Abstract
Selecting a suitable nano-liquid chromatography system (LC), ionization source and mass spectrometer for LC–tandem mass spectrometry (MS-MS) studies is complicated by numerous competing technologies. This study compares four popular nano-LC systems, four ionization sources and three MS facilities that use completely different LC–MS-MS systems. Statistically significant differences in LC performance were identified with similarly performing Proxeon, Waters and Eksigent nanoLC-Ultra systems [retention time routinely at 0.7–0.9% relative standard deviation (RSD)], and all outperformed the Eksigent nanoLC-2D (RSD ∼2%). In addition, compatibility issues were identified between the Bruker HCT ion trap mass spectrometer and both the Eksigent nanoLC-2D and the Bruker nanoelectrospray source. The electrospray source itself had an unexpected and striking effect on chromatographic reproducibility on the Bruker HCT ion trap. The New Objective nanospray source significantly outperformed the Bruker nanospray source in retention time RSD (1% RSD versus 14% RSD, respectively); and the Bruker nebulized nanospray source outperformed both of these traditional, non-nebulized sources (0.5% RSD in retention time). Finally, to provide useful benchmarks for overall proteomics sensitivity, different LC–MS-MS platforms were compared by analyzing a range of concentrations of tryptic digests of bovine serum albumin at three MS facilities. The results indicate that similar sensitivity can be realized with a Bruker HCT-Ultra ion trap, a Thermo LTQ-Velos Linear ion trap and a Thermo LTQ-Orbitrap XL-ETD.
Introduction
Proteomics plays a central role in characterizing post-translational modifications and in biomarker discovery, protein identification and protein–protein interaction (1–4). These studies typically involve liquid chromatography (LC) coupled to nanoelectrospray ionization mass spectrometry (MS) (5–7). Proteomics studies (8–10) depend upon LC for both sample cleanup and to increase the dynamic range of analysis through chromatofocusing of dilute analytes and separating abundant and rare analytes (11–12). Proteomics depends upon MS and tandem MS (MS-MS) for determining protein/peptide sequences and molecular composition.
Previous proteomics benchmarking efforts: Sample preparation, ionization sources and chromatography
Different segments of the proteomics platform (LC, ionization source and mass spectrometer) affect the quality of proteomics data. Previous studies evaluated sample loading conditions (13), column chemistry or stationary and mobile phases (14), spray stability (15), lower detection limits (16–17), upper limits for protein identifications in terms of peak capacity (18), chromatographic reproducibility (19), mass spectrometer platforms (20–21), database scoring algorithms (20, 22) and the sequence coverage of various platforms (23). Sample preparation is the sine qua non of proteomics (24–25), including quantitative proteomics (16), and has also been optimized in recent studies. Burgess et al. examined five electrospray ionization (ESI) tip emitters for sensitivity and reproducibility in ion intensity and found that the stainless steel nanospray needle provided the highest overall sensitivity, but lacked reproducibility for the number of identified proteins (23). In addition, Smith and coworkers examined optimal electrospray voltages during LC–MS gradients with increasing organic concentration and developed algorithms monitoring spray feedback (15). Two-dimensional (2D)-LC approaches were evaluated by Gilar and coworkers, who found that SCX-RP, HILIC-RP and RP-RP 2D systems provided orthogonality (26). In comparing matrix-assisted laser desorption/ionization (MALDI) versus ESI, Shirran and Botting found that more accurate quantitative proteomics measurements resulted when using nanoLC–MALDI-MS-MS than nanoLC–ESI-MS-MS (22).
Previous proteomics benchmarking efforts: Mass spectrometers
Both Han et al. (27) and Syka et al. (28) compared the performance of first generation Paul-type ion traps (LCQ) to second generation linear ion traps (LTQ) and found that linear ion traps performed with marked superiority, yielding 4–6-fold increases in the number of peptide and protein identifications. Smith and coworkers also demonstrated improved performance of the LTQ when studying the S. oneidensis proteome, achieving approximately twice the proteome sequence coverage than the LCQ (11). This difference was attributed to the higher sensitivity of the LTQ, because doubling the MS-MS analysis time did not significantly increase proteome coverage. More recently, a large-scale analysis by Tabb et al. evaluated four Thermo LTQs and four Orbitrap instruments, and found that Orbitraps normally demonstrate higher reproducibility and repeatability, but paradoxically, had irregular performance (29). They also found that among the same platforms, reproducibility among Orbitraps lagged behind repeatability. Gygi and coworkers compared the performance of hybrid quadrupole time-of-flight instruments with linear ion traps, demonstrating similar performances in protein identification, and improved protein and proteome coverage using these complementary MS instruments (20).
Previous proteomics benchmarking efforts: Acquisition parameters and data analysis
The effects of acquisition and search parameters upon performance cannot be overemphasized; these effects confound interplatform comparisons. For example, the difference in performance following the optimization of a given platform can exceed interplatform differences. Kalli and Hess performed systematic investigations of MS instrument settings during data-dependent acquisition mode on a Thermo LTQ-Orbitrap to evaluate their influences on peptide and protein identification rates, such as the automatic gain control target value for MS and MS-MS and maximum ion injection time for MS-MS (30). Andrews and coworkers used the design of experimental platform for evaluation of nine LTQ-Orbitrap MS-MS instrument parameters, ultimately affording an increase of ∼60% in proteome coverage, with parameters such as ionization time, tandem MS event monoisotopic precursor selection, capillary temperature and tube lens voltage showing the largest impact. (31). Researchers from Human Proteome Organization (HUPO) published the results of a multi-laboratory study to illustrate methodological challenges for MS-based proteomics studies, and their centralized data analysis revealed missed protein identifications and database matching problems, illustrating that data analysis, perhaps more than any other factor, limits reproducibility (32).
Benchmarking LC systems
The limited quantity of material present in many biological samples, coupled with the sensitivity gains associated with nano-ESI, necessitate nanoflow chromatography. Moreover, limited solvent availability and environmental effects have made splitless flow LCs invaluable. The majority of recent benchmarking studies, including those described previously, considered the back-end analyses of the mass spectrometer or data processing. This study presents efforts to examine and optimize the front-end chromatography and ionization source while concurrently evaluating MS platforms. Specifically, four splitless flow nano HPLC systems were considered: Eksigent nanoLC-2D and nanoLC-Ultra, Proxeon EASY nLC and Waters Corporation nanoACQUITY UPLC; and four ionization sources: Bruker Daltonics ESI, nebulized nanoFlow ESI, and nano ESI, and New Objective nano ESI. Finally, three mass spectrometers were evaluated: Bruker Daltonics HCT-Ultra PTM Discovery ion trap, Thermo Scientific LTQ-Velos Linear ion trap and Thermo Scientific LTQ-Orbitrap XL-ETD. It is demonstrated that differences do exist in terms of sensitivity and retention time reproducibility between the LCs, and especially ionization sources. MS platforms did not reveal striking differences.
Experimental
Instrumentation and reagents
For LC studies, the nano-LC systems were coupled online with an HCT-Ultra PTM Discovery ion trap (Bruker Daltonics, Billerica, MA); the nanosource used for these LC studies was the PicoView 200 (New Objective, Woburn, MA). LCs included: (i) Eksigent nanoLC-2D (currently part of AB SCIEX, Dublin, CA); (ii) Proxeon EASY-nLC (now Thermo Scientific, West Palm Beach, FL); (iii) Eksigent nanoLC-Ultra; and (iv) Waters nanoACQUITY UPLC (Milford, MA). For ionization source studies, three sources from Bruker Daltonics, nano ESI (PN: 554189), nebulized nanoFlow ESI sprayer (PN: 255780) and ESI source (PN: G1548A), and the New Objective nano source (PN: PV-200) were all attached to the Bruker HCT-Ultra PTM Discovery ion trap. The LC used for all source experiments was the Waters nanoACQUITY ultra-performance liquid chromatography (UPLC). For facility comparison studies, Facility A used Agilent 1200 HPLC (Santa Clara, CA) coupled to an LTQ-Velos Linear ion trap with the standard nano source (Thermo Scientific); Facility B used Proxeon EASY-nLC (Thermo Scientific) HPLC connected to a hybrid LTQ-Orbitrap XL-ETD mass spectrometer through the standard nano source (Thermo Scientific); the authors' group administrates Facility C, the Brandeis University Mass Spectrometry Facility (BUMS), which used a Waters nanoACQUITY UPLC coupled to HCT-Ultra PTM Discovery ion trap with nanoFlow ESI sprayer.
LC mobile phase solvents and sample dilutions used 0.1% formic acid in water (Buffer A) and 0.1% formic acid in acetonitrile (Buffer B) (Chromasolv LC–MS grade; Sigma-Aldrich, St. Louis, MO). Tryptic digested bovine serum albumin (BSA) (lyophilized) (Bruker Daltonics) was prepared to 1 pmole/μL stock concentrations. BSA stock solution was diluted to the following amounts of sample for 1 µL injection: 250, 125, 63 and 16 fmole (LC and MS platform studies); 50, 25, 10, 5, 1 and 0.5 fmole (ionization source studies).
Methods
LC studies
Five replicates were run at each quantity of BSA digest for LC comparisons, unless otherwise noted. The columns used for the LC studies were a C18-reversed phase EASY-Column (10 cm × 75 µm i.d., 3 µm beads, 120 Å pore size from Thermo Fisher Scientific) or an in-house packed C18 column [10 cm × 75 µm i.d., 5 µm beads, 300 Å pore size using TARGA C18 packing material (The Nest Group, Inc., Southborough, MA)] used in the Waters LC study.
For the LC studies, the flow rate was 325 nL/min with the following gradient: 3% Buffer B at 0 min, linearly increased to 35% B at 48 min, followed by 5 min washing at 95% B from 49 to 54 min (for the Eksigent nanoLC-2D, Proxeon EASY-nLC and Waters nanoACQUITY UPLC); and 3% Buffer B at 0 min, linearly increased to 35% B at 30 min, followed by 5 min washing at 95% B from 32 to 37 min (Eksigent nanoLC-Ultra); all followed by more than 15 column volume re-equilibrations. The ion trap capillary temperature was set to 180°C and the dry gas flow was 8 L/min. Capillary voltage was set to the lowest voltage required to obtain a stable spray at 30% B (dependent on LC used), which ranged from 950–1,250 V. The ion trap was set to acquire in positive ion mode, scanning in the manufacturer-specified standard enhanced mode (8,100 m/z/s) between m/z 300 and 1,400 for MS, averaging five spectra, and accumulated either 200,000 charges [by ion charge control (ICC)] or for 200 ms, whichever came first. Collision induced dissociation (CID) fragmentation was performed on the four most intense ions within m/z 300–1,200, with the threshold for precursor ion selection set at an absolute intensity of 20,000. Singly charged ions were excluded and doubly charged ions were preferred. Strict active exclusion was used and a precursor ion was excluded after one spectrum, and then released after 0.1 min. MS-MS spectra were scanned from m/z 100–2,800, averaging three spectra.
Ionization source studies
The columns used for the ionization source studies were in-house packed C18-reversed phase column (TARGA C18 packing material): ∼15 cm × 75 µm i.d., 5 µm beads, 300 Å pore size. The sample was loaded over a heated column at 35°C in the heating and trapping module of the nanoACQUITY UPLC system; samples were run in triplicate. The flow rate was 350 nL/min (nanoESI tests) or 3 µL/min (ESI tests) with the following gradient: 0 min 5% Buffer B, 55 min 60% B, followed by 11 column volume re-equilibrations. The ion trap parameter setup was the same as described previously, with the following differences: the drying temperature set to 300°C, the dry gas was set to 5 L/min and the nebulizer gas was set to 10 psi for both the ESI source and the nebulized nanoFlow ESI sprayer. The CID fragmentation threshold for precursor ion selection was an absolute intensity of 10,000 and between m/z 300–1,400. The active exclusion set for precursor ion excluded after two spectra and released after 1 min, averaging three spectra for MS-MS.
Facility comparison studies
The gradient used to elute peptides at Facility A was 5% Buffer B (97% acetonitrile–2.9% water–0.1% formic acid) at 0 min to 35% B at 17min; Buffer A was 3% acetonitrile–96.9% water–0.1% formic acid. Facility B eluted peptides at a flow rate of 275 nL/min with a PicoFrit column [15 cm × 75 µm i.d. × 15 µm (tip); New Objective], self-packed with Magic C18 resin (Michrom Bioresources). The gradient began after equilibration with Buffer A (0.1% formic acid–0.9% acetonitrile–99% water) from 5% Buffer B (acetonitrile) to 38% B, followed by 95% B for washing. The authors' group administrates Facility C, the BUMS, which used an in-house packed C18 column (∼15 cm × 75 µm i.d., 5 µm beads, 300 Å pore size) (TARGA C18 packing material) with the following gradient: 0 min 5% Buffer B, 55 min 60% B, followed by 11 column volume re-equilibrations (Buffer A: 0.1% formic acid in water; Buffer B: 0.1% formic acid in acetonitrile).
Data analysis
All mass spectra were processed by DataAnalysis V. 4.0 and BioTools V. 3.0 (Bruker Daltonics), including an in-house Mascot server (Matrix Science Inc., Boston, MA) database search against SwissProt database, version 9.6. In DataAnalysis, compound lists were generated by setting the compound detection intensity threshold to 10,000 and reporting a maximum of 250 detected compounds. Compound lists were exported as Mascot generic files (mgf) for further processing in BioTools, in which data were subjected to a Mascot search (33) against the SwissProt database for mammals, which has ∼65,549 sequences, with less than 5% false discovery rate at the peptide level [enzyme: trypsin, allowed up to one missed cleavage; fixed modification: Carboxymethyl (C); peptide tolerance: ±1.5 Da; MS-MS tolerance: ±0.7 Da]. High-scoring peptide ions were selected for further retention time analyses. Statistical analysis was conducted using Microsoft Excel V. 2003 and 2007 (Redmond, WA) and weighted linear regressions graphed in SPSS V. 18 (IBM, Somers, NY). Analysis of variance (ANOVA) data swarm plots were graphed in http://www.physics.csbsju.edu/stats/anova_pnp_NGROUP_form.html.
Analysis for data from other facilities
To standardize the interpretation of raw data from all of the facilities, the raw data files were loaded into Mascot Distiller and generated mgf files by selecting “peak pick all scans” with default parameters for searching via Mascot Distiller V. 2.3.2.0, picking 2,800–4,000 compounds per raw file. Generated files were then subjected to the same parameters described previously for Mascot searches.
Results and Discussion
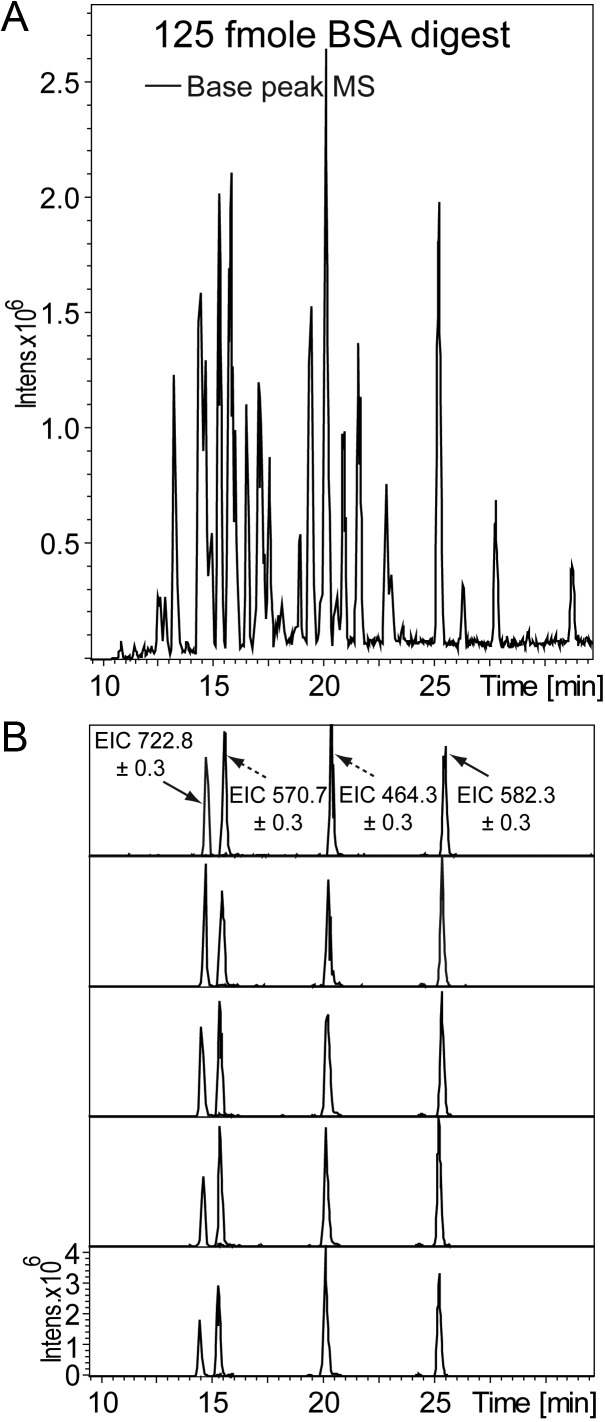
Recognizing that splitless flow LC systems, by virtue of their decreased solvent consumption, are both environmentally friendly and less expensive to operate, four commercially available LCs were evaluated (Eksigent nanoLC-2D, Proxeon EASY-nLC, Eksigent nanoLC-Ultra and Waters nanoACQUITY UPLC). As a means of assessing the relative performance of these four LCs, the data obtained from LC–MS-MS analyses of trypsin digested BSA (Uniprot accession number P02769) were compared. A commercially available BSA tryptic digest was chosen because BSA is among the most common benchmarks for LC–MS-MS systems for retention time analysis, MS mass accuracy and MS-MS functionality and calibration checks. Reversed–phase (RP) chromatography (34) was selected to separate the complex peptide mixture, which is commonly used to desalt, concentrate and fractionate proteomics samples (12). The field service engineers for each LC manufacturer installed and serviced the HPLCs before analysis to ensure that the instruments were performing optimally. BSA was confidently identified by Mascot when the p-value was <0.05 and ≥2 peptides were characterized. Figure 1A illustrates a typical nano-chromatography base peak chromatogram, consisting of ∼30 identified and unique BSA peptides. Five replicates of 125 fmole BSA analyzed on a Proxeon EASY-nLC coupled to a New Objectives nano source illustrate the repeatability, with 61% of the total identified peptides observed in every run, and ∼87% in at least three of five runs. Repeatability is also emphasized by extracted ion chromatograms (EIC) of four representative peptides in Figure 1B. The four peptides (m/z 570.7 = CCTESLVNR, m/z 464.3 = YLYEIAR, m/z 582.3 = LVNELTEFAK, m/z 722.8 = YICDNQDTISSK, all with ion charge 2+) were selected for retention time analysis because they had highly confident ion scores and well separated retention times that spanned the chromatography gradient.
Figure 1.
NanoLC repeatability: An example of a typical LC run shows peptide separation for trypsin digested BSA; 125 fmole digested BSA was analyzed on a Proxeon EASY-nLC coupled to a New Objectives nano source; and base peak chromatogram of MS (solid line) is shown (A); illustration of repeatability for LC runs (spectra adapted from replicates of 125 fmole digested BSA analyzed on the setup described in Figure 1A); shown here are EICs for four peptides graphed on the same intensity axis, using the same quantity of BSA as in Figure 1A (B). These EICs were used in the total evaluation of the retention times, with examples shown in Supplementary Table I. Details of MS and MS-MS for m/z 570.7 are shown in Supplementary Figure 1.
HPLC retention times
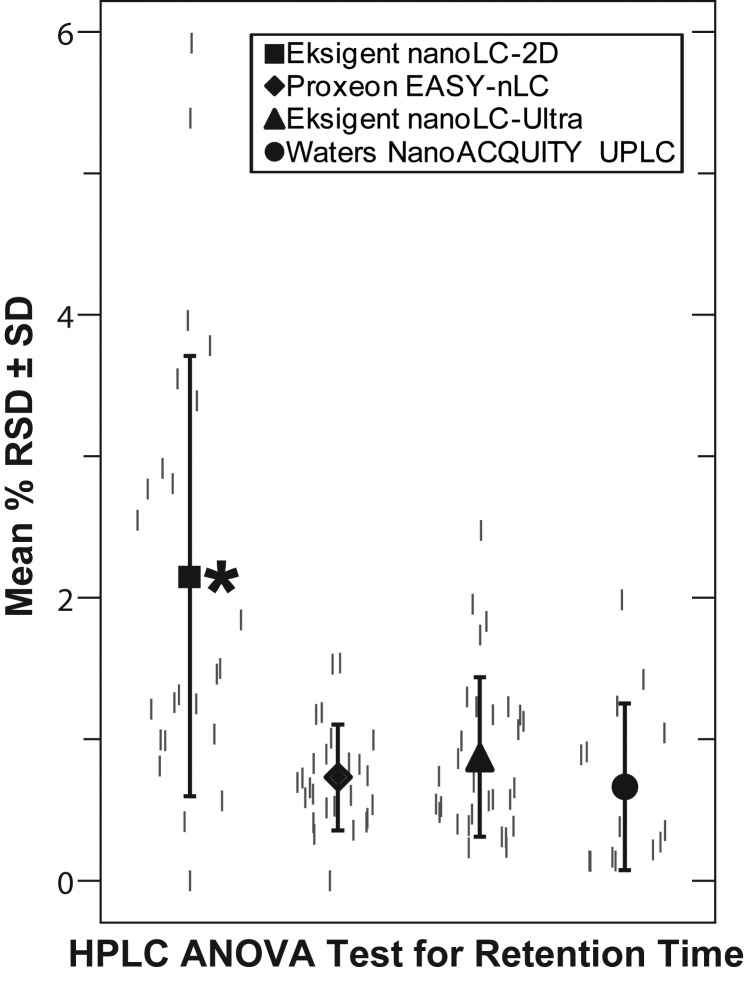
The principle metric of LC performance is retention time repeatability. LC retention times were processed to relative standard deviation (RSD) to compare the different systems because the retention times varied based upon system void volumes. The RSDs (%) of the Eksigent nanoLC-Ultra, Proxeon EASY-nLC and Waters nanoACQUITY UPLC systems were 0.87, 0.73 and 0.66, respectively, and were statistically indistinguishable (Figure 2). These three LCs, however, outperformed the Eksigent nanoLC-2D (denoted by an asterisk), which operated with the least reproducibility (2.2% RSD confirmed by ANOVA). The retention time RSDs were within the range of other proteomics analyses (Supplementary Table I). For example, normalized elution times for all observed peptides of multiple, large scale proteomics analyses of A. thaliana chloroplast, D. radiodurans and S. oneidensis were between 2.4–2.7% RSD using one-dimensional (1D) chromatography (19, 35–37). Using 2D chromatography, Smith and coworkers were able to achieve 0.56% RSD on virion peptides (38). Manufacturers and many researchers tend to report the RSD of only a single peak. The best single-peak RSDs in this study for m/z 582.3 were 0.14% for Waters nanoACQUITY UPLC, 0.33% for Proxeon EASY-nLC, 0.23% for Eskigent nanoLC-Ultra and 0.42% for Eksigent nanoLC-2D, which were all within the manufacturers' specifications and similar to the manufacturers' application notes. For example, the Waters nanoACQUITY UPLC reported a result of 0.14% RSD [median value using 1.7 µm packing material (39)—the measured value in this study was 0.14%]; Proxeon reported results of 0.27% and 0.30% RSDs for two peptide features [m/z 585.8 and 740.4 (40)—the measured value in this study was 0.33%]; Eksigent reported results of <0.3% RSD (41) for four peptide features (Eksigent nanoLC-Ultra—the measured value in this study was 0.23%) and an average of 0.35% RSD [Eksigent nanoLC-2D (42)—the measured value in this study was 0.42%]. A potential caveat is that the Waters LC study was conducted using a different C18 column. However, the Eksigent nanoLC-2D was re-tested using the same column as used with the Waters study and similar sensitivity to previous analyses was noted (data not shown). Additionally, due to time constraints (instrument on-loan during a demo) the gradient change for the Eksigent nanoLC-2D Ultra was increased to 1.06% B/min, whereas the gradients for the other LCs were 0.66%B/min. To address this, the Waters LC gradient was changed from 0.66 to 1%, which had the following effect upon RSDs: 0.66% RSD at 0.66%/min versus 0.99% RSD at 1%/min.
Figure 2.
Comparison retention time reproducibility of HPLCs. The percentage RSDs of four selected peptides' (shown in Figure 1B) retention times were compared among the LCs. ANOVA demonstrates that the Eksigent nanoLC-2D/Bruker HCT combination (denoted by an asterisk) had significantly less reproducibility than other LCs. This lack of reproducibility resulted from the occasional (but repeatable) catastrophic run in which all elution times shift by ∼10 min and is not the result of the nanoLC-2D per se, but from the combination of the nanoLC-2D and Bruker Ion Trap. It was accompanied by electrical etching of the autosampler stator, and therefore, appears to involve an improper path to the ground. Small gray lines represent individual LC retention time data points around the mean RSD on the data swarm plot.
Lack of compatibility between Bruker HCT Ultra and Eksigent nanoLC-2D
After this study was performed, a progressive and reproducible decline was noted in the compatibility of the Eksigent nanoLC-2D and the Bruker HCT-ion trap, which was proposed to be caused by less effective sample ionization and entry into the mass spectrometer resulting from insufficient grounding. Consistent with this interpretation, electrical etching of the stator face was observed within the 6-port valve of the Eksigent autosampler, suggesting an issue with path-to-ground. This is not strictly related to the Eksigent nanoLC-2D, because it continues to function on the facility's Fourier transform mass spectrometer (also from Bruker Daltonics), and it is not strictly an MS issue, because the facility's HCT-Ultra continues to function with equal performance with the Waters nanoACQUITY UPLC. Specifically, reproducible and catastrophic declines in sensitivity were observed when using the Eksigent nanoLC-2D with the Bruker HCT-Ultra and using identical column, ionization source and instrument conditions that worked with other LCs. The same lack of compatibility was experienced with two different Eksigent nanoLC-2D instruments; the experiments presented here were acquired using a replacement of the first unit. Data analyzed for the Eksigent nanoLC-2D shown in Figure 2, therefore, represents a best-case scenario. The comparison of ionization sources discussed in the following involved only the Waters LC.
Ionization source sensitivity and effects upon chromatographic reproducibility
The advantages of nano ESI include the high sensitivity resulting from efficient ionization (43) and the low solvent consumption resulting from low solvent flow rates (100–400 nL/min). Online nano ESI spray sources were developed to couple a mass spectrometer with a capillary nano-LC, which typically require significant optimization of the x, y and z position of the spray needle and the spray voltage. This study evaluated two nano ESI sources (nanospray source from Bruker Daltonics and nano ESI source from New Objective), one electrospray source (ESI source from Bruker Daltonics) and one hybrid nano-electrospray source (nebulized nanoFlow ESI source from Bruker Daltonics) for their sensitivity and associated chromatography. The nebulized nanoFlow ESI source from Bruker Daltonics needs to be connected to a pressurized nebulizing gas source. It operates as a direct mechanical replacement for the standard ESI sprayer, works at the same flow rates as an online nano ESI source, and does not require x, y, z optimization.
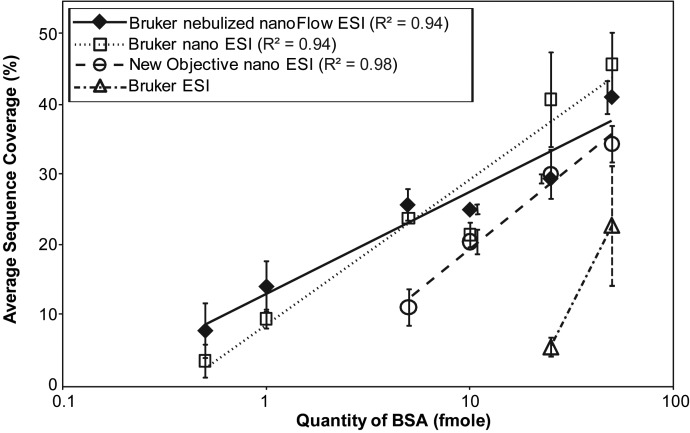
Figure 3 illustrates the comparison of sensitivity for the four tested ionization sources. Linear regression analysis indicated the Bruker nebulized nanoFlow-ESI and nano-ESI sources performed similarly and were slightly more sensitive than the New Objective source. As expected, all three nano-flow rate sources outperformed the higher flow rate ESI. This is the first literature report for a hybrid ESI-nanospray source, and by virtue of its similarity to nanospray (and not ESI) sensitivity, it is proposed that this source has nano-spray-like operating principles. The retention times of selected peptides were compared (m/z 570.7 = CCTESLVNR, m/z 464.3 = YLYEIAR, m/z 582.3 = LVNELTEFAK, m/z 722.8 = YICDNQDTISSK, all with ion charge 2+) to check the quality of the chromatography (Supplementary Figure 2) and a marked lack of reproducibility was observed in the Bruker nano ESI source (14% RSD) when compared to the other nano sources (<1.4% RSD on average). These results were consistent with the authors' qualitative experiences from at least 10 different attempts. Nonetheless, these experiments were repeated and ∼11–17% RSD was observed.
Figure 3.
Comparison of the sensitivity of ionization sources. The average protein sequence coverage of ionization sources is compared at respective amounts of BSA (in fmole). The data points at each concentration represent the average of three replicates. Comparisons were graphed on a log scale and error bars (95% confidence interval of the standard error) are offset to the sides for easier visualization of data points.
Mass spectrometry platform sensitivity
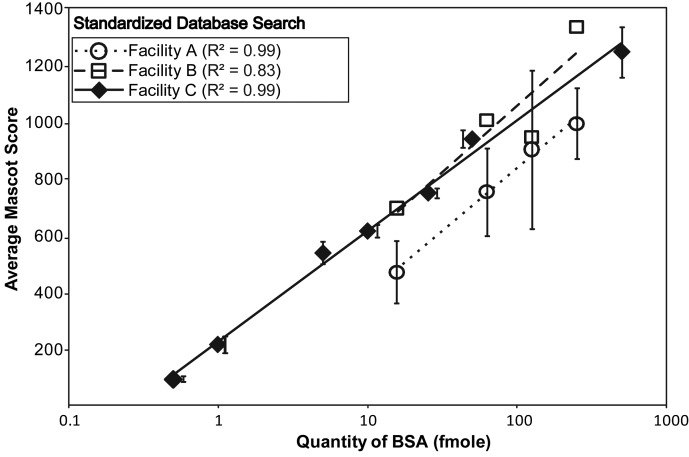
To compare entirely different LC–MS-MS platforms, three facilities were provided with various concentrations of identical BSA standards. When raw Sequest (44–45) data was provided, it was converted to the same format as the Bruker HTC-Ultra ion trap data using Mascot Distiller. Several studies (33, 46–47) have determined that when comparing data from different platforms, Mascot scoring trends can vary from the sequence coverage, and so both are shown in Figure 4 and Supplementary Figure 3. No statistically significant difference was observed between the Agilent 1200-LTQ-Velos Linear ion trap (Facility A), the Proxeon-LTQ-Orbitrap XL-ETD (Facility B) and the Waters nanoACQUITY UPLC-Bruker HCT-Ultra (Facility C) (Figure 4). Prior to the standardization of data processing, significantly lower sequence coverage was observed for Facility B in Supplementary Figure 3B, confirming the importance of data processing in overall sensitivity, as described in the recent study by HUPO (32).
Figure 4.
Comparison of the sensitivity of facilities and different mass spectrometer platforms. Using standardized data analysis, Mascot scores of data from Facility A (Agilent 1200 HPLC coupled to LTQ-Velos Linear ion trap with standard nano source), Facility B (Proxeon EASY-nLC coupled to hybrid LTQ-Orbitrap XL-ETD mass spectrometer with standard nano source) and Facility C (Waters nanoACQUITY UPLC coupled to HCT-Ultra PTM Discovery ion trap with nebulized nanoFlow ESI source) are compared. There are no replicates for data from Facility B.
Conclusions
The authors' group has invested thousands of hours and considerable resources in assessing numerous nanoflow LCs, ionization sources and MS instrument platforms. Given the improved performance of certain systems, and especially the lack of compatibility of certain LCs and sources with certain mass spectrometers, it was necessary to share this analysis. For example, the coupling of either the Eksigent nLC-2D or the Bruker nano ESI source to a Bruker HCT ion trap led to irregular performance. The Bruker source is unusual because it operates with the opposite polarity compared to most sources (the electrospray tip is ground) and it contains proprietary components. Therefore, it is unlikely that these compatibility issues translate to other mass spectrometers. Overall, the Eksigent nLC-Ultra, Proxeon EASY-nLC and Waters nanoACQUITY UPLC performed similarly, with each outperforming the Eksigent nanoLC-2D. For the tested ionization sources, the Bruker nano ESI had similar sensitivity to the other three sources, but highly irreproducible retention times. The overall preference for was the nebulized nanoFlow ESI (nL/min flow), which had similar sensitivity to other nanospray sources but did not require adjustment or optimization. This evaluation of three LC–MS-MS platforms should provide useful sensitivity benchmarks and indicates that the Bruker HCT-Ultra PTM Discovery ion trap, Thermo Scientific LTQ-Velos Linear ion trap and Thermo Scientific LTQ-Orbitrap XL-ETD offer similar sensitivities. This study should enable other researchers to make better informed decisions about their choices for available HPLCs, ionization sources and LC–MS-MS platforms.
Funding
The results described were supported by funding from the National Institute of Health Award Number R01NS065263 of Neurological Disorders and Stroke.
Supplementary Material
Acknowledgments
We thank members of the Agar lab for helpful comments on the manuscript. The authors gratefully acknowledge the help of the following individuals: Fred Mannarino from Eksigent (now AB SCIEX); Russ Constantineau and Michael Barrett Andersen from Proxeon (now Thermo Fisher Scientific); Vladimir Binshok and Robin Andreotti from Waters Corporation; Vic Fursey, Paul Speir, Desmond Kaplan, and Christiane Mueller from Bruker Daltonics; Amanda Berg from New Objective; Ross Tomaino; and John Asara.
The authors have no conflicts of interest.
References
- 1.Lenz E.M., Wilson I.D. Analytical strategies in metabonomics. Journal of Proteome Research. 2006;6:443–458. doi: 10.1021/pr0605217. [DOI] [PubMed] [Google Scholar]
- 2.Lescuyer P., Hochstrasser D., Rabilloud T. How shall we use the proteomics toolbox for biomarker discovery? Journal of Proteome Research. 2007;6:3371–3376. doi: 10.1021/pr0702060. [DOI] [PubMed] [Google Scholar]
- 3.Vitzthum F., Behrens F., Anderson N.L., Shaw J.H. Proteomics: from basic research to diagnostic application. A review of requirements & needs. Journal of Proteome Research. 2005;4:1086–1097. doi: 10.1021/pr050080b. [DOI] [PubMed] [Google Scholar]
- 4.Wilson I.D., Plumb R., Granger J., Major H., Williams R., Lenz E.M. HPLC-MS-based methods for the study of metabonomics. Journal of Chromatography B. 2005;817:67–76. doi: 10.1016/j.jchromb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 5.Comisarow M.B., Marshall A.G. Fourier transform ion cyclotron resonance spectroscopy. Chemical Physics Letters. 1974;25:282–283. [Google Scholar]
- 6.Haynes P. A., Fripp N., Aebersold R. Identification of gel-separated proteins by liquid chromatography-electrospray tandem mass spectrometry: Comparison of methods and their limitations. Electrophoresis. 1998;19:939–945. doi: 10.1002/elps.1150190609. [DOI] [PubMed] [Google Scholar]
- 7.Stone K.L., Deangelis R., LoPresti M., Jones J., Papov V.V., Williams K.R. Use of liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) for routine identification of enzymatically digested proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Electrophoresis. 1998;19:1046–1052. doi: 10.1002/elps.1150190620. [DOI] [PubMed] [Google Scholar]
- 8.Aebersold R., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher N.L. Top-down proteomics. Analytical Chemistry. 2004;76:196A–203A. [PubMed] [Google Scholar]
- 10.Mann M., Kelleher N.L. Precision proteomics: The case for high resolution and high mass accuracy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y., Zhang R., Moore R.J., Kim J., Metz T.O., Hixson K.K., et al. Automated 20 kpsi RPLC-MS and MS/MS with chromatographic peak capacities of 1000-1500 and capabilities in proteomics and metabolomics. Analytical Chemistry. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 12.Moon D.-C., Kelley J.A. A simple desalting procedure for fast atom bombardment mass spectrometry. Biological Mass Spectrometry. 1988;17:229–237. doi: 10.1002/bms.1200170312. [DOI] [PubMed] [Google Scholar]
- 13.Peterson A., Hohmann L., Huang L., Kim B., Eng J.K., Martin D.B. Analysis of RP-HPLC loading conditions for maximizing peptide identifications in shotgun proteomics. Journal of Proteome Research. 2009;8:4161–4168. doi: 10.1021/pr9001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wehr C.T.C., Abbott S.R. Evaluation of stationary and mobile phases for reversed-phase high performance liquid chromatography of peptides; Journal of Chromatographic Science. 1982;20:114–119. [Google Scholar]
- 15.Marginean I., Kelly R.T., Moore R.J., Prior D.C., LaMarche B.L., Tang K., et al. Selection of the optimum electrospray voltage for gradient elution LC-MS measurements. Journal of the American Society for Mass Spectrometry. 2009;20:682–688. doi: 10.1016/j.jasms.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turck C.W., Falick A. M., Kowalak J. A., Lane W. S., Lilley K. S., Phinney B. S., et al. The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 Study. Molecular and Cellular Proteomics. 2007;6:1291–1298. doi: 10.1074/mcp.M700165-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., et al. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 18.Gilar M., Daly A.E., Kele M., Neue U.D., Gebler J.C. Implications of column peak capacity on the separation of complex peptide mixtures in single- and two-dimensional high-performance liquid chromatography. Journal of Chromatography A. 2004;1061:183–192. doi: 10.1016/j.chroma.2004.10.092. [DOI] [PubMed] [Google Scholar]
- 19.Strittmatter E.F., Ferguson P.L., Tang K., Smith R.D. Proteome analyses using accurate mass and elution time peptide tags with capillary LC time-of-flight mass spectrometry. Journal of the American Society for Mass Spectrometry. 2003;14:980–991. doi: 10.1016/S1044-0305(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 20.Elias J.E., Haas W., Faherty B.K., Gygi S.P. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nature Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 21.Paulovich A.G., Billheimer D., Ham A.J., Vega-Montoto L., Rudnick P.A., Tabb D.L., et al. Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS platform performance. Molecular & Cellular Proteomics. 2010;9:242–254. doi: 10.1074/mcp.M900222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirran S.L., Botting C.H. A comparison of the accuracy of iTRAQ quantification by nLC-ESI MSMS and nLC-MALDI MSMS methods. Journal of Proteomics. 2010;73:1391–1403. doi: 10.1016/j.jprot.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess K.E.V., Lainson A., Imrie L., Fraser-Pitt D., Yaga R., Smith D.G.E., et al. Performance of five different electrospray ionisation sources in conjunction with rapid monolithic column liquid chromatography and fast MS/MS scanning. Proteomics. 2009;9:1720–1726. doi: 10.1002/pmic.200800200. [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 25.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 26.Gilar M., Olivova P., Daly A.E., Gebler J.C. Orthogonality of separation in two-dimensional liquid chromatography. Analytical Chemistry. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 27.Mayya V., Rezaul K., Cong Y.-S., Han D. Systematic comparison of a two-dimensional ion trap and a three-dimensional ion trap mass spectrometer in proteomics. Molecular and Cellular Proteomics. 2005;4:214–223. doi: 10.1074/mcp.T400015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz J.C., Senko M.W., Syka J.E.P. A two-dimensional quadrupole ion trap mass spectrometer. Journal of the American Society for Mass Spectrometry. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 29.Tabb D.L., Vega-Montoto L., Rudnick P.A., Variyath A.M., Ham A.-J.L., Bunk D.M., et al. Repeatability and reproducibility in proteomic identifications by liquid chromatography–tandem mass spectrometry. Journal of Proteome Research. 2009;9:761–776. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalli A., Hess S. Effect of mass spectrometric parameters on peptide and protein identification rates for shotgun proteomic experiments on an LTQ-orbitrap mass analyzer. Proteomics. 2012;12:21–31. doi: 10.1002/pmic.201100464. [DOI] [PubMed] [Google Scholar]
- 31.Andrews G.L., Dean R.A., Hawkridge A.M., Muddiman D.C. Improving proteome coverage on a LTQ-Orbitrap using design of experiments. Journal of the American Society for Mass Spectrometry. 2011;22:773–783. doi: 10.1007/s13361-011-0075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell A.W., Deutsch E.W., Au C.E., Kearney R.E., Beavis R., Sechi S., et al. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nature Methods. 2009;6:411–423. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins D.N., Pappin D.J.C., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Howard G.A., Martin A.J.P. The separation of the C12-C18 fatty acids by reversed-phase partition chromatography. Biochemical Journal. 1950;46:532–538. doi: 10.1042/bj0460532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipton M.S., Paša-Tolić L., Anderson G.A., Anderson D.J., Auberry D.L., Battista J.R., et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petritis K., Kangas L.J., Ferguson P.L., Anderson G.A., Paša-Tolić L., Lipton M.S., et al. Use of artificial neural networks for the accurate prediction of peptide liquid chromatography elution times in proteome analyses. Analytical Chemistry. 2003;75:1039–1048. doi: 10.1021/ac0205154. [DOI] [PubMed] [Google Scholar]
- 37.Masselon C.D., Kieffer-Jaquinod S., Brugière S., Dupierris V., Garin J. Influence of mass resolution on species matching in accurate mass and retention time (AMT) tag proteomics experiments. Rapid Communications in Mass Spectrometry. 2008;22:986–992. doi: 10.1002/rcm.3447. [DOI] [PubMed] [Google Scholar]
- 38.Manes N.P., Estep R.D., Mottaz H.M., Moore R.J., Clauss T.R.W., Monroe M.E., et al. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. Journal of Proteome Research. 2008;7:960–968. doi: 10.1021/pr070432+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. NanoACQUITY UPLC SYSTEM: Your Direct Nano Flow Solution. http://www.waters.com/webassets/cms/library/docs/720001147en.pdf. (accessed November 16, 2010) [Google Scholar]
- 40. Proxeon EASY-nLC II: Effortless, split-free nano-LC for top performance in LC-MS. http://www.proxeon.com/productrange/nano_LC/introduction/index.html. (accessed November 16, 2010) [Google Scholar]
- 41. NanoLC-Ultra™ system Data Sheet: The new NanoLC-Ultra is Eksigent's third generation system, delivering superior gradient precision at pressures up to 10,000 psi. http://www.eksigent.com/pdf/NanoLCUltra_DataSheet_0208_LR.pdf. (accessed November 16, 2010) [Google Scholar]
- 42. Microfluidic flow control allows for splitless nanoLC with unprecedented retention time repeatability; application note # n-03. http://www.eksigent.com/pdf/NanoLC_MFC_AN.pdf. (accessed November 16, 2010) [Google Scholar]
- 43.Karas M., Bahr U., Dülcks T. Nano-electrospray ionization mass spectrometry: addressing analytical problems beyond routine. Fresenius' Journal of Analytical Chemistry. 2000;366:669–676. doi: 10.1007/s002160051561. [DOI] [PubMed] [Google Scholar]
- 44.Moore R.E., Young M.K., Lee T.D. Qscore: An algorithm for evaluating SEQUEST database search results. Journal of the American Society for Mass Spectrometry. 2002;13:378–386. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J., Li J., Xie H., Zhu Y., He F. A new strategy to filter out false positive identifications of peptides in SEQUEST database search results. Proteomics. 2007;7:4036–4044. doi: 10.1002/pmic.200600929. [DOI] [PubMed] [Google Scholar]
- 46.Koenig T., Menze B.H., Kirchner M., Monigatti F., Parker K.C., Patterson T., et al. Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. Journal of Proteome Research. 2008;7:3708–3717. doi: 10.1021/pr700859x. [DOI] [PubMed] [Google Scholar]
- 47.Ma J., Zhang J., Wu S., Li D., Zhu Y., He F. Improving the sensitivity of MASCOT search results validation by combining new features with Bayesian nonparametric model. Proteomics. 2010;10:4293–4300. doi: 10.1002/pmic.200900668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.