P values < 0.05 were considered significant.
Results
Study population.
A total of 5,361 positive sputum specimens were collected from 3,020 pulmonary TB patients who were referred to TB diagnostic centers in different provinces of Iran. Of these patients, only 1,242 (41.1%) had smear, culture, and DNA results positive for M. tuberculosis. The remaining specimens were excluded because they were either smear positive/culture negative (985 of 3,020, 32.6%) or had no suitable DNA to perform molecular genotyping (312 of 3,020, 10.3%). In addition, 445 (14.7%) of 3,020 samples were contaminated or contained Mycobacterium spp. other than M. tuberculosis (36 of 3,020, 1.1%).
The mean age of patients was 46.9 years (range = 1–16 years). Six hundred eighty-five patients (55.1%) of 1,242 were female and 557 (44.8%) of 1,242 were male. A total of 1,172 patients were from native populations (1,172 of 1,242, 94.3%) and the remainder were nationals from neighboring countries (Afghani = 62; Iraqi = 3; Azerbaijani = 5).
Most (1,007 of 1,242, 81.0%) patients had isolates with wild-type rpoB, KatG and inhA mutations. Overall, 73 (5.8%) had MDR-TB, 92 and 70 had isolates with RMP (7.4%) or INH (5.6%) monoresistant TB, respectively (Table 2) (repeat isolates were been excluded). Most (57 of 73, 78.0%) MDR and (69 of 92, 75%) RMR isolates had a single mutation detected in the rpoB gene. This substitution were seen more frequently at positions 531 (MDR 35 of 73, 47.9% and RMR 31 of 92, 33.6%), and 526 (MDR 19 of 73, 26% and RMR (21 of 92, 22.8%). Overall, 140 (84%) isolates had single rpoB mutation (P < 0.05), 20 (12%) had double mutations, and 5 (3%) had triple mutations in the rpoB gene. For INH resistance the most prevalent mutation in MDR (48 of 73, 65.7%) and INH monoresistant isolates (48 of 70, 68.5%) were substitutions in the codon for katG 315. In MDR-TB isolates, 30 (41%) of 70 strains had katG 315 and rpoB 531 mutations.
Table 2.
Results of genotyping and molecular drug susceptibility testing for 1,242 pulmonary tuberculosis patients with smear, culture and positive tuberculosis, Iran*
| Province | Spoligotype patterns | Susceptibility patterns | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAS | H | T | Beijing | Manu | U | LAM | Bovis | EAI | X | Orphan | Susceptible | MDR | RMR | INHR | Total | |
| Tehran | 107 | 85 | 47 | 35 | 14 | 6 | 2 | 4 | 1 | 23 | 246 | 32 | 26 | 20 | 324 | |
| Sistan and Balouchastan | 59 | 65 | 16 | 3 | 2 | 2 | – | – | – | 1 | 17 | 146 | 1 | 10 | 8 | 165 |
| Khouzestan | 67 | 7 | 13 | 5 | 2 | 6 | 7 | 1 | 2 | – | 9 | 103 | 6 | 3 | 7 | 119 |
| Khorasan | 34 | 42 | 19 | 12 | 3 | 2 | 1 | – | – | – | 4 | 96 | 2 | 9 | 10 | 117 |
| Ardebil | 16 | 10 | 29 | 7 | 1 | – | – | 2 | – | – | – | 55 | 4 | 4 | 2 | 65 |
| Qom | 18 | 20 | 8 | 10 | – | – | – | 2 | – | – | 3 | 51 | 4 | 3 | 3 | 61 |
| Golestan | 23 | 8 | 8 | 6 | – | – | – | – | – | – | 2 | 40 | 2 | 2 | 3 | 47 |
| Esfahan | 17 | 2 | 12 | 6 | – | – | 2 | – | – | – | 3 | 32 | 2 | 3 | 5 | 42 |
| Gilan | 18 | 13 | 2 | – | – | – | 1 | – | – | – | 5 | 31 | 3 | 2 | 1 | 39 |
| Fars | 16 | 14 | 1 | 5 | – | – | 1 | – | – | – | 3 | 28 | 5 | 5 | 2 | 40 |
| Hormozgan | 19 | 8 | 4 | 5 | 1 | – | 1 | – | – | – | – | 29 | 3 | 3 | 3 | 38 |
| Mazandarn | 4 | 6 | 13 | 1 | – | – | – | – | – | 1 | 1 | 23 | 1 | 1 | 1 | 26 |
| Semnan | 7 | 8 | 5 | 1 | – | – | – | – | – | – | – | 21 | 0 | 0 | 0 | 21 |
| Lorestan | 15 | 7 | 2 | – | – | – | – | – | – | – | – | 19 | 0 | 5 | 0 | 24 |
| Hamadan | 7 | 5 | 3 | 1 | 1 | – | – | – | – | – | 4 | 18 | 0 | 2 | 1 | 21 |
| Kerman | 10 | 8 | 2 | – | – | 1 | – | – | – | – | 3 | 17 | 3 | 3 | 1 | 24 |
| Kurdistan | 4 | 9 | 3 | – | – | – | – | – | – | – | – | 14 | 0 | 0 | 2 | 16 |
| Kermanshah | 9 | 2 | 4 | 1 | – | – | – | – | – | – | – | 12 | 1 | 2 | 1 | 16 |
| Markazei | 9 | 4 | 3 | – | – | – | – | – | – | – | 7 | 2 | 3 | 3 | 15 | |
| Yazd | 6 | 2 | 4 | – | – | – | – | – | – | – | – | 11 | 0 | 1 | 0 | 12 |
| Qazvin | 6 | 2 | – | – | 1 | – | – | 1 | – | – | – | 7 | 2 | 0 | 1 | 10 |
| Total | 471 | 326 | 195 | 101 | 25 | 17 | 15 | 10 | 3 | 2 | 77 | 1,007 | 73 | 92 | 70 | 1242 |
| % | 37.9 | 26 | 16 | 81.1 | 2 | 13 | 1.2 | 0.8 | 0.2 | 0.16 | 61.1 | 81.0 | 5.8 | 7.4 | 5.63 | |
There was no statistical difference between spoligotypes of susceptible and rifampin-monoresistant tuberculosis isolates. The CAS and H lineages are the most frequent superfamilies in Iran. MDR = multidrug resistant; RMR = rifampin monoresistant; INHR = isoniazid resistant.
Genotyping of total population.
Spoligotyping identified 10 genotypes (Table 1). The largest lineages were CAS (471, 37.9%) and Haarlem (326, 26.2%) families, followed by the T lineage (15.7%), the Beijing lineage (101, 8.1%), the Manu lineage (25;2%), the U lineage (17; 1.3%), LAM (15, 1.2%), and M. bovis (10, 0.8%). Only 3 (0.2%) strains of EAI were identified. The remaining isolates (77, 6.1%) were unique and could not be identified using SpolDB4[16].
Retrospective study of RMP-monoresistant isolates.
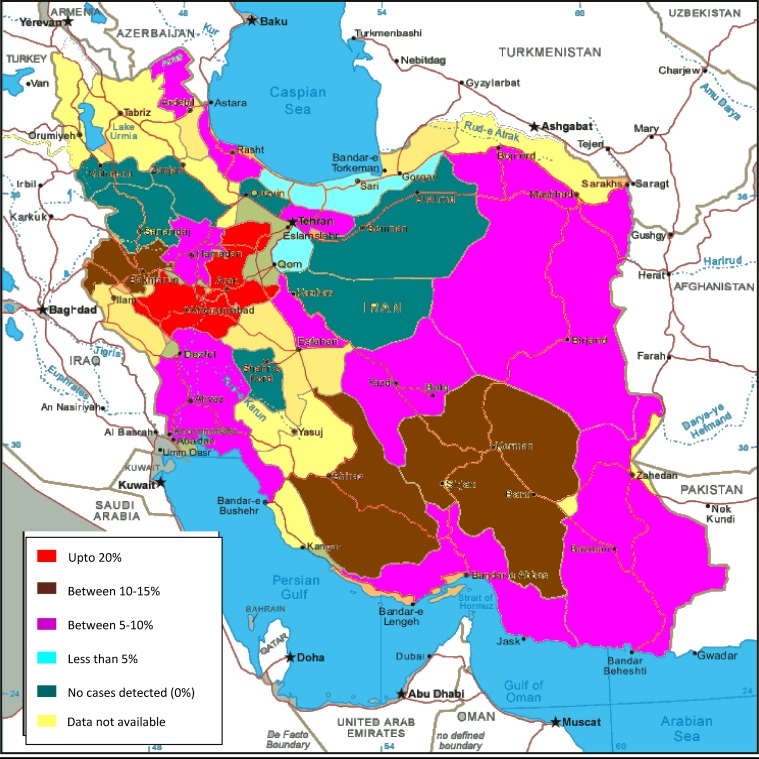
Of 1,242 TB patients who remained smear and culture positive, 92 (7.4%) of 1,242) had RMR isolates. As shown in (Figure 1), these patients were detected in different regions of Iran. Thirty-five (38.0%) were female and 57 (61.9%) were male. Thirty-one (33.6%) of 92 patients (P < 0.05) were immigrants (Afghani = 28, Azerbaijani = 2, and Iraqi = 1) and 61 were from the native Iranian pulmonary TB populations (Table 3). The mean ± SD age was 44.3 ± 17.6 years for Iranian case-patients and 38.1 ± 19.7 years for Afghani case-patients.
Figure 1.
Cases of rifampin-monoresistant tuberculosis in Iran.
Table 3.
Demographic characteristic of RMR-TB patients, Iran*
| Group receiving (DOTS) | Nationality | Mean ± SD age, years | M/F | History of TB | Close or family contact | Spoligotypes | ||
|---|---|---|---|---|---|---|---|---|
| Iranian | Afghani | Other | ||||||
| A (positive after 9 months) | 20 | 6 | 3 | 47.2 ± 12.5 | 18/11 | 4 | 7 | Beijing (6), CAS1_KILI(10), H4 (5), H3 (1), T1 (3), MANU2 (1), Unknown (1), S (1), LAM (1) |
| B (positive after 9 months) | 18 | 9 | – | 39.7 ± 6.9 | 19/8 | 2 | 5 | Beijing (1), CAS1_KILI (7), CAS1_DELHI (3), H4 (4), H3 (1), T1 (2), MANU2 (1), Unknown (5), LAM (1), S (1) |
| C (positive after 4 months) | 17 | 8 | – | 40.5 ± 11.3 | 13/12 | 3 | 3 | CAS1_KILI (8), H4 (6), H3 (1), T1 (7), Unknown (1), U (2) |
| D (positive after 2 months) | 6 | 5 | – | 37.0 ± 16 | 7/4 | 4 | 3 | CAS1_KILI (6), U (3), H4 (2) |
| Total | 61 (66.3%) | 28 (30.4%) | 3 (3.2%) | 41.1 ± 11.6 | 57/35 | 13 (14.1%) | 18 (19.5%) | CAS (32; 34.7%), H (20; 21.7%), T1(14; 15.2%), Beijing (7; 11.1%), U (5; 5.4%, Unknown (7; 7.6% LAM (2; 2.1%), MANU 2 (2; 2.1%), S (2; 2.1%) |
RMR-TB = rifampin-monoresistant tuberculosis.
Based on RMR detection rate, the studied provinces were subdivided into 3 main categories: provinces with a high level of RMR detected (Lorestan, 5 of 24; 20%; Markazei, 3 of 15, 20%; Fars, 5 of 40, 12.5%, Kerman, 3 of 24, 12.4%), and Kermanshah, 2 of 16, 12.5%), provinces with a with moderate level of RMR detected (47.5%; P < 0.05) (Tehran, 26 of 324, 8.0%; Khorasan, 9 of 119, 7.6%; Sistan-Balouchestan, 10 of 165, 6%), and provinces with a low level of RMR detected (Qom, 3 of 61, 4.9% and Golestan, 2 of 47, 4.2%). In some provinces that had only a few examined isolates, no RMR TB were detected (4 of 18, 22.2%; P < 0.05). These provinces were Semnan (n = 21 patients), Kurdistan (n = 16 patients), Qazvin (n = 10 patients), and Shahrood (n = 2 patients). Provinces near Afghanistan and Pakistan had moderate frequencies of RMR TB patients (5–10%). Unfortunately, the data from other provinces near Turkey, Turkmenistan, and Azerbaijan were not available.
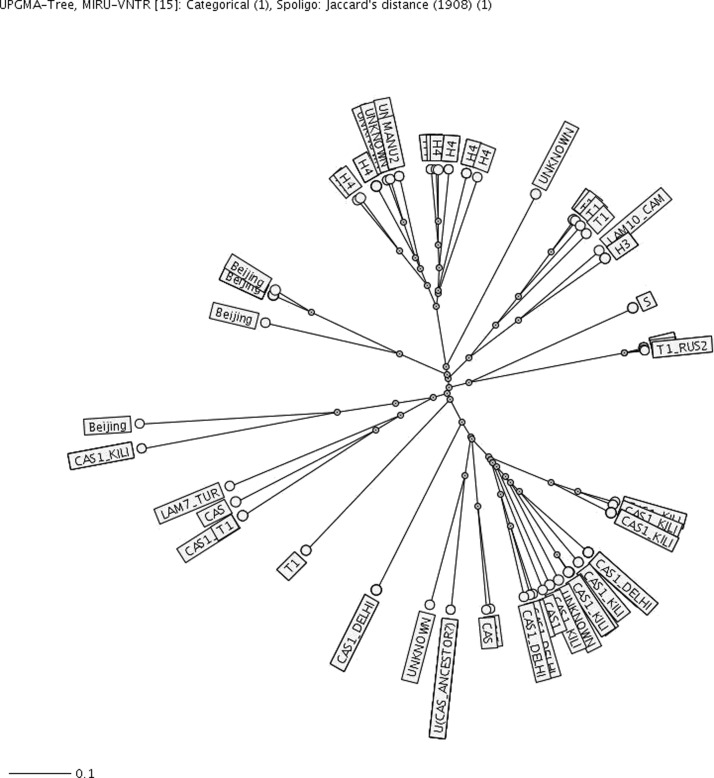
To identify transmission among RMR patients, molecular epidemiology patterns of isolates were reviewed. Spoligotyping classified 85 patients into 8 clusters. Major super families were CAS (32, 34.7%), H (20, 21.7%), T (14, 15.2%) and Beijing (7, 7.6%) (Table 3).The remaining isolates were U (5, 5.4%), LAM (2.1%), MANU2 (2.1%), and S lineages (2.1%). Seven (7.6%) isolates had orphan patterns. Further combination of spoligotyping and MIRU-VNTR for 15 loci classified 63 patients into 23 clusters. Three clusters of U (n = 5), CAS (n = 5), and Beijing (n = 4) lineages had the higher number of patients. Twelve and eight clusters consisted of two (52.1%) and three (34.7%) patients, respectively. The clustering rate was 0.22463769 by MIRU-VNTR and 0.5 by spoligotyping (Figure 2).
Figure 2.
Spanner diagram showing clusters of rifampin-monresistant isolates of Mycobacterium tuberculosis classified by spoligotying and mycobacterial interspersed repetitive unit–variable number tandem repeat in Iran. Scale bar indicates nucleotide substitutions per site.
Review of patient questionnaires identified a transmission link for seven patients (13%) in three clusters (H3 from Kerman, T from Tehran, and CAS lineage from Khuzestan). Allelic diversity for each MIRU-VNTR locus based on the Hunter-Gaston Index showed 13 loci with a higher discrimination value of 0.6, whereas the loci MIRU04 and Mtub30 had values of 0.42 and 0.57, respectively. Clinically, TB patients with RMR isolates were classified into four groups depending on their smear and culture positivity: group A (positive after 9 months), group B (positive after 6 months) group C (positive after 4 months), and group D (positive after 2 months) (Table 3). Prevalence of RMR in smear-positive TB patients after the second month of therapy was low (11 of 92, 11.9%) in comparison with other groups (P < 0.05). Most (7 of 11, 63.6%) of these patients had a history of TB or close contact with TB patients. In addition, two patients in group D were positive for HIV. A total of 31 patients (11 [37.9%] of 29 in group A, 7 [25.9%] of 27 patients in group B, 6 [24%] in group C, and 7 [63.6%] in group D had a family or previous history of TB. We found no other correlated risk factor for 59 (64.1%) of 92 patients (P < 0.05).
Discussion
Introduction of new algorithms for rapid identification of drug-resistant TB by using molecular test is one major advancement in TB laboratory diagnostics.6,7,22 Application of these molecular-based detection assays (PCR, PCR–restriction fragment length polymorphism, or multiplex PCR) could effectively help in screening and identifying large numbers of suspected TB cases throughout Iran. As a result, during 2010–2011, we examined 3,020 smear-positive TB patients for genotyping. Retrospective analysis of these data identified important epidemiologic aspects of TB in different provinces of Iran. Findings showed that 985 (32.6%) of 3,020 smear-positive TB patients had negative culture results during therapy. A prevalence of 32.6% smear-positive, culture-negative patients may underline the importance of culture examination during follow-up studies.
Genotyping identified 238 (18.9%) of 1,242 patients with drug-resistant isolates. Because the rate of RMR (7.4%) was higher than that for MDR (5.6%), precautions must be taken in interpreting the data. Relative frequencies of rpoB gene mutations was high in codons 531 (105 of 165, 63.3%), 526 (68 of 165, 41.2%), 516 (43 of 165, 26.0%), followed by 518 (9 of 165, 5.4%) 513 (7 of 165, 4.2%), and 511(3 of 165, 1.8%). Worldwide, frequencies of mutation at codon 531 and 526 ranged from 29% to 74% and from 0% to 43%, respectively.23–25 Thus, our reported frequencies were consistent with those of previous reports from other locations. The mean rate of RMP-monoresistant TB ranged from 5% to 10%, which was observed in 11 studied provinces (47.6%; P < 0.05). In large metropolitan cities such as Tehran (26 of 324, 8.0%), Khorasan (9 of119, 7.6%), and Sistan and Balouchestan (10 of 165, 6%), the mean RMR was 6–8% (Figure 2). However, in a few provinces, where the number of collected specimens was low, the detected rate was not fully reliable, either high (up to 20%) or 0%.
Generally, RMR-TB occurred when there is erratic use of drugs, omission of one or more prescribed drugs, suboptimal dosages, poor drug absorption, or an insufficient number of active agents in a drug regimen.2,4,5,25,26 Review of classical and clinical information showed no single therapy or suboptimal dosage in studied populations, although 31 (33%) of 92 isolates (P < 0.05) were collected from immigrant or other nationals who either visited Iran for TB treatment (10 of 28, 53.7%) or stayed in Iran < 10 years. Because immigrants enter Iran without been given physical examinations, collected information is limited to patient interviews.
Munsiff and Friedan,27 Ridzon and others,28 and Sandman and others29 described in detail clinical conditions in which RMR-TB might have occurred. They found strong associations between RMP monoresistance and infection with HIV and previous or extrapulmonary TB. In HIV-seropositive patients, malabsorption or drug–drug interactions could lead to altered active levels of anti-TB drugs. More recently, an additional risk factor (excessive alcohol use) has been reported in surveys for Brazil, South Africa, Burundi, and Russia.30–33 In the present study, 33.6% of RMR patients had a history of TB or had close contact with a known TB patient. This rate reached 58% in immigrants. However, in many cases, no risk factors were found.
Identification of RMR-TB by detection of INH resistance showed poor sensitivity. However, analysis of isolates by the Swedish Supranational Reference Laboratory indicated that most samples were accurately categorized as RMR-TB (the remaining isolates were categorized as MDR-TB). Therefore, a significant occurrence of RMP resistance without an association with INH resistance was verified.
To identify clonal expansion of these specific strains, spoligotyping and MIRU-VNTR analysis was conducted. A combination of these typing methods identified 63 (66.33%) of 92 patients in 23 clusters. Cluster sizes were small, ranging from 2 to 5 isolates. A retrospective epidemiologic analysis identified a transmission link for seven patients in three clusters (3 of 23, 13%). In two clusters, patients were family members (H3 and T lineages) and in another cluster the two patients were in close contact (prison inmates in Khousaztan Province). The CAS (34.7%), Haarlem (21.7%), and Beijing lineages (7.6%) were the most dominant super families among RMR-TB cases. However, CAS (471 isolates, 37.9%) and Haarlem (326 isolates, 26.2%) lineages were also found among susceptible isolates (Table 1). Therefore, these genotypes may be the most common strains circulating34,35 and are not necessary associated with RMR.
Overall, we showed that RMR-TB was strongly associated with close contact or previous history of TB. However, this estimate did not include use of free TB drugs. It has been more than 15 years that the ban on free sale of TB drugs was implemented in Iran, but enforcement of this ban is questionable.
Another limitation of this study was the low number of collected specimens in a few provinces, which need further investigation. In conclusion, if one considers the higher sensitivity of detection of resistance to RMP than to INH, the true level of INH resistance and MDR-TB might be underestimated. However, our results identified RMP resistance without an association with INH resistance. Therefore, the level of RMR-TB in any specific setting should be identified before new diagnostic algorithms are developed and implemented.
ACKNOWLEDGMENTS
We thank the directly observed treatment, short course chief medical officers and the staff of TB laboratories throughout Iran for their assistance.
Footnotes
Financial support: This study was supported by National Manager of TB and Leprosy Control Programs (Ministry of Health and Medical Education) and the Mycobacteriology Research Centre (MRC-6011-M-00-5505-01).
Authors' addresses: Ali Akbar Velayati, Parissa Farnia, Mohadese Mozafari, Maryam Fatemeh Sheikholeslami, Mona Afraei Karahrudi, and Payam Tabarsi, Mycobacteriology Research Centre, National Research Institute of Tuberculosis and Lung Disease, World Health Organization Collaborating Centre, Masih Daneshvari Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran, E-mails: velayati@theaasm.org, farina@theaasm.org, mycopf@hotmail.com, sheikholeeslam@theaasm.org, karahrudi@theaasm.org, and tabarsi@theaasm.org. Sven Hoffner, World Health Organization Suranational Reference Laboratory for Tuberculosis, Swedish Institute for Communicable Disease Control, Stockholm, Sweden, E-mail: sven.hoffner@smi.se.
References
- 1.Mirsaeidi SM, Tabarsi P, Khoshnood K, Pooramiri MV, Rowhani-Rahbar A, Mansoori SD, Masjedi H, Zahirifard S, Mohammadi F, Farnia P, Masjedi MR, Velayati AA. Treatment of multiple drug-resistant tuberculosis (MDR-TB) in Iran. Int J Infect Dis. 2005;9:317–322. doi: 10.1016/j.ijid.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison DS, Nunn AJ. Influence of initial drug resistance on the response to short-course chemotherapy of pulmonary tuberculosis. Am Rev Respir Dis. 1986;133:423–430. doi: 10.1164/arrd.1986.133.3.423. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard JS. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis. Annu Rev Biochem. 1996;65:215–239. doi: 10.1146/annurev.bi.65.070196.001243. [DOI] [PubMed] [Google Scholar]
- 4.Iseman MD, Madsen LA. Drug- resistant tuberculosis. Clin Chest Med. 1989;10:341–353. [PubMed] [Google Scholar]
- 5.Vos M, Quince C, Pijl AS, de Hollander M, Kowalchuk GA. A comparison of rpoB and 16S rRNA as markers in pyrosequencing studies of bacterial diversity. PLOS One. 2012;7:e30600. doi: 10.1371/journal.pone.0030600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams DL, Waguespack C, Eisenach K, Crawford JT, Portaels F, Salfinger M, Nolan CM, Abe C, Sticht-Groh V, Gillis TP. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Beenhouwer H, Lhiang Z, Jannes G, Mijs W, Machtelinckx L, Rossau R, Traore H, Portaels F. Rapid detection of rifampin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 8.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 9.Hanrahan CF, Dorman SE, Erasmus L, Koornhof H, Coetzee G, Golub JE. The impact of expanded testing for multidrug resistant tuberculosis using genotype MTBDR plus in South 15 Africa: an observational cohort study. PLoS ONE. 2012;7:e49898. doi: 10.1371/journal.pone.0049898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization . Anti-Tuberculosis Drug Resistance in the World. Geneva: World Health Organization; 1997. Report no. 1. WHO/TB/97.229. [Google Scholar]
- 11.Masjedi MR, Farnia P, Sorooch S, Pooramiri MV, Mansoori SD, Zarifi AZ, Akbarvelayati A, Hoffner S. Extensively drug resistant tuberculosis: 2 years of surveillance in Iran. Clin Infect Dis. 2006;43:641–847. doi: 10.1086/507542. [DOI] [PubMed] [Google Scholar]
- 12.Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, Ziazarifi AH, Hoffner SE. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest. 2009;136:420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 13.Khazaei HA, Rezaei N, Bagheri GR, Dankoub MA, Shahryari K, Tahai A, Mahmoudi M. Epidemiology of tuberculosis in the southeastern Iran. Eur J Epidemiol. 2005;20:879–883. doi: 10.1007/s10654-005-2152-y. [DOI] [PubMed] [Google Scholar]
- 14.Rafiee S, Besharat S, Jabbari A, Golalipour F, Nasermoaadeli A. Epidemiology of tuberculosis in northeast of Iran: a population-based study. IJMS. 2009;34:193–197. [Google Scholar]
- 15.Velayati AA, Farnia P, Masjedi MR, Hoffner S. Detection of and treatment protocol for rifampicin-monoresistant tuberculosis: what is the role of isoniazid. Int J Tuberc Lung Dis. 2013;17:849–845. doi: 10.5588/ijtld.13.0066. [DOI] [PubMed] [Google Scholar]
- 16.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium. tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ho ML, Martin C, Martin C, Mokrousov I, Narvskaïa O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, Singh UB, Somoskovi A, Skuce RA, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren RM, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTR plus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:326–331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and user-strategy of MIRU-VNTR plus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Durmaz R, Yang D, Gunal S, Zhang L, Foxman B, Sanic A, Marrs CF. Simultaneous detection of isoniazid, rifampin, and ethambutol resistance of Mycobacterium tuberculosis by a single multiplex allele-specific polymerase chain reaction (PCR) assay. Diagn Microbiol Infect Dis. 2005;53:201–208. doi: 10.1016/j.diagmicrobio.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Kazumi Y, Mitara S. The evaluation of an identification algorithm for Mycobacterium species using the 16S rRNA coding gene and rpoB. Int J Myco. 2012;1:21–28. doi: 10.1016/j.ijmyco.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Buyankhishig B, Oyuntuya T, Tserelmaa B, Sarantuya J, Lucero MG, Mitara S. Rapid molecular testing for multi-resistant tuberculosis in Mongolia: a diagnostic accuracy study. Int J Myco. 2012;1:40–44. doi: 10.1016/j.ijmyco.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Xin J, Zhang L, Jiang L, Cao H, Li L. Rapid detection of rpoB mutations in rifampin resistant M. tuberculosis from sputum samples by denaturing gradient gel electrophoresis. Int J Med Sci. 2012;9:148–156. doi: 10.7150/ijms.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma X, Wang H, Deng Y, Liu Z, Xu Y, Pan X, Musser JM, Graviss EA. rpoB gene mutations and molecular characterization of rifampin-resistant Mycobacterium tuberculosis isolates from Shandong Province, China. J Clin Microbiol. 2006;44:3409–3412. doi: 10.1128/JCM.00515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani C, Selvakumar N, Narayanan S, Narayanan PR. Mutation in the rpoB gene of multidrug resistant Mycobacterium tuberculosis clinical isolates from India. J Clin Microbiol. 2001;39:2987–2988. doi: 10.1128/JCM.39.8.2987-2990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchison DA. How drug resistance emerges as a result of poor compliance during short course chemotherapy for tuberculosis. Int J Tuberc Lung Dis. 1998;2:10–15. [PubMed] [Google Scholar]
- 27.Munsiff SS, Friedan TR. Mono-rifampin resistant tuberculosis, New York City, 1993–1994 (abstract no.C118). Abstracts of the 36th International Conference on Antimicrobial Agents and Chemotherapy; New Orleans, LA. Washington, DC: American Society for Microbiology Press; 1996. [Google Scholar]
- 28.Ridzon R, Whitney CG, McKenna MT, Taylor JP, Ashkar SH, Nitta AT, Harvey SM, Valway S, Woodley C, Cooksey R, Onorato IM. Risk factors for rifampin mono-resistant tuberculosis. Am J Respir Crit Care Med. 1998;157:1881–1884. doi: 10.1164/ajrccm.157.6.9712009. [DOI] [PubMed] [Google Scholar]
- 29.Sandman L, Schluger NW, Davidow AL, Bonk S. Risk factors for rifampin-monoresistant tuberculosis: a case–control study. Am J Respir Crit Care Med. 1999;159:468–472. doi: 10.1164/ajrccm.159.2.9805097. [DOI] [PubMed] [Google Scholar]
- 30.Pereira Pinto W, Hadad DJ, Silva Telles MA, Mizuka Ueki SY, Palaci M, Basile MA. Tuberculosis and drug resistance among patients seen at an AIDS reference center in Sao Paolo, Brazil. Int J Infect Dis. 2001;5:94–100. doi: 10.1016/s1201-9712(01)90034-7. [DOI] [PubMed] [Google Scholar]
- 31.Mukinda FK, Theron D, van der Spuy GD, Jacobson KR, Roscher M, Streicher EM, Musekiwa A, Coetzee GJ, Victor TC, Marais BJ, Nachega JB, Warren RM, Schaaf HS. Rise in rifampicin-monoresistant tuberculosis in Western Cape, South Africa. Int J Tuberc Lung Dis. 2012;16:196–202. doi: 10.5588/ijtld.11.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders M, Van Deun A, Ntakirutimana D, Masabo JP, Rukundo J, Rigouts L, Fissette K, Portaelst F. Rifampicin mono resistant Mycobacterium tuberculosis in Bujumbura, Burundi: results of a drug resistance survey. Int J Tuberc Lung Dis. 2006;10:178–183. [PubMed] [Google Scholar]
- 33.Drobniewski F, Balabanova Y, Ruddy M, Weldon L, Jeltkova K, Brown T, Malomanova N, Elizarova E, Melentyey A, Mutovkin E, Zhakharova S, Fedorin I. Rifampin and multidrug-resistant tuberculosis in Russian civilians and prison inmates: dominance of the Beijing strain family. Emerg Infect Dis. 2002;8:1320–1326. doi: 10.3201/eid0811.020507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velayati AA, Farnia P, Mirsaeidi M, Reza Masjedi M. The most prevalent Mycobacterium tuberculosis superfamilies among Iranian and Afghan TB cases. Scand J Infect Dis. 2006;38:463–468. doi: 10.1080/00365540500504117. [DOI] [PubMed] [Google Scholar]
- 35.Farnia P, Masjedi MR, Mirsaeidi M, Mohammadi F, Jallaledin-Ghanavi, Vincent V, Bahadori M, Velayati AA. Prevalence of Haarlem I and Beijing types of Mycobacterium tuberculosis strains in Iranian and Afghan MDR-TB patients. J Infect. 2006;53:331–336. doi: 10.1016/j.jinf.2005.12.020. [DOI] [PubMed] [Google Scholar]