Abstract
Human dipeptidylpeptidase IV (hDPPIV) is an enzyme that is in hydrolase class and has various roles in different parts of human body. Its deficiency may cause some disorders in the gastrointestinal, neurologic, endocrinological and immunological systems of humans. In the present study, hDPPIV enzyme was expressed on Spodoptera frugiperda (Sf9) cell lines as a host cell, and the expression of hDPPIV was obtained by a baculoviral expression system. The enzyme production, optimum multiplicity of infection, optimum transfection time, infected and uninfected cell size and cell behavior during transfection were also determined. For maximum hDPPIV (269 mU mL−1) enzyme, optimum multiplicity of infection (MOI) and time were 0.1 and 72 h, respectively. The size of infected cells increased significantly (P < 0.001) after 24 h post infection. The results indicated that Sf9 cell line was applicable to the large scale for hDPPIV expression by using optimized parameters (infection time and MOI) because of its high productivity (4.03 mU m L−1 h−1).
Keywords: Spodoptera frugiperda (Sf9), Dipeptidyl peptidase IV (DPPIV), Baculovirus, Transfection
Introduction
Human dipeptidyl peptidase IV (hDPPIV, EC 3.4.14.5) is a multifunctional type II transmembrane serine protease that catalyzes the release of N-terminal dipeptides from proline sites by hydrolysis (Bergmann and Bohuon 2002; Thoma et al. 2003). The enzyme is also known as an important signaling molecule CD26 on T and B lymphocytes and it is expressed in a variety of cell types, microbial (Demuth and Heins 1995; Cunningham and O’Connor 1997; Pérez-Guzmán et al. 2004), insect cells and nearly all mammalian tissues. hDPPIV regulates various physiological processes including immune system, endocrine functions, central nervous system, gastrointestinal system, inflammation, rheumatoid arthritis and cell adhesion. The enzyme removes dipeptides from N-terminus of regulatory peptides such as chemokines, neuropeptides and peptide hormones (Reichelt et al. 1981; Shattock et al. 1990; Urade et al. 2006; Iwaki-Egawa et al. 1998; Mentlein 1999; Brudnak 2001; Brudnak et al. 2002; Langford 2003; Lambeir et al. 2002; Reichelt and Knivsberg 2003; Aertgeerts et al. 2004; Reinhold et al. 2009).
Recently, hDPPIV has been used as a dietary supplement alone or in combination with other nutritional enzymes by the patients who are suffering from lack of hydrolysis enzyme. It has been commercially produced by some Aspergillus strains (Tachi et al. 1992; Doumas et al. 1998; Wilkinson and Houston 2001; Jalving et al. 2005; Ota et al. 2005). Recombinant protein production from baculovirus-insect cell system has some advantages like very high level of recombinant protein through post translational processing and modification of higher eukaryotes. However, large scale production of the enzyme from insect cell is very limited because of the commercial concerns like the cost of experimental tools and productivity (Effio et al. 2003; Ikonomou et al. 2003; Hitchman et al. 2009).
In the current study, the expression of hDPPIV was performed by using baculovirus infected insect cell line (Spodoptera frugiperda, Sf9). Monitoring the effect of transfection reactions on the cell characteristics is scarce in the literature. Therefore, the efficiency of transfection, optimum multiplicity of infection (MOI), optimum transfection time, cell behavior and cell size were monitored to determine optimal harvest time for large scale production of Sf9 cell line and expression of hDPPIV.
Determining the characteristics of transfected Sf9 cells might be useful to find out the point of peak protein production and to predict the maximum protein yield, thus stable and economical recombinant protein expression can be achieved. These characteristics may be differentiated according to cell line. Therefore, there are no certain signals about the transfection and/or infection effect on the insect cells. This current study may be a resource for those interested in a large scale production of recombinant hDPPIV enzyme on Sf9 cell line.
Materials and methods
Cell line and baculovirus
The insect cell line was Spodoptera frugiperda, Sf9 (Invitrogen, USA) derived from the pupal ovarian tissue of the fall army worm, and Autographa californica nucleopolyhedrovirus was used as a recombinant baculovirus. The transfer vector was pFastBac1, competent Escherichia coli DH10Bac cells, Sf 900 II SF medium, Cellfectin II and antibiotics were purchased from Invitrogen (Gibco BRL USA); Bluo-gal, IPTG and protease inhibitor mixture were from Sigma (USA).
Plasmid construction
The full-length cDNA of hDPPIV/CD26 was obtained from pEBFP-N1-DPPIV (Madgeburg University, Germany), and hDPPIV gene was amplified by PCR using forward and reverse primers. The primers used are shown below:Forward primer: 5′-CGCGGATCCATGAAGACACCGTGGAAGGTT-3′Reverse primer: 5′-GCGACTAGTCTAAGGTAAAGAGAAACATTG-3′. PCR was performed for KOD plus protocol and under following conditions: 1 μM Forward primer, 1 μM Reverse primer, 0.1 μg DNA template, 5 μl dNTPs, 2 μl MgSO4, Milli-Q water to a final volume of 50 μl. Amplification was achieved upon 2 min at 94 °C, 30 cycles of at 60 °C, and an extension step, at 68 °C, for 2.31 min. The PCR products were loaded to agarose gel (0.8 %, w/v) for confirmation. The PCR products containing the hDPPIV, were digested with BamH1 and Spe1 restriction enzymes in order to insert the DNA, in frame, into the pFastBacl transfer vector (Gibco BRL, USA) of the Baculovirus Expression System (Invitrogen, USA). The recombinant plasmids were used to transform competent E.coli DH10B and then DH10Bac (containing the wild-type baculovirus genome in bacmid form) to generate the corresponding recombinant bacmids upon transposition. Cloning steps were monitored by using sequence analysis and controlled by Genetyx-Win Program.
Transformation of baculoviral hDPPIV to E.coli DH10Bac
One vial of DH10Bac competent cells (100 μl) were thawed on ice for each transformation. 1 ng hCD26/pFastBac1 plasmid DNA were added to the cells and mixed gently. The cells were incubated on ice for 30 min. Heat shock was applied to the cells for 45 s at 42 °C without shaking and immediately transferred the tubes to ice and chilled for 2 min. SOC (Super Optimal broth with Catabolite Repression) medium was added 900 μl at room temperature and shake at 37 °C at 225 rpm for 4 h. Serial dilutions of the cells (10−1, 10−2, 10−3) was prepared with SOC medium. Each dilution was plated on LB (Luria Broth) agar that contained 50 μg mL−1 kanamycin, 7 μg mL−1 gentamicin, 10 μg mL−1 tetracycline, 100 μg mL−1 Bluo-gal and 40 μg mL−1 IPTG. The plates were incubated for 48 h at 37 °C. White colonies, which include recombinant bacmid E. coli clones, were picked by blue/white-screening (Invitrogen).
Production of recombinant baculovirus
After transformation of E. coli DH10Bac cells, Spodoptera frugiperda cells were cultured at 27 °C in Sf900-II SFM medium, supplemented with gentamicin (50 μg mL−1). For transfection, 9 × 105 cells were plated in 6 well tissue culture dishes and incubated for 1 h in 2 mL Sf900-II SFM without antibiotics to allow adhesion of the cells to surface. Recombinant bacmid DNA had been preincubated for 45 min at room temperature with Cellfectin II (6 μl). Cells were incubated with the liposome–DNA complex for 5 h at 27 °C. The transfection medium was removed and 2 mL of Sf900-II SFM medium, containing antibiotics was added. The DNA was transfected into Sf9 cells. Transfected cells were incubated at 27 °C for 144 h allowing baculovirus production.
The recombinant virus was amplified twice to obtain virus stocks of the highest titer and harvested. Classical plaque assay was applied for virus titration (Summers and Smith 1987).
Optimization of human DPPIV protein expression and transfection
Sf9 cells in 6 well plates (Sf-900 II SF medium, 10 % v/v heat-inactivated FBS and gentamicin (w/v, 50 μg mL−1) were infected by recombinant virus with MOI of 0.1, 0.25, 0.5, 0.75, 1 and 10 plaque-forming units per cell. Cells were harvested 24, 48, 72, 96, 120 and 144 h post transfection and centrifuged for 5 min at 100 g. The cells were washed three times with phosphate-buffered saline (PBS) and then resuspended on ice in 300 μl of sonication buffer (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 % (w/v) glycerol, IPEGAL CA-630) for 9 × 105 cells mL−1. After sonication three times at 20 s pulses with a Bandelin model sonicator at 45 % power, samples were centrifuged at 4 °C for 10 min at 12,000g. DPPIV activities were measured to determine optimum MOI with the maximum hDPPIV expression.
Determination of virus replication and cytopathic effects
Viral transfection may lead to cytopathic effects and structural changes in the host cell. The effects of transfection with MOI 0.1 on the cells were measured by using mitochondrial tetrazolium test [MTT, 3-(4,5-dimetiltiazol-2-il)-2,5- difenil-tetrazolium bromid assay] in every 6 h for 72 h post transfection (Celis et al. 2005). The viability of uninfected and infected cells was calculated, and the results were compared with the Student’s t test (Statistical Package for the Social Science for Windows, SPSS 17.0).
Measuring of Sf9 cell size during transfection
Transfection may cause changes in cell properties by the introduction of DNA, and the most distinctive change is the size of infected cells. After transfection with the MOI 0.1, 100 μl samples were placed into cover glass, and the cell sizes were measured under an inverse phase microscope (Olympus, Japan) for 100 cells from 10 different areas. The arithmetic averages of the cell sizes were calculated and the results were compared with the Student’s t test.
Monitoring hDPPIV expression and baculovirus replication with immunofluorescence method
The immunofluorescence method was used to detect the location and relative abundance of baculovirus and hDPPIV enzyme. CD26 rabbit polyclonal antibody (CD 26-H270:sc-9153; Santa Cruz, USA) and gp64 monoclonal antibody (gp 64-AcV5-ab91214; Abcam, UK) were used as primer antibodies, also goat antirabbit (594; Invitrogen, UK) and goat antimouse (488; Invitrogen, UK) IgG were seconder antibodies.
Determination of protein concentration, enzymatic activity and western blot
After transfection, cells were harvested at 24 h intervals for Western blot and enzymatic analysis. Enzymatic activity was determined with Gly-Pro-p-nitroanilide as a substrate as described previously (Pérez-Guzmán et al. 2004). Western blot samples (1.5 mL) were centrifuged at 16,000g for 2.5 min. After removing the supernatant, the cell pellets were stored at −25 °C. The protein concentration was determined with the BCA Protein Assay Reagent Kit (Pierce, USA).
Results
Optimization of hDPPIV protein expression and transfection parameters
Production of recombinant baculovirus
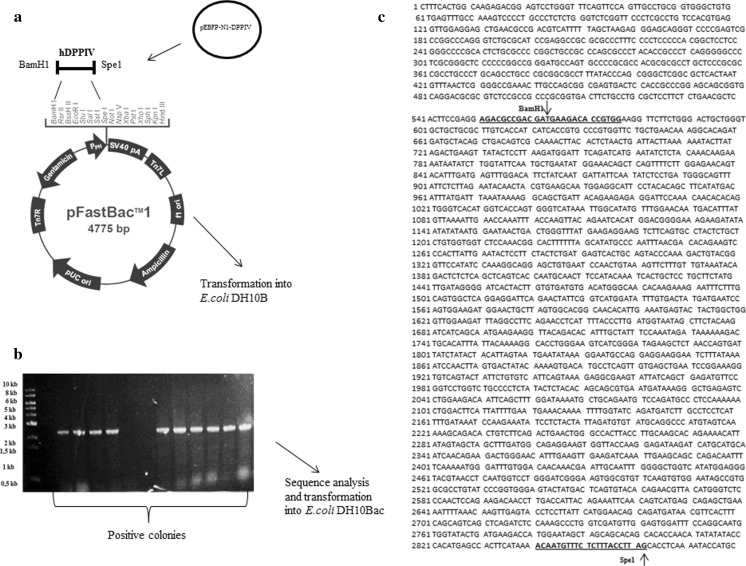
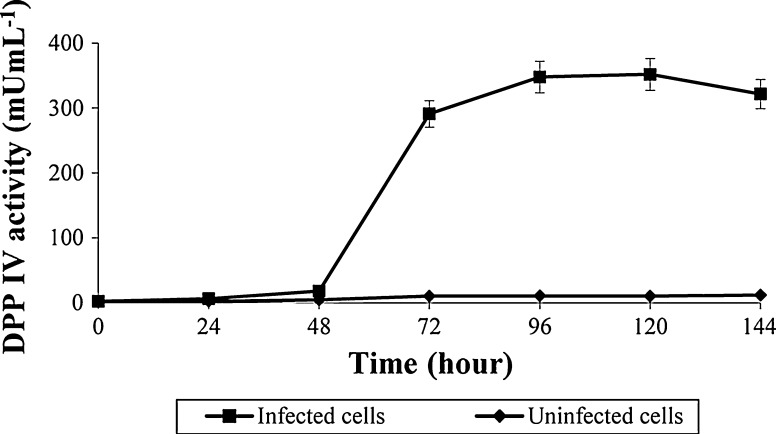
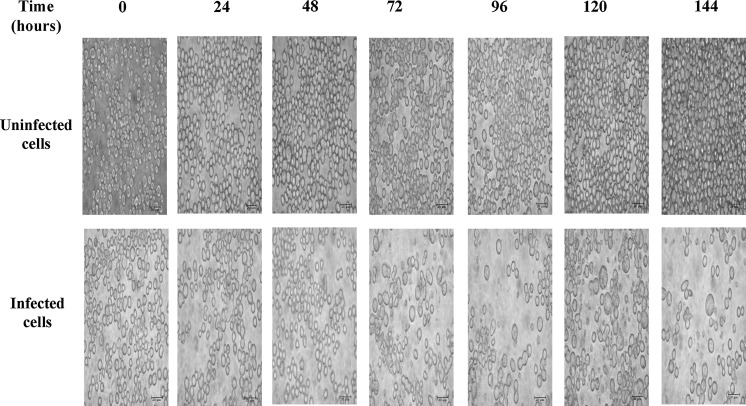
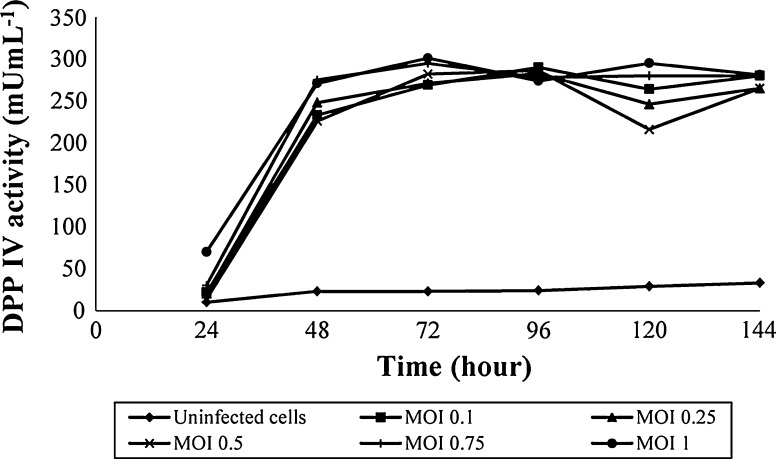
The plasmid construct was successfully transfected into E.coli DH10B and then DH10Bac, which resulted in the generation of the bacmid DNA. A detail of the cloning strategy and nucleotid sequence of hDPPIV is described in Fig. 1. Sf9 cells were transfected with this bacmid DNA by using cationic lipid agent for protein expression. Transfection was monitored daily by measuring the enzyme activity for post transfection. Expression of hDPPIV enzyme was similar in uninfected and infected Sf9 cells for 24 h, but it increased after 48 h post infection, and finally the maximum enzyme activity was determined for 96 h post infection. The expression of this enzyme decreased slightly after 120 h post infection (Fig. 2). Changes in cell shape, cell size and density of the cells are indicated in Fig. 3. The cells became bigger and more rounded while the number of cells decreased, and cells lost adherence ability to the culture plate obviously after 72 h post transfection. These typical changes in the morphological properties of cells at post transfection were reported in the literature (Radner et al. 2012).
Fig. 1.
Shematic of the cloning strategies and nucleotid sequence of hDPPIV. a The hDPPIV gene was obtained from pEBFP-N1-DPPIV inserted into thepFastBac1 cloning system, and constructed plasmid was transformed E.coli DH10B. b PCR screening of the positive E.coli DH10B colonies containing the pFastBac1hDPPIV plasmid. c nucleotid sequence of hDPPIV (ncbi. NM-001935 mRNA linear homo sapiens dipeptidyl peptidase IV, CD26)
Fig. 2.
Comparison of DPPIV expression with bacmid DNA by using cationic lipid agent (Cellfectin II)
Fig. 3.
Time dependent morphological differentiations of insect cells after transfection
The maximum enzyme activity was obtained at 96 h post transfection. However, when the transfection productivity (enzyme activity per hours) consider, it can clearly be seen that 72 h post transfection (4.03 mU mL−1 h−1) could be better than 96 h (3.62 mU mL−1 h−1) for optimum transfection.
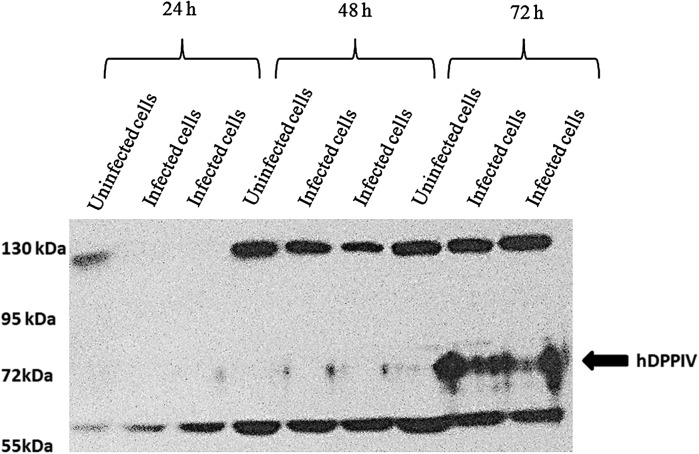
Transfected Sf9 cells were harvested and loaded into Western blot gel. The Western blot results implied that the expression of recombinant hDPPIV on infected Sf9 cells was increased at 72 h post transfection, while expression of hDPPIV had insignificant changes in uninfected cells (Fig. 4). The molecular mass of hDPPIV was about 70 kDa according to the marker on the gel. This molecular mass had already been predicted from the amino acid sequence of hDPPIV (2,300 bp) (Tanaka et al. 1992; Thoma et al. 2003). The result was in consistence with previous studies (Chang et al. 1999; Slack et al. 2001; Slack and Lawrence 2002; Ogay et al. 2006).
Fig. 4.
Western blot analysis of DPPIV expression from uninfected and infected cells
The enzyme activities of the samples, which were loaded to the Western blot, for uninfected cells were 2.62 ± 0.1 mU mL−1 at 24 h, 7.88 ± 0.24 mU mL−1 at 48 h and 12.79 ± 2.01 mU mL−1 at 72 h, while the activities for infected cells at 24, 48 and 72 h increased to 2.86 ± 0.14, 8.36 ± 0.34 and 291.91 ± 5.73 mU mL−1, respectively.
Next to the transfection time for standard recombinant enzyme production, MOI was another important parameter. First, a logarithmic pattern was formed for MOI as 0.1, 1 and 10 for 24, 48, 72, 96, 120 and 144 h post transfection. hDPPIV expression levels were monitored by measuring the enzyme activity. The enzyme activity time course is shown in Fig. 5.
Fig. 5.
Transfection of Sf9 cells with MOI 0.1, 1 and 10
As shown in Fig. 5, the expression of hDPPIV with MOI 0.1 was similar to MOI 1 and three times higher than MOI 10. The results indicated that the range of MOI 0.1–1 should be studied further for the maximum hDPPIV expression. Therefore, different ratios of MOI were performed as 0.1, 0.25, 0.5, 0.75 and 1 in the range for 24, 48, 72, 96, 120 and 144 h post transfection, respectively (Fig. 6).
Fig. 6.
Transfection of Sf9 cells with MOI 0.1, 0.25, 0.5, 0.75 and 1
The amount of expressed hDPPIV from infected Sf9 cells was found similar from MOI 0.1 to 1.
Baculovirus replication and cytopathic effects
The transfected Sf9 cells showed typical cytopathic effects such as ceasing of cell growth and decreasing of the infected cell viability. The viability of infected and uninfected cells is given in Table 1. There was a significant difference between the number of viable uninfected and infected cells (P < 0.001). Moreover, irregular cell shapes, increasing cell size and lost attachment were observed under inverse phase microscope (Olympus, Japan). At the end of the 30 h post infection, 60 % of adherent cells lost attachment capability to the culture surface. Finally, at 66 h post transfection the 80–90 % of the cells were detached and formed clusters.
Table 1.
Measurement of the viability of infected (MOI 0.1) and uninfected Sf 9 cells by MTT assay
| Time (hour) | Number of the viable cells (cell.104.mL−1) | |
|---|---|---|
| Uninfected Sf9 cells | Infected Sf9 cells | |
| 0 | 1 | 1 |
| 6 | 2.21 | 2.1 |
| 12 | 2.89 | 1.92 |
| 18 | 3.16 | 1.81 |
| 24 | 5.13 | 3.01 |
| 30 | 5.19 | 3.61 |
| 36 | 5.32 | 2.5 |
| 42 | 7.11 | 2.71 |
| 48 | 9.25 | 2.89 |
| 54 | 10.40 | 3.64 |
| 60 | 11.70 | 3.43 |
| 66 | 12.40 | 3.30 |
| 72 | 14.70 | 2.89 |
P < 0.001
The effect of transfection on Sf9 cell morphology
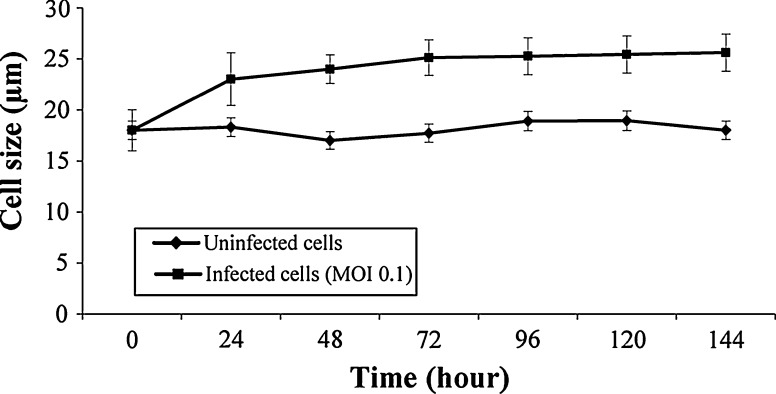
The most distinctive effect of transfection on Sf9 cell morphology was the cell size. As given in Fig. 7, the size of uninfected cells was 18.0 ± 0.67 μm. Significant differences were found between uninfected and infected cell sizes after 24 h post infection (P < 0.001). The infected cells (MOI 0.1) were monitored by using immunofluorescence method for 72 h post infection (Fig. 8). The immunofluoresence images revealed that the percentage of baculovirus increased gradually after 24 h post infection, and hDPPIV was expressed from the transfected Sf9 cells after 48 h post infection.
Fig. 7.
Effect of transfection on Sf9 cell size
Fig. 8.

Immunofluorescence images of baculovirus infected Sf9 cells. DAPI (column 1), gp64 (column 2), CD 26 (column 3) for uninfected cells (line 1), 24 h (line 2), 48 h (line 3) and 72 h post infection (line 4)
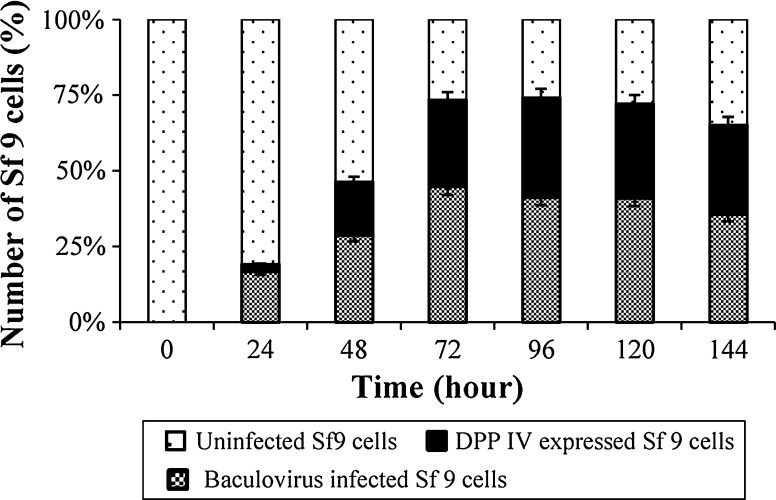
Correlation between the replication of baculovirus and the expression of hDPPIV were detected by using fluorescence intensity of infected cells and uninfected cells. By the way, it was demonstrated that although there was about 20 % increase in baculovirus infection, hDPPIV expression increased only 3 % after 24 h post infection. Expression of the hDPPIV from infected Sf9 cells rose gradually until 72 h post transfection before increasing slightly at 96 h post transfection.
The results indicated that the transfection efficiency, that is, percentage of transfected Sf9 cells in the population, was 81 %. Moreover, the ratio of the number of infected cells (MOI = 0.1), which expressed hDPPIV, to the total infected cells was 80 % (Fig. 9).
Fig. 9.
Correlation between uninfected Sf9 cells, baculovirus infected Sf9 cells and expression of DPPIV (from infected Sf9 cells) by using immunofluorescence images
Discussion
Recently, baculovirus expression system and Sf9 cell line have widely used for recombinant protein production. The hDPPIV gene was successfully expressed in the baculovirus-insect cell system in a related study (Dobers et al. 2002). Examined parameters, MOI and infection time, were expanded to determine optimum MOI value (0.1, 0.25, 0.5, 0.75, 1 and 10) and harvest time (0, 24, 72, 96, 120 and 144 h) in this study.
The entire process was monitored to observe the effects of this approach. In addition to hDPPIV activity and MOI against time, productivity of hDPPIV, and yield of the enzyme determination of the change in the cell size were also carried out. These parameters have been considered as potentially useful items for monitoring infected cell cultures to determine the maximum protein production and to calculate the maximum productivity.
The most critical parameter for maximizing the production is to find an optimal harvest time for transfection of the cells. Therefore, productivity of hDPPIV enzyme had been used to determine the time of maximum protein production (enzyme activity), and optimal harvest time hereby. As mentioned above, since productivity of hDPPIV from infected Sf9 at 72 h was 1.11 fold higher than at 96 h. Therefore, post infection of 72 h was performed as an optimum harvest time (Fig. 2). Related manuscript have been supported the data (Nishikawa et al. 2003; Sandhu et al. 2007).
Monitoring methods may indicate some correlations between cell density, enzyme activity or cell size and infection rate. The data represented that density of the infected cells was similar to the uninfected cells at 0 h. However, the infected cell density decreased gradually until 144 h (Fig. 3). It is thought that density of the infected cell may be related to recombinant baculovirus replication in the cells or the infected cells may change their metabolism in order that expressing of hDPPIV instead of their growth. Hence, the results indicate that there has been an opposite correlation between cell density and infection, namely protein production in infected cells. Whereas Nishikawa et al. (2003) have supported the data, Sander and Harrysson (2007) reported no strong connection between the cell density and infection.
Although a similar pattern was found for infected Sf9 cell line (with MOI = 0.1–1) in Fig. 6, the yields of hDPPIV enzyme per MOI were measured as 2.69 and 0.3 Ucell.pfu−1 mL−1 for MOI = 0.1 and 1, respectively. Therefore, MOI = 0.1 value was performed as optimum MOI for expression of hDPPIV from Sf9 cells. As it is well known, using low MOI is preferable for industrial production because of the low passage effects.
After infection with MOI = 0.1, infected cell viability was checked by MTT assay and any sharp decrease was not observed in viability of infected cells. Therefore, it was thought that baculovirus could not be a lytic virus, but could be a budded virus. The baculovirus species is the most frequently used for baculovirus studies. Brondyk (2009) remarked it as a non-lytic virus.
The average diameter of the cells in the uninfected cultures was 18.0 ± 0.67 μm at the beginning of infection. The average size of uninfected Sf9 cells was reported by Gotoh et al. (2008) as 18.5 ± 1.5 μm, which is in correlation with the current study.
In the current study; Sf9 cells infected with low MOI (0.1) swelled rapidly and reached to a point at average 23 μm diameter at 72 h likewise the study by Gotoh et al. (2008). The average diameters were similar at the beginning of infection for uninfected and infected cells, but the diameter of infected cells increased by 42 % at 72 h post infection (Fig. 7). Sander and Harrysson (2007) reported the increased diameter of infected cell as 28 % with MOI = 0.5 at the same time. It is clearly understood that the cell size of Sf9 could be considered as one of the infection signs.
Increased cell diameter leads to lack of membrane resistant and becomes more fragile against hydrodynamic forces like shear stress. Therefore, it is important to measure the uninfected and infected cell diameters, and calculate possible shear stress as preliminary calculations before large scale production.
The immunofluorescence images supported the findings from the cell diameter and cell density experiments, and also showed baculovirus infected Sf9 cells and expressed hDPPIV. In this study proliferation of transfected Sf9 cell ceased, and cell size increased after 24 h post infection. These findings indicate that viral replication gained acceleration after 24 h post infection, and this situation cause increasing in the number of infected Sf9 cells, which has been also supported by Luckow (1991).
A success in the expression studies depends on the efficiency of infection. Radner et al. (2012) found infection efficiency approximately 20–30 % of Sf9 and Sf21 cells at 72 h, while another related study gave the infection efficiency as 40–60 % (Pham et al. 2003). In the present study, the highest transfection efficiency of recombinant hDPPIV was achieved by 80 % transfected Sf9 at 72 h. High infection efficiency and the short incubation period have offered fast and high yield recombinant protein production by large scale infection of Sf9 cells.
In summary, the findings about optimum harvest time, productivity, infection efficiency, also some negative or positive correlations between cell density, hDPPIV activity and cell size have been manifested. The phenomenons might be a guide for baculoviral hDPPIV producers from Sf9 cells.
The results of research that are described in this report will be performed in the following study that is based on scaling up hDPPIV expression from infected Sf9 cell line.
Acknowledgments
This research was partly supported by grant from Japan Student Service Organization (JASSO).
References
- Aertgeerts K, Ye S, Tennant MG, Kraus ML, Rogers J, Sang BC, Skene RJ, Webb DR, Prasad GS. Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Sci. 2004;13:412–421. doi: 10.1110/ps.03460604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A, Bohuon C. Decrease of serum dipeptidylpeptidase activity in severe sepsis patients: relationship to procalcitonin. Clin Chim Acta. 2002;321:123–126. doi: 10.1016/S0009-8981(02)00042-6. [DOI] [PubMed] [Google Scholar]
- Brondyk W (2009) Selecting an appropriate method for expressing a recombinant protein. Methods Enzymol 463:131–147 [DOI] [PubMed]
- Brudnak M. Genomic multi-level nutrient-sensing pathways. Med Hypotheses. 2001;56:194–199. doi: 10.1054/mehy.2000.1300. [DOI] [PubMed] [Google Scholar]
- Brudnak MA, Rimland B, Kerry RE, Dailey M, Taylor R, Stayton B, Waickman F, Waickman M, Pangborn J, Buchholz I. Enzyme-based therapy for autism spectrum disorders–is it worth another look? Med Hypotheses. 2002;58:422–428. doi: 10.1054/mehy.2001.1513. [DOI] [PubMed] [Google Scholar]
- Celis JE, Carter N, Simons K, Small JV, Hunter T, Shotton D. Cell biology, four-volume set: a laboratory handbook. Waltham: Elsevier Academic Press; 2005. [Google Scholar]
- Chang MJ, Kuzio J, Blissard GW. Modulation of translational efficiency by contextual nucleotides flanking a baculovirus initiator AUG codon. Virology. 1999;259:369. doi: 10.1006/viro.1999.9787. [DOI] [PubMed] [Google Scholar]
- Cunningham DF, O’Connor B. Proline specific peptidases. Biochimica et Biophysica Acta (BBA)—Protein Struct Mol Enzymol. 1997;1343(2):160–186. doi: 10.1016/S0167-4838(97)00134-9. [DOI] [PubMed] [Google Scholar]
- Demuth H, Heins J (1995) Catalytic mechanism of dipeptidyl peptidase IV. Dipeptidyl Peptidase IV (CD26) In: Fleischer B (Ed) Metabolism and the immune response:pp 1–35
- Dobers J, Zimmermann-Kordmann M, Leddermann M, Schewe T, Reutter W, Fan H. Expression, purification, and characterization of human dipeptidyl peptidase IV/CD26 in Sf9 insect cells. Protein Expr Purif. 2002;25:527–532. doi: 10.1016/S1046-5928(02)00043-8. [DOI] [PubMed] [Google Scholar]
- Doumas A, Van Den Broek P, Affolter M, Monod M. Characterization of the prolyl dipeptidyl peptidase gene (dppIV) from the koji mold Aspergillus oryzae. Appl Environ Microbiol. 1998;64:4809–4815. doi: 10.1128/aem.64.12.4809-4815.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effio PC, Folgueras-Flatschart A, Montor W, Pernasetti F, Pueyo M, Sogayar M. Expression of functional Anopheles merusα-amylase in the baculovirus/Spodoptera frugiperda system. Insect Mol Biol. 2003;12:415–425. doi: 10.1046/j.1365-2583.2003.00423.x. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Fukuhara M, Kikuchi KI. Mathematical model for change in diameter distribution of baculovirus-infected Sf-9 insect cells. Biochem Eng J. 2008;40:379–386. doi: 10.1016/j.bej.2008.01.008. [DOI] [Google Scholar]
- Hitchman RB, Possee RD, King LA. Baculovirus expression systems for recombinant protein production in insect cells. Recent Pat Biotechnol. 2009;3:46–54. doi: 10.2174/187220809787172669. [DOI] [PubMed] [Google Scholar]
- Ikonomou L, Schneider YJ, Agathos S. Insect cell culture for industrial production of recombinant proteins. Appl Microbiol Biotechnol. 2003;62:1–20. doi: 10.1007/s00253-003-1223-9. [DOI] [PubMed] [Google Scholar]
- Iwaki-Egawa S, Watanabe Y, Kikuya Y, Fujimoto Y. Dipeptidyl peptidase IV from human serum: purification, characterization, and N-terminal amino acid sequence. J Biochem. 1998;124:428–433. doi: 10.1093/oxfordjournals.jbchem.a022130. [DOI] [PubMed] [Google Scholar]
- Jalving R, Godefrooij J, Veen WJ, Ooyen AJJ, Schaap PJ. Characterisation of the Aspergillus niger dapB gene, which encodes a novel fungal type IV dipeptidyl aminopeptidase. Mol Genet Genomics. 2005;273:319–325. doi: 10.1007/s00438-005-1134-9. [DOI] [PubMed] [Google Scholar]
- Lambeir AM, Proost P, Scharpé S, Meester ID. A kinetic study of glucagon-like peptide-1 and glucagon-like peptide-2 truncation by dipeptidyl peptidase IV, in vitro. Biochem Pharmacol. 2002;64:1753–1756. doi: 10.1016/S0006-2952(02)01415-6. [DOI] [PubMed] [Google Scholar]
- Langford WS (2003) A comprehensive guide to managing autism. The Autism File. Special Supplement. Slightly changed by Kees de Vries, Drunen, Holland
- Luckow V. Cloning and expression of heterologous genes in insect cells with baculovirus vectors. In: Prokop A, Bajpai RK, Ho C, editors. Recombinant DNA technology and applications. New York: McGraw-Hill; 1991. pp. 97–152. [Google Scholar]
- Mentlein R. Dipeptidyl-peptidase IV (CD26)-role in the inactivation of regulatory peptides. Regul Pept. 1999;85:9–24. doi: 10.1016/S0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- Nishikawa N, Yamaji H, Fukuda H, Dong X, Teruya K, Katakura Y, Zhang Y, Miura T, Daimon Y, Mori T. Preface 1–2 recombinant protein production by the baculovirus–insect cell system in basal media without serum supplementation. Cytotechnology. 2003;43:167–168. doi: 10.1023/B:CYTO.0000039894.27256.0f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogay I, Lihoradova O, Azimova SS, Abdukarimov A, Slack J, Lynn D. Transfection of insect cell lines using polyethylenimine. Cytotechnology. 2006;51:89–98. doi: 10.1007/s10616-006-9022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öngen G, Sargin S, Üstün Ö, Kutlu C, Yücel M. Dipeptidyl peptidase IV production by solid state fermentation using alternative fungal sources. Turk J Biol. 2012;36:665–671. [Google Scholar]
- Ota T, Itoh A, Tachi H, Kudoh K, Watanabe T, Yamamoto Y, Tadokoro T, Maekawa A. Synthesis of morphiceptin (Tyr-Pro-Phe-Pro-NH2) by dipeptidyl aminopeptidase IV derived from Aspergillus oryzae. J Agric Food Chem. 2005;53:6112–6116. doi: 10.1021/jf050420d. [DOI] [PubMed] [Google Scholar]
- Pérez-Guzmán AE, Cruz y Victoria T, Cruz-Camarillo R, Hernández-Sánchez H. Improvement of fermentation conditions for the production of X-prolyl-dipeptidyl aminopeptidase from Lactococcus lactis. World J Microbiol Biotechnol. 2004;20:413–417. doi: 10.1023/B:WIBI.0000033066.86658.34. [DOI] [Google Scholar]
- Pham PL, Perret S, Doan HC, Cass B, St-Laurent G, Kamen A, Durocher Y. Large-scale transient transfection of serum-free suspension-growing HEK293 EBNA1 cells: peptone additives improve cell growth and transfection efficiency. Biotechnol Bioeng. 2003;84:332–342. doi: 10.1002/bit.10774. [DOI] [PubMed] [Google Scholar]
- Radner S, Celie PHN, Fuchs K, Sieghart W, Sixma TK, Stornaiuolo M (2012) Transient transfection coupled to baculovirus infection for rapid protein expression screening in insect cells. J Struct Biol 179:46–55 [DOI] [PubMed]
- Reichelt K, Knivsberg A. Can the pathophysiology of autism be explained by the nature of the discovered urine peptides? Nutr Neurosci. 2003;6:19–28. doi: 10.1080/1028415021000042839. [DOI] [PubMed] [Google Scholar]
- Reichelt K, Hole K, Hamberger A, Saelid G, Edminson P, Braestrup C, Lingjaerde O, Ledaal P, Orbeck H. Biologically active peptide-containing fractions in schizophrenia and childhood autism. Adv Biochem Psychopharmacol. 1981;28:627. [PubMed] [Google Scholar]
- Reinhold D, Goihl A, Wrenger S, Reinhold A, Kühlmann UC, Faust J, Neubert K, Thielitz A, Brocke S, Täger M. Role of dipeptidyl peptidase IV (DP IV)-like enzymes in T lymphocyte activation: investigations in DP IV/CD26-knockout mice. Clin Chem Lab Med. 2009;47:268–274. doi: 10.1515/CCLM.2009.062. [DOI] [PubMed] [Google Scholar]
- Sander L, Harrysson A. Using cell size kinetics to determine optimal harvest time for Spodoptera frugiperda and Trichoplusia ni BTI-TN-5B1-4 cells infected with a baculovirus expression vector system expressing enhanced green fluorescent protein. Cytotechnology. 2007;54:35–48. doi: 10.1007/s10616-007-9064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Naciri M, Al-Rubeai M. Prediction of recombinant protein production in an insect cell–baculovirus system using a flow cytometric technique. J Immunol Method. 2007;325:104–113. doi: 10.1016/j.jim.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Shattock P, Kennedy A, Rowell F, Berney T (1990) Role of neuropeptides in autism and their relationships with classical neurotransmitters. Brain Dysfunction 3:328–345
- Slack JM, Lawrence SD. Purification of DNA for the transfection of a Spodoptera frugiperda cell line. Method Cell Sci. 2002;24:155–163. doi: 10.1023/A:1024413604663. [DOI] [PubMed] [Google Scholar]
- Slack JM, Dougherty EM, Lawrence SD. A study of the Autographa californica multiple nucleopolyhedrovirus ODV envelope protein p74 using a GFP tag. J Gen Virol. 2001;82:2279–2287. doi: 10.1099/0022-1317-82-9-2279. [DOI] [PubMed] [Google Scholar]
- Summers MD, Smith GE (1987) A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station Bulletin, No 1555
- Tachi H, Ito H, Ichishima E. An X-prolyl dipeptidyl-aminopeptidase from Aspergillus oryzae. Phytochemistry. 1992;31:3707–3709. doi: 10.1016/S0031-9422(00)97513-7. [DOI] [Google Scholar]
- Tanaka T, Camerini D, Seed B, Torimoto Y, Dang N, Kameoka J, Dahlberg H, Schlossman S, Morimoto C. Cloning and functional expression of the T cell activation antigen CD26 [published erratum appears in J Immunol 1993 Mar 1; 150 (5): 2090] J Immunol. 1992;149:481–486. [PubMed] [Google Scholar]
- Thoma R, Löffler B, Stihle M, Huber W, Ruf A, Hennig M. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure. 2003;11:947–959. doi: 10.1016/S0969-2126(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Urade M, Uematsu T, Mima T, Ogura T, Matsuya T. Serum dipeptidyl peptidase (DPP) IV activity in hamster buccal pouch carcinogenesis with 9, 10-dimethyl-1, 2-benzanthracene. J Oral Pathol Med. 2006;21:109–112. doi: 10.1111/j.1600-0714.1992.tb00992.x. [DOI] [PubMed] [Google Scholar]
- Üstün Ö, Öngen G (2012) Production and separation of dipeptidyl peptidase IV from Lactococcus lactis: scale up for industrial production. Bioprocess Biosyst Eng 35(8):1417–1427 [DOI] [PubMed]
- Wilkinson RE, Houston DB (2001) Compositions containing dipepitidyl peptidase IV and tyrosinase or phenylalaninase for reducing opioid-related symptons. U.S. Patent No 6,251,391. U.S. Patent and Trademark Office, Washington, DC