Abstract
HIV-1 entry into CD4+ target cells is mediated by cleaved envelope glycoprotein (Env) trimers that have been challenging to characterize structurally. Here, we describe the crystal structure at 4.7 Å of an antigenically near-native, cleaved, stabilized, soluble Env trimer (termed BG505 SOSIP.664 gp140) in complex with a potent broadly neutralizing antibody, PGT122. The structure shows a pre-fusion state of gp41, the interaction between the component gp120 and gp41 subunits, and how a close association between the gp120 V1/V2/V3 loops stabilizes the trimer apex around the three-fold axis. The complete epitope of PGT122 on the trimer involves gp120 V1, V3 and several surrounding glycans. This trimer structure advances our understanding of how Env functions and is presented to the immune system, and provides a blueprint for structure-based vaccine design.
The envelope glycoprotein (Env) trimer is the only virally encoded antigen on the surface of HIV-1, the pathogen responsible for the global AIDS epidemic, and is responsible for viral entry into host cells. The trimer is composed of gp120/gp41 heterodimers and is the target for neutralizing antibodies. Various structures of components of gp120 and gp41, alone and in complex with different ligands, have been determined (1-10). Cryo-electron microscopy (EM) and tomography have been integrated with core gp120 x-ray structures to visualize the Env trimer at resolutions that extend from 30 Å to below 10 Å, and thereby provide insights into its overall conformation before and after receptor binding (11, 12). However, determining an atomic-level structure of the Env trimer has been difficult. A higher resolution structure would not only help to understand how the trimer functions during virus-cell fusion, but also guide HIV-1 vaccine design by delineating the key antigenic sites recognized by the humoral immune system and the defenses evolved by the virus as a counter-measure.
During Env synthesis, gp160 precursors trimerize and are subsequently cleaved by proteases of the furin family into gp120 and gp41 subunits, which associate non-covalently before the native complex reaches the surface of infected cells and is then packaged onto virions (13). Cleavage is obligatory for Env trimers to function in viral infection of target cells (14). Virus-cell fusion is a multistep process involving three major Env conformations, each with distinct roles: 1) pre-fusion (interacts with CD4 receptor); 2) extended gp41 intermediate (interacts with CCR5 or CXCR4 co-receptors); and 3) gp41 six-helix bundle (hemi-fusion of viral and cell membranes) (15).
The requirement for the cleaved, native Env trimer to undergo conformational changes during receptor binding and fusion makes it metastable, which has substantially hindered both structure determination and vaccine development. The extensive N-linked glycosylation (on average 81 sites/trimer) creates additional complications for x-ray structural studies. Moreover, membrane-associated forms of Env are more difficult to express and purify in appropriate quantities and qualities than soluble versions. Our approach to these various problems has been to express soluble (i.e., truncated prior to the gp41 transmembrane domain), cleaved forms of trimeric Env (SOSIP gp140) that are engineered to improve their stability and homogeneity. Specifically, a disulfide bond (termed SOS) between gp120 residue 501 (HXB2 numbering) and gp41 residue 605 covalently links these subunits, while an Ile to Pro change at position 559 (termed IP) strengthens gp41-gp41 associations (16). A recent version of the SOSIP gp140 trimer, based on a Tier-2 subtype A virus (BG505) (17), was further engineered to delete all but 4 residues of the hydrophobic membrane proximal external region (MPER) of gp41 (17-20). Together, these various modifications allow the expression of a thermostable, non-aggregating and homogenous soluble Env trimer, BG505 SOSIP.664 gp140, suitable for structural characterization by x-ray crystallography (Fig. 1A). These trimers are reactive with a large panel of diverse broadly neutralizing antibodies (bnAbs), including those to quaternary epitopes, while being minimally reactive with non-neutralizing antibodies that preferentially recognize individual gp120/gp41 subunits and/or uncleaved, non-native trimer forms (17, 18). The near-native antigenic properties of the BG505 SOSIP.664 gp140 trimer suggest that its structure resembles the native viral spike, although we cannot completely rule out slight conformational differences resulting from engineered features, such as truncation of the gp41 MPER and transmembrane domain (19). Here, we show that the BG505 SOSIP.664 gp140 trimers could be successfully crystallized with a highly potent bnAb, PGT122, that targets the glycan-dependent Asn332 (N332) supersite of vulnerability on gp120 (21). These crystals allowed the structure of an Env trimer to be determined at a resolution of 4.7 Å.
Fig. 1. Overall architecture of a soluble, cleaved, recombinant HIV-1 Env trimer in complex with bnAb PGT 122.
(A) Schematic of the soluble, cleaved, recombinant HIV-1 Env BG505 SOSIP.664 construct in comparison to full-length gp160. N-linked glycans are shown and numbered on their respective Asn residues. The constant (C1-C5) and variable (V1-V5) regions in gp120 and the fusion peptide (FP), HR1 and HR2 helices, membrane proximal external region (MPER), transmembrane (TM) and cytoplasmic (CT) elements in gp41 are indicated. The SOSIP mutations are shown in red, as well as the added N332 glycan site. The color coding is as in B. (B) Side view of the soluble Env trimer complex with PGT122 showing two of the three Env gp140 protomers associated with PGT122 Fab (blue). A 2Fo-Fc electron density map contoured at 1.0 σ is shown as a gray mesh around the leftmost gp140 protomer. The membrane to which gp41 is attached would be at the bottom of the figure. (C) Side view of the Env trimer. For one of the three protomers on Env, core gp120 is shown in yellow, whereas V1/V2 and V3 regions are highlighted in orange and red, respectively. The main gp41 helical elements are colored in different shades of green. Protein components are rendered according to their secondary structure and glycans are depicted as spheres. (D) View of Env down the trimer axis. Loops of high variability in gp120 (V1-V5) all map to the periphery of the trimer and are labeled. Glycans have been omitted for clarity. Dashed lines indicate the location of gp120 V2 and V4 loops for which electron density was absent or ambiguous. The figure was generated with Pymol (63).
Structure determination
The BG505 SOSIP.664 construct was expressed in HEK 293S GnTI−/− cells, yielding trimers enriched for homogeneous oligomannose (Man5-Man9) glycans (see supplemental material (22)). Following incubation of the purified trimers with a 6-fold molar excess (2-fold for the binding sites) of PGT122 Fab, the complex was treated with EndoH glycosidase (fig. S1, A and B) to truncate any accessible N-linked glycans (i.e., not buried or occluded by PGT122) to a single N-acetyl glucosamine (NAG) moiety that remains covalently attached to the Asn side chain. Crystals of the purified complex diffracted well, albeit anisotropically, to 3.7 Å along the c-axis, but to lower resolution along the other two axes (fig. S1, C and D). Merging diffraction data from two crystals resulted in a complete dataset to a maximum resolution of 4.7 Å (table S1). Phases were obtained by molecular replacement using integrative approaches in which a search model was generated from crystal structures of the unliganded PGT122 Fab variable region (PDB ID: 4JY5, (23)) and CD4-bound gp120 core (PDB ID: 3JWD, (5)), docked into our previous ~14 Å EM reconstruction of the same complex (EMDB: 5624, (23)). Only one complex was present in the asymmetric unit with 82% solvent, which simplified structure determination (fig. S2). Initial phases resulted in a well-defined electron density map that enabled subsequent placement of the high-resolution PGT122 Fab constant domains, and the gp120 V1/V2 (PDB ID: 3U4E, (4)) and V3 (PDB ID: 2ESX, (24)) structures. Previously uncharacterized elements in the trimer, such as the gp41 helices, the V1/V2/V3 loops and various Man5-Man9 glycans were visible in the electron density maps, as were residual NAG moieties attached to their respective Asn residues (fig. S3). Together, these identifiable features, along with aromatic side chains, aided in model building and refinement at this moderate resolution. In addition, the prominent new features for the Env trimer that we observed in the electron density maps are the same as those visualized in the accompanying 5.8 Å cryo-EM reconstruction of the same trimer in complex with bnAb PGV04 (fig. S4) (25).
Architecture of the Env trimer
The soluble BG505 SOSIP.664 trimer adopts a compact mushroom shape reminiscent of the Env trimer pre-fusion closed conformation determined at 20 Å resolution by Electron Microscopy (11, 26). Three PGT122 Fabs protrude vertically from the membrane-distal gp120 subunits, whereas the gp41 components are membrane-proximal and interspersed with the gp120 C1 and C5 elements (Fig. 1, B-D). Previous high-resolution crystal structures of core gp120 (PDB ID: 3JWD, (5)), scaffolded V1/V2 (PDB ID: 3U4E, (4)) and the variable domains of PGT122 Fab (PDB ID: 4JY5, (23)) fit well into the electron density of the trimer complex (Figs. 1 and 2 and fig. S5), and have Cα root mean square deviations (r.m.s.d.'s) of 1.3 Å, 2.9 Å and 0.8 Å with the final trimer model, respectively. Thus, the core gp120 elements in the trimer adopt conformations similar to those observed in unliganded gp120 (2) or gp120 in complex with various ligands (specifically CD4 (3, 5), b12 (9), VRC01 (8), PGT128 (7), PGT135 (1) and PG9/PG16 (4, 6)). In addition, all previously described disulfide bonds in gp120 (3) are present and all of the variable loops (V1-V5) are on the outside of the structure (Fig. 1D).
Fig. 2. Comparison of gp120 and components as observed in high-resolution crystal structures and in the soluble HIV-1 Env trimer.
(A) High-resolution crystal structure of core gp120 (PDB ID: 3JWD, (5), pale green) is superimposed on the gp120 component of the soluble, cleaved SOSIP.664 trimer crystal structure (yellow). A longer α1 helix (gp120 residues 99-117) likely contributes to rearrangement in the bridging sheet, particularly in β2 and β3, which extend into V1/V2 atop the trimer. The gp120 β2-proximal residues 115-125 are highlighted in blue and brown in core gp120 and trimeric gp140, respectively. A 2Fo-Fc electron density map contoured at 1.0 σ is shown as a gray mesh around the α1 helix. (B) Superimposition of the scaffolded gp120 V1/V2 crystal structure (PDB ID: 3U4E, pale green) on V1/V2 in the trimer crystal structure (orange). There are differences in V1 and in the β2 and β3 connecting strands. Electron density for carbohydrates at gp120 N156 and N160 is shown as a 2Fo-Fc gray mesh contoured at 1.0 σ. (C) Structural arrangement of gp120 V1/V2 (orange) and V3 (red) in the context of the trimer. A 2Fo-Fc electron density map contoured at 1.0 σ is shown as a gray mesh around V1/V2 and V3 elements. All structures are depicted according to secondary structure elements, with glycans depicted as yellow spheres and the Cys126-Cys196 disulfide bond colored blue. The figure was generated with Pymol (63).
Our Env trimer structure contrasts markedly with one recently described for an uncleaved, membrane-bound JR-FL Env trimer (27). Compared to our soluble cleaved structure, the uncleaved trimer EM structure differs substantially even in the gp120 core, as well as in the arrangement of the gp41 helices. Our structure also does not contain the large hole that has been reported to be present in the uncleaved Env trimer (27-29). It remains to be determined whether these differences are attributable to the use of different forms of trimer (soluble, truncated, cleaved vs. detergent-solubilized, almost full-length, uncleaved), or reflect concerns about the cryo-EM methodology used to derive the uncleaved trimer structure (30-34).
Comparison with component crystal structures
High-resolution crystal structures of monomeric gp120 have been largely obtained using a core construct stabilized in the CD4-bound conformation and with the V1/V2/V3 elements truncated (1, 3, 8-10). We observe only a few small deviations from these gp120 crystal structures, mostly in elements leading to and from V1/V2 (Fig. 2A and fig. S6). The differences are expected because V1/V2 undergoes significant conformational changes when gp120 binds CD4 (2, 11, 12). In the trimer structure, clear density for an additional helical turn (residues 114-117) is observed in the gp120 α1 helix (residues 99-113), which leads into V1/V2 (Fig. 2A). In addition, the β2 (119-123) and β3 (199-201) strands are only partially involved in forming the bridging sheet with β20 and β21; instead, compared to gp120 monomeric cores, β2 and β3 flip and translocate slightly towards the trimer apex to connect with strands A and D of V1/V2, respectively (Fig. 2A). This elongated α1 helix-bridging sheet arrangement is also observed in the accompanying cryo-EM structure of the SOSIP gp140 trimer in complex with bnAb PGV04 (25). Whether this β-strand inversion at the V1/V2 base in the trimer structures compared to the CD4-bound (3, 5), antibody-bound (8-10, 35) and unliganded core monomeric crystal structures (2) can be attributed to oligomer-monomer differences, to changes induced by CD4-binding, or to the truncation of V1/V2 in core gp120, is not yet known.
Electron density at the trimer apex indicates that V1/V2 adopts a 4-stranded Greek-key β-sheet arrangement similar to that observed in the high-resolution crystal structures of V1/V2-scaffolds in complex with bnAbs PG9 ((4), Fig. 2B) and PG16 (6). The V1 loop (132-140) connecting strands A and B adopts a more flattened structure that is parallel to the four-stranded β-sheet topology, differing slightly from its more vertical conformation in the V1/V2 scaffolded structure (4) and in the accompanying high-resolution cryo-EM trimer structure in complex with a receptor binding site antibody (25). It is unclear whether this difference in V1 orientation is a consequence of PGT122 binding or arises because V1 is a flexible loop. In the trimer crystal structure, the Cys126-Thr128 and Leu193-Cys196 segments diverge from one another, while still maintaining the Cys126-Cys196 disulfide bond, and they no longer contribute to V1/V2 strands A and D (Fig. 2B). Instead, they interact atop the Env spike with V1/V2 strands B and C and the conserved crown of the V3 loop from a neighboring protomer (Fig. 2C). We observe no clear electron density for V2 loop residues 178-190, which were also disordered in the V1/V2 scaffolded structures (4, 6).
The gp120 V3 loop completes the trimer apex and forms a β-hairpin with its two anti-parallel β-strands nestling directly below V1/V2 strands B and C of the same protomer (Fig. 2C). V3 peptides complexed with Abs adopt similar β-hairpin structures (24, 36, 37), as does the V3 region of monomeric gp120 (38) (fig. S5). The V3 crown is buried under a NAG moiety from the N197 glycan at the C-terminal end of V2 strand D from an adjacent protomer (Fig. 2C). Removing this glycan from other viruses increases nAb sensitivity (39) and can confer CD4-independent entry into CCR5-expressing cells (40). Our structure suggests that the N197 glycan helps to stabilize native Env by occluding V3 and inhibiting its premature release prior to CD4 binding. Any heterogeneity in the presence or composition of the N197 glycan may contribute to the reactivity of non-neutralizing V3 antibodies with BG505 SOSIP.664 trimers in some binding assays (17). Overall, the trimer structure is compatible with both intra- and inter-trimeric (cis-trans) V3 shielding by V1/V2 (41, 42).
gp41 architecture
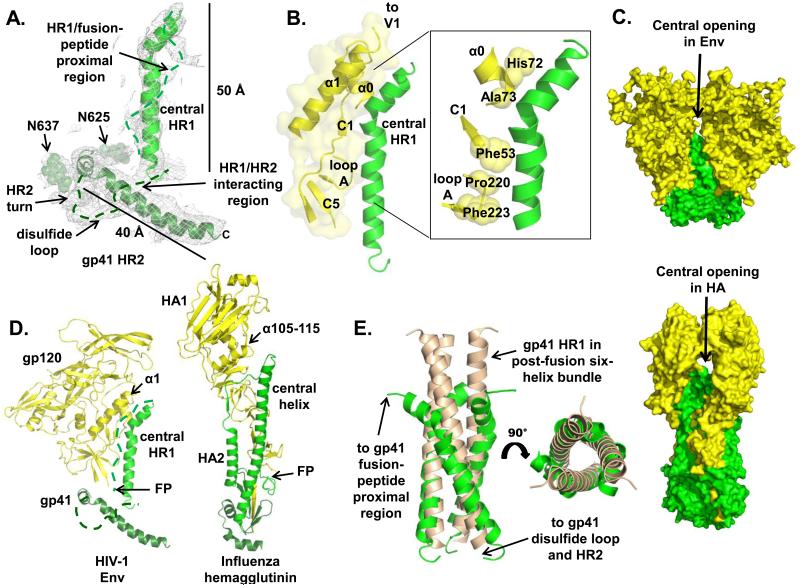
Strong electron density for helices attributable to gp41 is clearly visible in the trimer structure (Fig. 3A). Three central helices (6 turns) extend ~30 Å along the trimer axis, perpendicular to the viral membrane, and are ascribed to gp41 heptad-repeat 1 (HR1). Similar helices were recently observed in an ~9 Å cryo-EM reconstruction of the KNH1144 SOSIP gp140 trimer (12). An additional short helix (2.5 turns) extends from the central helix bundle but is kinked away from the trimer axis. The two HR1 helices pack against hydrophobic gp120 residues in C1 and C5 as well as loop A (Fig. 3B), and are capped by the gp120 α1 helix (Fig. 3B). Overall, these topological features are consistent with previous mutagenesis and structural studies that suggest gp41-gp120 interactions may propagate long-range effects upon receptor binding (5, 43).
Fig. 3. Structural organization of gp41 in the soluble cleaved HIV-1 Env trimer.
(A) Overall arrangement of gp41 elements from one protomer is shown in a gray 2Fo-Fc electron density map contoured at 1.0 σ. Carbohydrates are shown as spheres. Dashed lines delineate connecting electron density for which a chain trace and secondary structure determination was ambiguous at this moderate resolution. (B) Regions of contact between the gp120 inner domain and the gp41 central helix. The inset shows hydrophobic residues in the gp120 high-resolution crystal structure from C1, α0 and loop A that line the interface with gp41 HR1. (C) Surface rendering of two of the three HIV-1 Env and Influenza hemagglutinin (HA) (PDB ID: 4FNK) protomers (back protomer omitted) emphasizes the similarity in position and size of a small, central inter-protomer opening that presumably facilitates conformational changes during the fusion process. (D) Comparison between the structures of HIV-1 Env and Influenza HA (PDB ID: 4FNK), the prototype type I fusion protein. There are striking similarities in the position of structural elements in the two glycoproteins. (E) The trimeric arrangement of gp41 HR1 in the post-fusion conformation (PDB ID: 2X7R, (50), beige) superimposes closely with the central HR1 in the soluble HIV-1 Env trimer, which is similar to the retention of the three-helix bundle at the top of the long HA2 helix in Influenza HA. All structures are depicted according to secondary structure elements. The figure was generated with Pymol (63).
The weak and diffuse electron density corresponding to the FP and the residues that connect it to the HR1 central helix makes model building of this region particularly difficult at this resolution (fig. S7A), but is likely indicative of lack of regular secondary structure (44). Nonetheless, residues adjacent to the N-termini of the HR1 helices initially wrap around the gp120 α0 helix, forming a meta-stable loop-helix structure akin to similar elements in influenza HA2 (45) (Fig. 3C-D) that lead to an extension of the central helix in the post-fusion form (46). Overall, the central trimeric coiled-coil arrangement of the fusion protein (gp41) surrounded by three receptor subunits (gp120) is a characteristic of type-1 membrane-fusion proteins (47-49). We now extend that hallmark to the capping of the central helices by another helix (cf. α1 and α105-115) in both gp120 and HA. A small inter-protomer central opening is created at the top of the central helix in both fusion proteins (Fig. 3C), as well as in Ebola and PIV5/RSV fusion proteins, that presumably facilitates conformational rearrangements of the head domains (e.g. gp120, HA1) during the fusion process. The Env trimer structure also agrees well with what is known about the transition from a pre-fusion to a post-fusion conformation, in that the three HR1 central helices adopt a similar arrangement in the post-fusion six-helix bundle (Fig. 3E) (50).
A second, well-defined, ~40 Å long, helical element, at a ~60° angle relative to the HR1 central helix, is located at the bottom of the trimer, proximal to the membrane, where it wraps around the trimer base (Fig. 3A). We interpret this long helix (7 turns) as the C-terminal portion of the HR2 sequence, which is consistent with a cryo-EM reconstruction in the accompanying paper (25), where deletion of residues 651-664 from the BG505 SOSIP.664 trimer eliminated the density corresponding to the end of this helix (25, 51). Strong electron density between the bottom of HR1 and the middle of the HR2 helix is likely indicative of substantial intra-subunit gp41-stabilizing interactions. The residues that are N-terminal to the long HR2 helix appear to adopt a horseshoe conformation for the polypeptide backbone (Fig. 3A and fig. S4B); the NAG moieties at glycosylation sites 625 and 637 are clearly visible in the electron density maps and define the location of turns. Strong electron density, predominantly in a flat-extended shape characteristic of β-strands, is present between the C-terminus of HR1 and the N-terminus of HR2, where the gp41 disulfide loop (DSL; residues 585-609) is thought to be located (fig. S7B). However, we cannot determine the arrangement of these gp41 components with confidence at this resolution, and a further complication is the close proximity of gp120 C1 and C5 elements that most likely participate in tertiary structure interactions with residues connecting HR1 to HR2, including the DSL (5, 52). As such, we attribute the approximate location of the gp41 DSL and, hence, the engineered inter-subunit disulfide bond, to the base of the Env trimer. Finally, no conclusion can be drawn about the arrangement of the gp41 membrane-proximal-external-region (MPER) because it is not present in the BG505 SOSIP.664 gp140 trimer (19, 20). The gp41 HR2 may continue to extend in the same orientation as it enters the MPER, or could change direction near residue 664 (53, 54) so that the MPER could align parallel to the trimer axis.
Structural definition of the PGT122 epitope
The PGT121 family of antibodies potently neutralizes ~70% of circulating HIV-1 viruses (21, 55) and PGT121 is highly effective in protecting against mucosal challenge in a macaque model of HIV-1 infection (56). The crystal structure presented here agrees well with our lower resolution negative-stain EM reconstruction of the same PGT122-trimer complex (23) (fig. S8), but now allows a more complete description of the epitope. As initially predicted (21), the N332 Man8/9 glycan that is a central feature of the PGT122 epitope sits directly at the junction between the light chain complementarity determining region 2 (LCDR2) and the 26-residue heavy chain CDR3 (HCDR3) (Fig. 4A and fig. S9). Residues in the LCDR1, LCDR3 and light chain framework 3 (LFR3) regions of PGT122, which were predicted by alanine-scanning mutagenesis to be important for HIV-1 neutralization (23), interact directly with the gp120 V3 base near Ile323-Arg327, a key part of the epitope (21, 57). The trimer structure also helps explain why PGT121-class bnAbs depend on V1/V2 residues for locking gp120 into the pre-CD4 conformation (23); in fact, gp120 V1 residues 135-139, including the glycan at N137, come into close proximity to HCDR1, HCDR2, HCDR3 and LCDR3 of PGT122 (Fig. 4A and fig. S9). Indeed, the N137 oligomannose glycan in the trimer structure superimposes almost perfectly with the bi-antennary complex glycan that occupies the groove formed by the PGT121 heavy chain CDRs in previous crystal structures (23, 55, 58) (Fig. 4A and B). Alanine scanning mutagenesis of gp120 glycans clearly indicates that bnAbs of the PGT121 family depend on N137 for neutralizing the BG505 virus (Fig. 4C and fig. S10). Locating this key residue in the crystal structure has allowed us not only to delineate the full PGT122 epitope, but also to confidently determine the relative position of V1/V2 elements at the trimer apex. In addition to N137 and N332, the N156 and N301 glycans are also protected from EndoH deglycosylation by PGT122. Although the composition and nature of the glycoforms on native Env are still uncertain, PGT122 recognition is compatible with the presence of a complex/hybrid carbohydrate at position N156/N173, as observed in the crystal structure of a V1/V2 scaffold with bnAb PG16 (6) (Fig. 4B). The N156 and N301 glycans do have a clear involvement, albeit a minor one, in neutralization by PGT121-family bnAbs (Fig. 4C and fig. S10). Thus, the Env trimer structure reveals the complexity of the PGT122 epitope, which involves the V1 and V3 base and four glycans (N137, N156/N173, N301 and N332).
Fig. 4. Complete structural definition of the PGT122 epitope.
(A) In addition to the N332 glycan (yellow), the PGT122 bnAb recognizes both protein and glycan elements near the base of gp120 V1 (orange) and V3 (red) to mediate broad and potent HIV-1 neutralization. Heavy and light chain CDRs are shown as dark and light blue tubes, respectively. Electron density for oligomannose glycans (spheres) surrounding the PGT122 epitope is shown as a 2Fo-Fc gray mesh contoured at 1.0 σ. (B) Superimposition of the PGT122 epitope with glycans from PGT121 (PDB ID: 4JY4 and 4FQC, (23, 55)) and PG16 (PDB ID: 4DQO, (6)) liganded crystal structures. PGT122 binding is compatible with the involvement of complex/hybrid glycans at gp120 N137 and N156/173 (green) when the trimer is expressed in human cells capable of making this type of glycan. The figure was generated with Pymol (63). (C) The use of glycan knock-out mutants of HIV-1 BG505 pseudoviruses reveals the importance of glycans at N137, N156 and N301 for PGT122 recognition and neutralization. These glycans are part of the PGT122 epitope in the trimer crystal structure to varying extents.
Revisiting glycan-dependent bnAb epitopes in the context of the HIV-1 Env trimer
The recent isolation (21, 59) and structural characterization (1, 4, 6, 7) of potent glycan-reactive bnAbs has emphasized the need to precisely define the composition of various glycans that are sites of vulnerability on Env. The trimer structure allows us to reevaluate previous crystal structures of glycan-dependent bnAbs that were obtained with monomeric core gp120 or epitope scaffolds. When viewed in the context of the trimer, the gp120 epitopes for the N160-dependent (PG9/PG16) and N332-dependent (PGT122, PGT128 and PGT135) epitopes are in close proximity (Fig. 5A). Once considered separate sites of vulnerability (Fig. 5B), these two epitope clusters are larger than were first thought and overlap to some extent, helping to explain why some of these antibodies compete with one another in certain binding assays (21) (Fig. 5C). All of these glycan-dependent bnAbs use their six CDR loops, in addition to framework regions, when recognizing their epitopes. As previously suggested (18), PG9 and PG16 would interact with two N160 glycans across different protomers in the trimer apex (Fig. 5D). In addition, while one side of the elongated HCDR3 hammerhead inserts into the trimer apex, the other is available to interact with elements of V3 (Fig. 5D) and may help explain why V3 residues influence neutralization by PG16 (59). The trimer crystal structure also suggests that HCDR1 and HFR3 of PG9/PG16 may interact with the V3-protecting N197 glycan on an adjacent protomer (Fig. 5D). PGT128 would also make contacts with Env elements additional to those previously reported in the crystal structure with a gp120 outer domain lacking V1/V2 (7) (Fig. 5E). Thus, LCDR1, LCDR2 and LFR3 of PGT128, for which no previous role could be assigned, are now predicted to be in close proximity to V1 elements, including N137 and N156 (60), whereas a change in the V1 loop orientation is required to allow PGT135 binding (Fig. 5F). Indeed, a V1 orientation closer to that observed in the V1/V2 scaffold or in the high-resolution cryo-EM trimer structure (25) would remove a clash with N137 and facilitate PGT135 interaction with V1. A flexible role for V1 might help to explain the plasticity of PGT135-Env interactions seen in previous EM studies (1).
Fig. 5. Revisiting glycan-dependent epitopes of bnAbs in the context of the soluble Env trimer structure.
(A) Superimposition of the co-crystal structures of PG16 (green), PGT128 (red) and PGT135 (cyan) on the PGT122 (blue)-Env trimer crystal structure reveals that these glycan-dependent epitopes overlap. (B) Co-crystal structures of glycan-dependent bnAbs in complex with monomeric gp120 or scaffolded gp120 elements provide crucial, but incomplete, details of their epitopes and, hence, how they recognize Env. The various bnAb epitopes are colored individually on the surfaces, with overlapping elements in purple. (C) Superimposition of the PG9/16, PGT128 and PGT135 co-crystal structures on the soluble trimer-PGT122 co-crystal structure reveals that these glycan-dependent bnAbs have expanded epitopes, which creates overlapping sites of vulnerability on HIV-1 Env. (D) Model of the expanded PG16 epitope on Env. Antibody interactions additional to those previously described in the PG9/PG16-V1/V2-scaffold co-crystal structures (4, 6) are predicted to occur with gp120 V3, as well as with residues N160 and N197 of the adjacent protomer. Protomers are denoted with P1, P2, P3 subscripts. (E). Model of the expanded PGT128 epitope includes additional interactions with glycans at N137 and N156 mediated via the PGT128 light chain (light blue). (F) PGT135 would clash (yellow star) with the gp120 V1 conformation recognized by PGT122, based on superimposition of the PGT135-core gp120 co-crystal structure (1). A slight re-orientation of gp120 V1 (green) would allow PGT135 to interact, consistent with the hypothesis that the V1 loop is flexible enough to permit different modes of bnAb interaction. Heavy and light chains are shown as dark and light blue tubes, respectively. The figure was generated with Pymol (63).
Conclusion
The crystal structure of the soluble BG505 SOSIP.664 trimer is in excellent agreement (fig. S4) with the accompanying cryo-EM structure of the same Env construct as a complex with PGV04 Fab (25). Structures determined using these two independent techniques show complete concordance in the major new features that are visualized at the present level of 5-6 Å resolution (fig. S4). We have defined the overall architecture of the soluble trimer, as well as the secondary, tertiary and quaternary interactions between gp120 and gp41 that are involved in its assembly. The trimer is relatively tightly packed, especially in gp41, but with a small opening between the Env apex and the top of the central gp41 helices. The gp120 subunits are held together, at least in part, by association of the V1/V2/V3 regions at the apex of the trimer (11, 61, 62). The gp41 central helices provide the main stabilizing contact between gp41 and gp120. Finally, the complete definition of how neutralization epitopes are presented in the context of the trimer, here and in the accompanying manuscript (25), should help the design of new immunogens as candidate vaccines.
Materials and Methods
Fab expression and purification
Recombinant Fabs for co-crystallization experiments were expressed using a protocol similar to that previously described (23). Briefly, the heavy and light chain genes were co-transfected using 293Fectin (Invitrogen) into human embryonic kidney (HEK) 293F cells in a 2:1 ratio to minimize formation of light chain homodimers. Secreted Fabs were isolated from the expression media 6-days post-transfection by affinity purification via an anti-human λ/κ light chain affinity matrix (CaptureSelect Fab λ/κ; BAC), followed by cation exchange and gel filtration chromatography.
SOSIP gp140 expression and purification
To increase the probability of successful crystallization and structure determination, several soluble, cleaved trimers of the SOSIP.664 gp140 design were expressed and purified as described elsewhere (17, 18, 23, 64). Crystallization candidates based on env genes from HIV-1 isolates KNH1144, ADA, 1182.6.1D2, BG505 and 208.9.C10 were selected from an initial panel of 20, based on their expression and trimer-formation properties, as assessed by SDS-PAGE and BN-PAGE. Notably, for isolates that did not naturally contain residue N332, a point substitution was made to introduce it, thereby creating the 2G12 epitope to facilitate affinity purification. The Env constructs were co-transfected with the furin protease in HEK 293S GnTI−/− cells, which lack N-acetylglucosaminyltransferase I and, therefore, produce glycoproteins bearing only high-mannose (Man5-9) glycans. Secreted SOSIP.664 Env proteins were harvested from supernatants and affinity purified using a 2G12 MAb affinity column. Following a high salt elution process, trimers were purified to size homogeneity using a Superose 6 size exclusion chromatography (SEC) matrix (GE Healthcare).
Sample preparation and crystal screening
Purified SOSIP.664 gp140 trimers were tested for crystallizability either alone or as complexes with one or more ligands from the following list: soluble CD4, Fabs: PGT121, PGT122, PGT123, PGT127, PGT128, PGT135, PG9, PG16, PGT145, PGV04, 17b. In addition, purified trimers and complexes were in some cases treated with the following glycosidases, alone or in combination, for 3 h at temperatures ranging between 20-37°C: EndoH (New England Biolabs), EndoD (VWR), Endo F1 (EMD Chemicals) or α1,2,3,6 mannosidase (QA-Bio). The monodispersity of the resulting samples was measured by SDS-PAGE, BN-PAGE and SEC coupled in-line with multi-angle light scattering (SEC-MALS) using the following layout: a Superose 6 10/30 SEC column (GE Healthcare) operated on an AKTA Avant FPLC system (GE Healthcare) with the following calibrated detectors: 1) HP1 1050 Hewlett-Packard UV detector (Norwalk, CT); 2) MiniDawn Treos multi-angle light scattering (MALS) detector (Wyatt Corporation, CA); 3) quasi-elastic light scattering (QELS) detector (Wyatt Corporation, CA); and 4) Optilab T-reX refractive index (RI) detector (Wyatt Corporation, CA). The purified complexes were concentrated to approximately 2-6 mg/ml and subjected to extensive crystallization trials using either: 1) the Oryx8 crystallization robot (Douglas Instruments) with 96 different crystallization conditions at 16°C; or 2) the automated IAVI/JCSG/TRSI CrystalMation robotic system (Rigaku) at the Joint Center for Structural Genomics (www.jcsg.org), where 384 different crystallization conditions were tested at 4°C and 22°C.
In the above trials, only the partially deglycosylated SOSIP.664 gp140 trimers from various isolates in complex with antibodies of the PGT121-family (PGT121, PGT122 and PGT123) resulted in crystal hits. Surprisingly, for these complexes, hits were obtained from approximately 10% of the crystallization conditions, but the majority of crystals tested (>600) only diffracted to 8 Å or worse at various synchrotron sources (APS, SSRL, and CLS). Several pre- and post-crystallization strategies were explored in an effort to improve the x-ray diffraction properties of these crystals: however, the use of single-chain Fv instead of Fab, limited in-situ proteolysis, varying temperature of crystal growth, additives screening, crystal cross-linking, dehydration, and annealing all met with limited success. In addition, we attempted use of a SOSIP construct further truncated to gp41 position 650. Although crystals could be obtained with the trimer in complex with PGT122 Fab, these did not show improved diffraction properties compared to the majority of SOSIP.664-containing crystals.
X-ray data collection, structure solution and model building
Ultimately, only the thermostable BG505 SOSIP.664 trimers (18, 23) allowed us to achieve higher resolution x-ray diffraction. Two crystals obtained from crystallization condition 0.1 M CAPS, 2 M ammonium sulfate, 0.2 M lithium sulfate, pH 10.5, with the EndoH-treated, SEC-purified PGT122 Fab plus BG505 SOSIP.664 trimer complex showed a maximum visible diffraction of up to 3.7 Å along the c-axis at the Advanced Photon Source (APS) beamline 23-ID-D (fig. S1). However, these crystals showed severe anisotropic diffraction and diffracted to a maximum of only ~5 Å and ~6 Å along the a- and b-axes, respectively. In addition, due to radiation damage during data collection, it was necessary to merge the data from two crystals to obtain a dataset with sufficient redundancy and completeness. Integration and scaling were performed using the program XDS (65), and output files were subsequently corrected for anisotropy using the UCLA MBI — Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/anisoscale_xds/) (66), with truncations at 4.5 Å, 5.5 Å and 4.0 Å along the a, b and c axes, respectively. The overall data were then truncated to 4.7 Å based on an I/σ >2 criteria in the highest resolution shell. Only few higher resolution reflections to 3.7 Å met these I/σ >2 criteria and their inclusion in the dataset did not subsequently improve the quality of maps. Data statistics are reported in table S1.
Molecular replacement was performed using Phaser (67) with a composite search model comprised of: 1) the variable domain of PGT122 at 1.8 Å (PDB ID: 4JY5); 2) CD4-bound gp120 core at 2.6 Å (PDB ID: 3JWD); and 3) these components docked in a trimeric arrangement using the previously reported ~14 Å EM reconstruction of the PGT122 Fab in complex with the BG505 SOSIP.664 trimer (EMDB: 5624). The molecular replacement solution indicated only one copy of the trimer-antibody complex was present in the asymmetric unit with a TFZ=6.2 and an LLG=1,344 (resulting in an ~82% solvent content (Vm= 6.87 A3/Da) based on a molecular weight of ~400 kDa for the partially deglycosylated complex (68)). Additional components, such as the PGT122 Fab constant domain, gp120 V1/V2/V3, and gp41 helices were added manually based on clear electron density for these elements in the initial maps (fig. S3). Rigid-body, grouped-B-factor, XYZ, real-space and TLS refinements were carried out in PHENIX using NCS-restraints, as well as secondary structure and reference model restraints imposed from high-resolution structures (69). Model building was performed using Coot (70). Secondary structure was determined using STRIDE (71). Slightly higher crystallographic R-values for the trimer structure compared to the mean reported for other structures to the same resolution (72) likely results from the relatively poor diffraction properties of the crystals and the anisotropy of the resulting data. We also note that ~20% of the trimer is not modeled (N-terminus of gp120, V2, V4, C-terminus of gp120, N-terminus of gp41, gp41 disulfide loop region, N-terminus of gp41 HR2), which also contributes to higher R-values. BG505 SOSIP.664 trimer elements included in the deposited model are summarized in table S2. Structure validation was performed using Molprobity (73) and refinement statistics are reported in table S1.
Pseudovirus production and neutralization assay
The BG505 env gene used to make Env-pseudoviruses lacks the N332 glycosylation site, but the viruses are still neutralized by antibodies of the PGT121 family. To assess the importance of specific glycosylation sites on neutralization by these antibodies, site-directed mutations were introduced into the env gene using QuikChange according to the manufacturer's protocol (Stratagene, La Jolla, CA). Mutants were verified by DNA sequence analysis (Eton Bioscience, San Diego, CA). Pseudoviruses were generated by transfection of 293T cells for 72 h with an Env-expressing plasmid and an Env-deficient genomic backbone plasmid (pSG3ΔEnv) in a 1:2 ratio, using Fugene 6 (Promega). A single-cycle neutralization assay with TZM-bl target cells was performed as described previously (74). Briefly, TZM-bl cells were seeded in a 96-well flat bottom plate at a concentration of 20,000 cells/well. Pseudoviruses were preincubated with serial dilutions of antibody for 1 h at 37°C before infection of the cells. Luciferase reporter gene expression was quantified 48 h after infection by cell lysis and the addition of Luciferase substrate (Promega). To determine IC50 values, dose-response curves were fitted using nonlinear regression.
Supplementary Material
Summary.
The overall architecture of the structure involved in HIV entry provides a blueprint for vaccine design.
Acknowledgments
We thank Y. Hua, L. Kong, P.S. Lee, X. Zhu, R. Pejchal, H. Tien, T. Clayton, K. Saye and J. Korzun, for technical assistance, previous contributions to the Env trimer project, and insightful discussions and C.R. King and W. Koff for support and encouragement. This work was supported by NIH P01 AI82362 (J.P.M., I.A.W.), as well as the International AIDS Vaccine Initiative Neutralizing Antibody Consortium and Center (D.R.B., I.A.W., J.P.M., A.B.W.), CHAVI-ID UM1 AI100663 (D.R.B., I.A.W., AB.W.), NIH R01 AI084817 (I.A.W.), NIH R37 AI36082 (J.P.M.), NIH R01 AI33292 (D.R.B.), a Vidi grant from the Netherlands Organization for Scientific Research (R.W.S.), a Starting Investigator Grant from the European Research Council (R.W.S.), Canadian Institutes of Health Research fellowship (J.-P.J.), and the Ragon Institute. D.L. is supported by the U. S. NIH NIGMS Biomedical Technology Research Center program (GM103310). Use of the Advanced Photon Source for data collection was supported by the DOE, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. GM/CA CAT has been funded in whole or in part with federal funds from NCI (grant Y1-CO-1020) and NIGMS (grant Y1-GM-1104). Extensive crystal screening was also carried out at the Stanford Synchrotron Radiation Lightsource (SSRL) and at the Canadian Light Source (CLS). SSRL, a Directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility is operated for the U.S. Department of Energy (DOE) Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research; NIH's National Center for Research Resources, Biomedical Technology Program (P41RR001209); and the National Institute of General Medical Sciences (NIGMS). CLS is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. The content is the responsibility of the authors and does not necessarily reflect the official views of the NIGMS, NCI or NIH. Coordinates and structure factors have been deposited with the Protein Data Bank under accession code 4NCO. The International AIDS Vaccine Initiative (IAVI) has previously filed a patent relating to the BG505 SOSIP.664 trimer: U.S. Prov. Appln. No. 61/772,739, titled “HIV-1 envelope glycoprotein”, with inventors M. Caulfield, A.C., H. Dean, S. Hoffenberg, C.R.K., P.J.K., A. Marozsan, J.P.M., R.S., A.B.W., I.A.W., J.-P.J., but no patents have been filed on any work described here. Materials and information will be provided under an MTA. This is manuscript 25054 from The Scripps Research Institute.
References and Notes
- 1.Kong L, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwon YD, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A. 2012;109:5663. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;23:336. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107:1166. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pancera M, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran EE, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410:582. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCune JM, et al. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 15.Gallo SA, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614:36. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 16.Sanders RW, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. 2013;110:4351. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khayat R, et al. Structural characterization of cleaved, soluble human immunodeficiency virus type-1 envelope glycoprotein trimers. J Virol. 2013;87:9865. doi: 10.1128/JVI.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klasse PJ, et al. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol. 2013;87:9873. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Materials and methods are available as supplementary material on Science Online.
- 23.Julien JP, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen O, Sharon M, Quadt-Akabayov SR, Anglister J. Molecular switch for alternative conformations of the HIV-1 V3 region: implications for phenotype conversion. Proc Natl Acad Sci U S A. 2006;103:13950. doi: 10.1073/pnas.0606312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. doi: 10.1126/science.1245627. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The BG505 SOSIP.664 trimer possesses most, if not all, of the characteristics associated with the pre-fusion closed Env trimer conformation: the presence of a V1V2/V3 cap assembled at the membrane-distal apex; antigenic reactivity with antibodies of quaternary epitopes; the likely inability of gp41 to bind to free HR2 peptides; a coherent binding site for residues associated with binding of the small molecule inhibitor of HIV-1 entry, BMS806; and many residues of the inner domain previously analyzed by mutational substitution for which we can now explain their mutational phenotypes.
- 27.Mao Y, et al. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci U S A. 2013;110:12438. doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Y, et al. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol. 2012;19:893. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A hole was also reported in cleaved trimer structures determined by EM (11), but seems to result from the low resolution of the reconstructions (see discussion in (25)).
- 30.Cohen J. Structural biology. Is high-tech view of HIV too good to be true? Science. 2013;341:443. doi: 10.1126/science.341.6145.443. [DOI] [PubMed] [Google Scholar]
- 31.Henderson R. Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1314449110. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramaniam S. Structure of trimeric HIV-1 envelope glycoproteins. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1313802110. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Heel M. Finding trimeric HIV-1 envelope glycoproteins in random noise. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1314353110. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao Y, Castillo-Menendez LR, Sodroski JG. Reply to Subramaniam, van Heel, and Henderson: Validity of the cryo-electron microscopy structures of the HIV-1 envelope glycoprotein complex. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1316666110. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanfield R, et al. Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing Fabs. Structure. 1999;7:131. doi: 10.1016/s0969-2126(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 37.Jiang X, et al. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol. 2010;17:955. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- 38.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001;75:2041. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001;75:3435. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rusert P, et al. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J Exp Med. 2011;208:1419. doi: 10.1084/jem.20110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Cimbro R, Lusso P, Berger EA. Intraprotomer masking of third variable loop (V3) epitopes by the first and second variable loops (V1V2) within the native HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci U S A. 2011;108:20148. doi: 10.1073/pnas.1104840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finzi A, et al. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell. 2010;37:656. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stronger electron density reappears for a loop prior to the N-terminus of the gp41 HR1 central helix, near the gp120/gp41 interface. We assign this electron density to the gp41 fusion peptide proximal region (FPPR) elements. Indeed, gp41 and gp120 elements in this region (gp120 residues 73-84, glycans at gp120 N88 and gp41 N625, and the C-terminal HR2 helix from an adjacent protomer) are particularly hydrophobic and could easily interact with and occlude the hydrophobic gp41 FP. In addition, this ascribed FP region is in close relative proximity to the C-terminus of gp120 C5, to which it was connected in the uncleaved gp160 precursor prior to furin cleavage.
- 45.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 46.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 47.Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A slight difference in HR2 helix orientation is observed between the trimer crystal structure and the cryo-EM structure reported in the accompanying manuscript (25). Crystal packing interactions at the gp41 C-terminus probably account for this small shift (fig. S2).
- 52.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Julien JP, Bryson S, Nieva JL, Pai EF. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J Mol Biol. 2008;384:377. doi: 10.1016/j.jmb.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 55.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moldt B, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109:18921. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadjadpour R, et al. Emergence of gp120 V3 variants confers neutralization resistance in an R5 simian-human immunodeficiency virus-infected macaque elite neutralizer that targets the N332 glycan of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 2013;87:8798. doi: 10.1128/JVI.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The complex biantennary glycan observed in the PGT121 paratope in crystal structures (23, 55) is fortuitous, as the glycan comes from a symmetry-related PGT121 Fab molecule in the crystal. Indeed, Fab PGT121 is glycosylated and was expressed in mammalian cells. PGT121 also reacts on the glycan array with complex biantennary glycans possessing α1-6 sialylated ends (23), which make substantial contacts with the paratope in the crystal structures. Although the N137 glycoform remains uncharacterized on virus-derived HIV-1 Env, PGT121 reactivity with the glycan array, and the liganded crystal structures, suggest that N137 is probably a biantennary complex glycan. The relatively weaker electron density for the oligomannose N137 glycan in the trimer structure also suggests that, when it is not an α1-6 sialylated complex carbohydrate, the N137 glycan lacks significant putative sites of interaction with PGT122. Hence, it might not be fully protected from glycosidase treatment, or it may be slightly disordered.
- 59.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The PGT122 and PGT128 bnAbs appear to recognize a closely related N332-dependent epitope, but they approach gp120 with an inverted disposition of their light and heavy chains. Indeed, while the PGT128 heavy chain predominantly interacts with the gp120 V3 base, PGT122 recognizes this region mainly through its light chain. Conversely, N137 is recognized by the PGT122 heavy chain, but is putatively contacted by the PGT128 light chain.
- 61.Sullivan N, Thali M, Furman C, Ho DD, Sodroski J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J Virol. 1993;67:3674. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang SH, et al. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol. 2010;84:3147. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.The PyMOL Molecular Graphics System . Version 1.6.0.0. Schrödinger, LLC.; [Google Scholar]
- 64.Depetris RS, et al. Partial enzymatic deglycosylation preserves the structure of cleaved recombinant HIV-1 envelope glycoprotein trimers. J Biol Chem. 2012;287:24239. doi: 10.1074/jbc.M112.371898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 69.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 71.Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32:W500. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Urzhumtseva L, Afonine PV, Adams PD, Urzhumtsev A. Crystallographic model quality at a glance. Acta Crystallogr D Biol Crystallogr. 2009;65:297. doi: 10.1107/S0907444908044296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Madani N, et al. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol. 2004;78:3742. doi: 10.1128/JVI.78.7.3742-3752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 77.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.