Abstract
Genetic analysis of single cells is emerging as a powerful approach for studies of heterogeneous cell populations. Indeed, the notion of homogeneous cell populations is receding as approaches to resolve genetic and phenotypic variation between single cells are applied throughout the life sciences. A key step in single-cell genomic analysis today is the physical isolation of individual cells from heterogeneous populations, particularly microbial populations, which often exhibit high diversity. Here, we detail the construction and use of instrumentation for optical trapping inside microfluidic devices to select individual cells for analysis by methods including nucleic acid sequencing. This approach has unique advantages for analyses of rare community members, cells with irregular morphologies, small quantity samples, and studies that employ advanced optical microscopy.
1. INTRODUCTION
The ability to amplify very small amounts of DNA, even single molecules, has been a reality for more than a decade (Dean, 2002; Dean, Nelson, Giesler, & Lasken, 2001; Sykes et al., 1992; Vogelstein & Kinzler, 1999). This approach was eventually applied to individual microbial genomes (Kvist, Ahring, Lasken, & Westermann, 2006) and ultimately coupled with high-throughput sequencing technologies to analyze genomes of both laboratory-cultured organisms (Zhang et al., 2006) and uncultured environmental microbes (Marcy, Ouverney, et al., 2007). From this early work, single-cell whole-genome sequencing (SC-WGS) expanded from proof-of-concept to an established and popular technique that is applied widely in biology to supplement culture-based isolation/genomic approaches (Fig. 4.1). SC-WGS has been used extensively to access the genomes of yet-uncultivated organisms in environmental microbiology (Blainey, Mosier, Potanina, Francis, & Quake, 2011; Malmstrom et al., 2013; Marcy, Ouverney, et al., 2007; Stepanauskas & Sieracki, 2007; Swan et al., 2011; Woyke et al., 2009; Youssef, Blainey, Quake, & Elshahed, 2011) and problems in medicine, for example, studies of pathogens and components of the human microbiome (Marcy, Ouverney, et al., 2007; Pamp, Harrington, Quake, Relman, & Blainey, 2012). Applications of single-cell genomics to human genetics are also becoming more wide-spread (Fan, Wang, Potanina, & Quake, 2010; Wang, Fan, Behr, & Quake, 2012; Zong, Lu, Chapman, & Xie, 2012), and single-cell genetics has also been used for lineage analysis (Frumkin et al., 2008). While nearly all single amplified genomes (SAGs) have been produced using the multiple displacement amplification (MDA) chemistry, a variety of different approaches to isolating cells for SC-WGS have been implemented with success (Blainey, 2013).
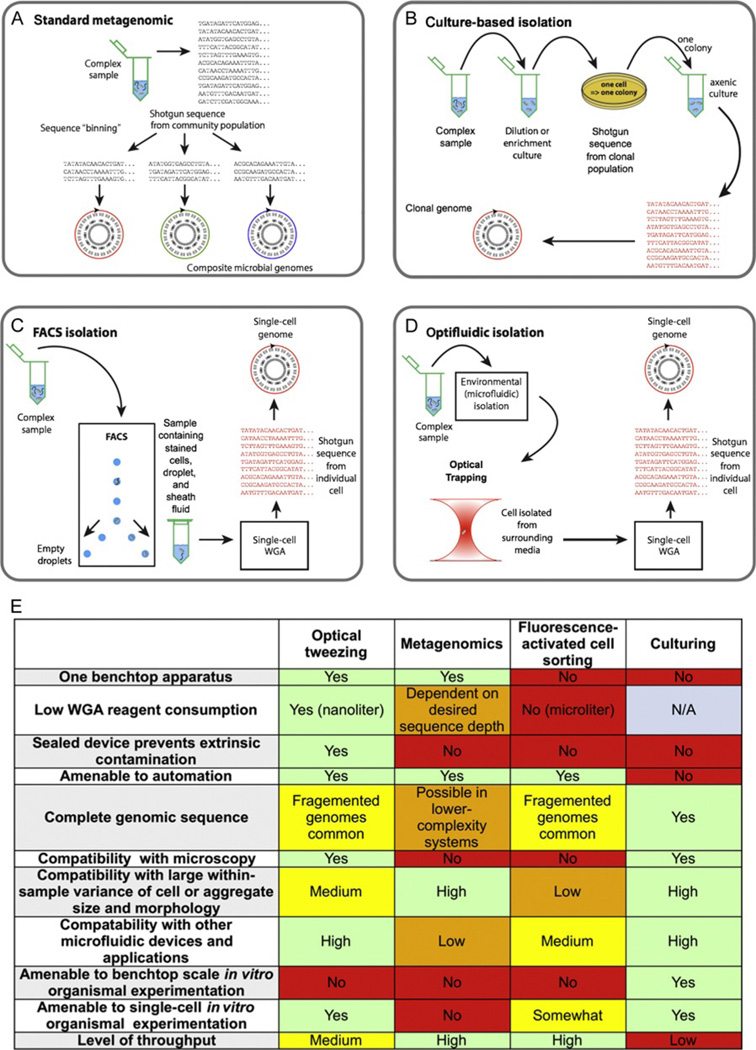
Figure 4.1.
Schematic diagram contrasting of methods to obtain genome sequences from yet-uncultured microbial species. Established techniques such as metagenomics (A) or culturing (B), and the two newer methods of single-cell whole-genome amplification by fluorescence-activated cell sorting (C), or the optofluidic approach discussed in this chapter (D). Advantages and features of each methodology are summarized in the table (E). Parts A–D were adapted with permission from Blainey (2013), FEMS Microbiology Reviews 2013.
Molecular techniques have assumed an increasingly important role in microbial ecology in recent decades, beginning with the application of gene sequencing to environmental DNA (Olsen, Lane, Giovannoni, Pace, & Stahl, 1986). Subsequently, it became clear that many species prevalent in the environment have no known relatives in culture (Rappé & Giovannoni, 2003). This presented the grand challenge of identifying the role of these unknown taxa in the environment, a task that remains unfinished today. With the rise of metagenomics, the extent of gene diversity became known for many environments (Venter et al., 2004). However, evolution acts on assemblages of genes, and the reconstruction of genomes, and thus the metabolism of cells, was hampered by the difficulty of assembling genomes from any but lower-complexity microbial systems (Tyson et al., 2004; Wrighton et al., 2012).
In recent years, SAGs have provided valuable genomic information for dominant uncultured microorganisms (Dupont et al., 2011; Pamp et al., 2012; Swan et al., 2011). In one example, SC-WGS revealed that the abundant SAR324 group of marine deltaproteobacteria may couple carbon fixation to sulfur oxidation, a prediction that was consistent with subsequent labeling of cells with radioisotope tracers (Swan et al., 2011). This information has important implications for the global ocean sulfur and carbon cycles. It is expected that similar advances in many underexplored environments will result from wider application of SC-WGS. Genome sequences have also paved the way for culturing previously uncultured strains in the laboratory by providing insight into unanticipated metabolic requirements (Renesto et al., 2003).
Fluorescence-activated cell sorting (FACS) is a popular method for producing SAGs today; however, optofluidic approaches, the topic of this chapter, offer advantages for many applications. Microscopic sorting makes it possible for the operator to sort cells visually and offers advantages in situations where (1) cells or assemblages of cells have distinctivemorphology, (2) fluorescent signatures are too weak or too complex for resolution by FACS, (3) only very small volumes of sample are available, or (4) large quantities of contaminating DNA are present, such as in the isolation of intracellular parasites from cell lysates. Other examples of problems that meet these criteria are small aggregates of cells such as “marine snow” that may be too fragile to study with FACS, epiphytic interactions, and syntrophic assemblages, such as anaerobic methane-oxidizing archaeal/bacterial consortia (Boetius et al., 2000).
2. OPTICAL TWEEZING FOR CELL ISOLATION
The production of SAGs requires two fundamental steps. First, the isolation of individual cells from a bulk sample, and second, the amplification of the genomic contents of the isolated cell by WGA. Various approaches to each of these steps and the impact of different technologies on contamination, the major challenge in single-cell genomics, have been reviewed previously (Blainey, 2013). We have implemented a particular cell isolation approach, optical tweezing (Ashkin, 1992, 1997; Ashkin & Dziedzic, 1989; Ashkin, Dziedzic, & Yamane, 1987; Ashkin & Gordon, 1983), to select individual microbial cells (Ashkin et al., 1987; Huber et al., 1995) and separate them for individual analysis inside custom microfluidic devices, where WGA can be carried out in nanoliter volumes while excluding extrinsic contaminants (Blainey et al., 2011). Figure 4.2 provides a granular breakdown of the component steps in the generation and sequencing of the genome of single cells using the microfluidic/optical tweezing approach.
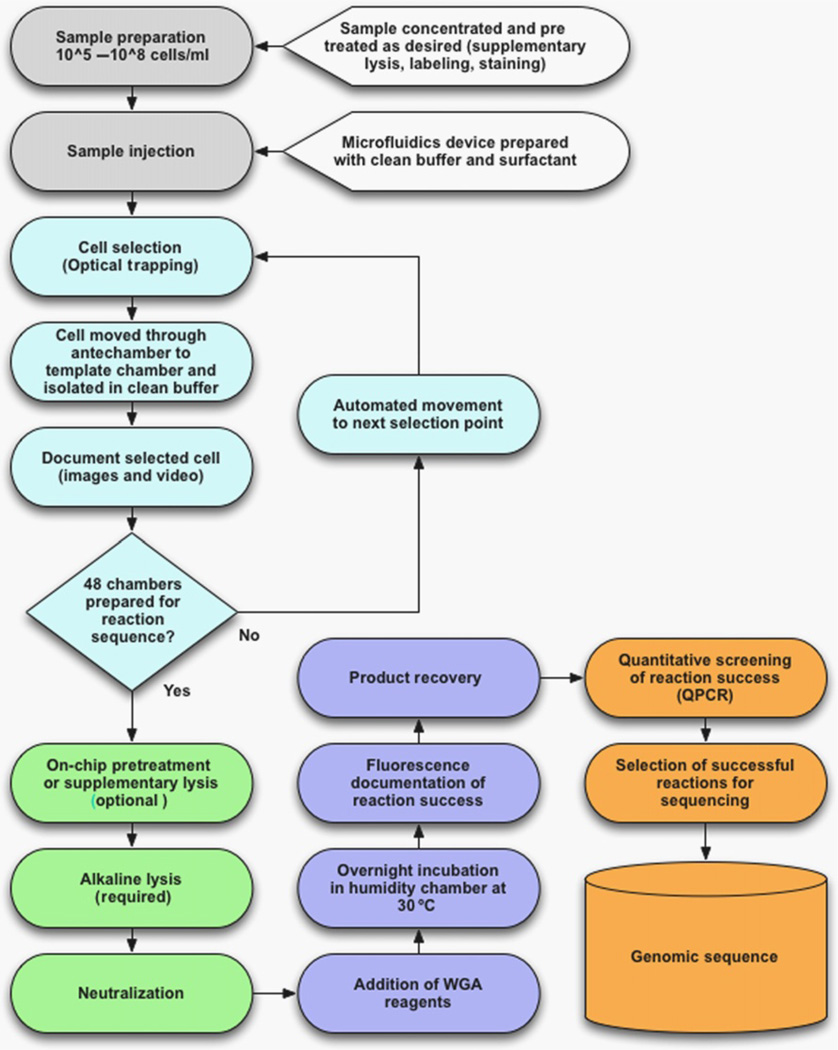
Figure 4.2.
Flowchart for single-cell whole-genome amplification optofluidics. Sample preparation and cell selection steps are shown in white; lysis-related steps are shown in light gray; amplification and product recovery are shown in dark gray; postprocessing steps are shown in black.
Optical tweezing presents two unique advantages that distinguish it from other methods of cell isolation that derive from a unique feature of the tweezing approach. First, movement of the cell is uncoupled from movement of the cell’s immediate surrounding environment. This is to say that the cell itself rather than a voxel (volumetric pixel) from the sample is selected. This specificity for objects within the sample can be understood based on the mechanism by which laser tweezers operate.
2.1. Applying forces to objects in solution with light
Optical tweezing (the terms optical/laser and tweezing/trapping are used interchangeably here) relies on a difference in refractive index between the cell and the surrounding solution. As such, cells (or other structures with a boundary defined by a refractive index gradient), but not the solution itself, can be trapped using this method. High-performance microscope objectives can focus light very tightly to create an optical trap that has a capture radius comparable to the wavelength of light used to create the trap.
These properties allow an optical trap to be used with surgical precision to move one cell when many others are nearby, while minimizing the possibility of carrying forward an extraneous cell or foreign DNA. Contamination by environmental DNA or double sorting is a significant practical concern in single-cell genomics, particularly in de novo applications, where reference sequence to identify contaminating reads is not available. By contrast, other popular methods such as fluorescence-activated flow cytometry and micromanipulation work by subdividing a liquid sample, and require a high dilution of cells in clean buffer to reduce the chance of contamination.
While idealized representations often depict spherical objects being trapped, in practice, cells of essentially any morphology can be moved using an optical trap. Tiny coccoid cells with a maximum dimension of about 0.5 µm can be trapped (Blainey et al., 2011), as can high aspect ratio rods (Dodsworth et al., 2013), or even huge filaments over 100 µm in length (Pamp et al., 2012). Cells much larger than the dimensions of the optical trap are effectively moved by tugging at the cell boundary.
2.2. Action at a distance
The second unique property of the optical trap is its ability to apply forces to cells at a distance, without bringing instrumentation for manipulation into contact with the cells or the sample solution. This property enables manipulation of cells inside a closed (transparent) container. The notion of carrying out single-cell genomics inside a sealed microfluidic device is extremely appealing for the purpose of eliminating contamination from extrinsic sources and automating the subsequent lysis and whole-genome amplification of the sorted cells in tiny nanoliter volumes. The unique features of the trap allow the order of operations to be reversed compared with other common single-cell workflows. In flow cytometry and micromanipulation, the sample is first subdivided into isolate cells and then sealed in separate vessels where lysis and WGA are carried out. The optical tweezer enables an inversion of steps: the sample can first be introduced to a sealed vessel prior to the isolation of individual cells.
2.3. Practical approach to SC-WGS with lower resources
One of the largest hurdles to overcome when attempting single-cell genomics is the issue of reagent cost. Kits marked for WGA, including MDA, currently cost on the order of $1 per reaction microliter. The cost of reagents can be prohibitive for even moderate-scale studies when conventional reaction volumes of 10–50 µl are used. Although ultra-clean WGA reagents can be produced in-house at lower cost (Blainey et al., 2011), reducing reaction volumes to the nanoliter scale on a microfluidic device (1 nl = 0.001 µl) is an attractive alternative to sourcing low-cost reagents. For example, the series of devices developed at Stanford carry out WGA in 60 nl volumes, using about $0.06 worth of retail reagents per reaction (Marcy, Ishoey, et al., 2007; Marcy, Ouverney, et al., 2007). If 40 conventional-scale (20 µl) reactions normally requiring $800 worth of WGA reagents can be reduced to the nanoliter scale, the reagent cost becomes insignificant and the incremental cost for single-cell genomics becomes dominated by the cost of the microfluidic device. Two-layer custom polydimethylsiloxane (PDMS) devices of the type presented in this review cost $50–100 each in low-volume manufacturing (e.g., from the Stanford Microfluidics Foundry), yielding a net cost savings for moderate scale single-cell genomics projects.
When capabilities for sorting and reaction setup are integrated in a single benchtop microdevice, the specialized infrastructure for single-cell genomics sample prep collapses to a single apparatus that sits on a bench in a standard lab environment. An advanced laser-trapping microscope can be built from about $100,000 in parts (far less if a suitable microscope is already available, which is the case in many labs) and turn-key laser-tweezing microscopes are priced around $200,000. These costs compare favorably with those for flow cytometry equipment and cleanroom space. Paired with a modern benchtop sequencing instrument, the optofluidic approach to single-cell genomics is well suited to individual research labs, teaching labs, and portable operation. Figure 4.2 gives an overview of the single-cell genomics process using the optical trapping/microfluidic amplification approach from sample to data.
2.4. Avoiding contamination
Reducing reaction volume pays an additional dividend in the reduction of reagent-borne contamination (Blainey et al., 2011). In the example moving from 20 µl to 60 nl reactions, the amount of contamination from reagents is expected to drop more than 300-fold.
Overall, optical tweezing provides a means to isolate cells that intrinsically selects against carry-over contamination and can be used to pluck cells from concentrated cell suspensions while leaving behind sample-borne contaminants. If paired with a capability for lysis and WGA in small volumes, a great savings in reagent cost and reduced reagent contamination can also be achieved.
2.5. Integrating microscopy and cell sorting
Optical trapping approaches to single-cell genomics retain all the characteristics of classical microscopy: the ability to identify and characterize cells or cell aggregates according to morphology as well as providing the option of integrating a number of staining or labeling techniques. At present, microfluidic approaches have lower throughput rates than single-cell genomics by flow cytometry and might not be suitable for broadly assessing biodiversity. However, the unique attributes of microfluidics open the door for the application of novel screens for cell selection. When implemented in a semimanual approach where a human user selects cells under high-resolution imaging (typically 100 × phase contrast microscopy), the selection of cells allows for direct interaction of the user with the sample in a notional “microbial safari” where the user observes a variety of fauna, occasionally “bagging” a trophy. Cells can straightforwardly be selected on the basis of any observable characteristic such as size, morphology, intracellular structure, or even faint fluorescence from native chromophores or reagents such as fluorescent in situ hybridization probes (Yilmaz, Haroon, Rabkin, Tyson, & Hugenholtz, 2010). Screens based on complex morphological characteristics such as the selection of certain microaggregates are also feasible and are likely to blur the distinction between single-cell genomics and metagenomics in ways that reveal new information about communities of interest.
2.6. Low required sample volume
Finally, microfluidic approaches are extremely parsimonious with the sample. In principle, a sample containing a single cell can be introduced to a device and intercepted with the laser trap for sorting. In practice, a 5-µl sample containing a few thousand cells in total is quite usable, indicating the suitability of this approach to single-cell genomics for especially rare or precious samples, where the volume required for other types of analyses may present a significant commitment or risk.
2.7. Technical considerations for sorting using an optical trap
For objects larger than the wavelength of impinging light (often the case when trapping cells with light, but not a necessary condition), a ray optics analysis of the optical trap is valid (Rocha, 2009). An object captured in a laser trap can be considered as a small lens that refracts (bends) and scatters the light from the optical trap. When light moves through an interface from a medium with one refractive index to a different medium with a different refractive index, the direction of the light is changed and the cell experiences a small reactive force in the opposite direction of this change. This happens because photons passing through the object or scattering off of it can exchange momentum with the trapped object, and in this way, apply force to it. When the object is centered in the trap, the optical forces on the object are balanced, but when the object is displaced from the center of the trap, the applied forces are unbalanced, and due to the optical geometry of the trap, the object experiences a force pushing it toward the center of the trap (in three dimensions). The amount of momentum carried by each photon is small, so a large number of photons must interact with the cell for any appreciable force to be applied. This necessitates the use of milliwatts of optical power for many applications, which corresponds to a very high power density when focused to a diffraction-limited spot to form a laser trap. The power density at the focus of an optical trap commonly exceeds the optical power density at the surface of the sun by several orders of magnitude. Surprisingly, this is not necessarily harmful to cells, which can easily survive continuous trapping when near-infrared light is used (Ericsson, Hanstorp, Hagberg, Enger, & Nystrom, 2000; Neuman, Chadd, Liou, Bergman, & Block, 1999).
3. OPTICAL HARDWARE SETUP
3.1. Specifying the laser and microscope
A complete list of parts for the optical setup and microfluidic controller is available in Supplement 2 (http://dx.doi.org/10.1016/B978-0-12-407863-5.00004-6). Selection of a light source for minimally perturbative manipulation of biological materials is limited to the near-infrared wavelengths where water and the stuff of life absorb only weakly. The choice of wavelength is especially important as even a small amount of absorption can be devastating to a cell exposed to the high optical power in an optical trap. Although, in principle, any collimated light source of sufficient power can be used for trapping, high-powered lasers are used almost exclusively. Recommended wavelengths are 980 nm, which has been shown experimentally to be one of the least biologically active wavelengths for use (Liang et al., 1996; Neuman & Block, 2004; Neuman et al., 1999; Zakharov & Thanh, 2008), and 1064 nm, a wavelength used in most commercially available trapping systems (also accepted to be compatible with cells). The 1064 nm lasers are readily available in high wattages with excellent beam quality from $5000 to $15,000 in the 0.5–3W range often used for optical trapping.
Considerations other than power and wavelength are relevant to trapping. Solid-state, continuous-wave lasers (as opposed to pulsed lasers) are recommended for trapping to avoid high peak powers and unwanted multiphoton photophysical phenomena. To form a single, tight focus (helpful for small cells), the beam must have a Gaussian transverse mode (single round spot brightest in the center with Gaussian intensity distribution across the beam when projected, also known as “TEM-00”). Lastly, power is a consideration. Although not the only factor in determining trapping force, a higher power light source will help increase trapping capabilities. A minimum of 100 mW reaching the sample is necessary for practical trapping, with higher powers allowing the application of more force to cells (e.g., faster movement of cells). The optics used to condition the beam for trapping attenuate the beam by absorption and back reflection. The laser setup described in this chapter outputs a 1-W, 976-nm beam from the head of the laser module, with about 250 mW reaching the sample (the largest single loss in the optical train occurs at the microscope objective). An additional safety consideration when purchasing a laser source is the laser power supply. A variable power supply is desirable, as it allows alignment of the laser source at lower (less dangerous) power levels, while prolonging the life of the laser.
When taking an automated or semiautomated microscopy approach to microfluidic single-cell genomics, microscope and objective selection are paramount. By far, the most convenient implementation entails an inverted “fluorescence” microscope with an automated (motorized) stage. Unwanted beam attenuation can be mitigated by careful selection of optical components. Although many groups build simple microscopes for optical trapping from individual components, a commercial inverted microscope is desirable when one desires to use many different imaging configurations (e.g., different magnifications and different techniques like phase, fluorescence, etc.) or would like to automate configuration changes. Typically, the laser beam will be introduced to the microscope via the “fluorescence excitation” path. Importantly, some microscopes have internal optics that may need to be accounted for and possibly modified. Specifically, any diffusing optics will need to be removed (preferably by the manufacturer) to achieve a tight focus, as was necessary in the Leica model DMI6000B automated scopes used for our applications. Preferably, the microscope presents a port with a clear path (no optics) to the dichroic mirror that reflects the fluorescence excitation up to enter the microscope objective. Figure 4.3 shows an idealized (simplest) optical diagram (part A) and two as-configured layouts specific to the Leica DMI6000B microscope (parts B and C).
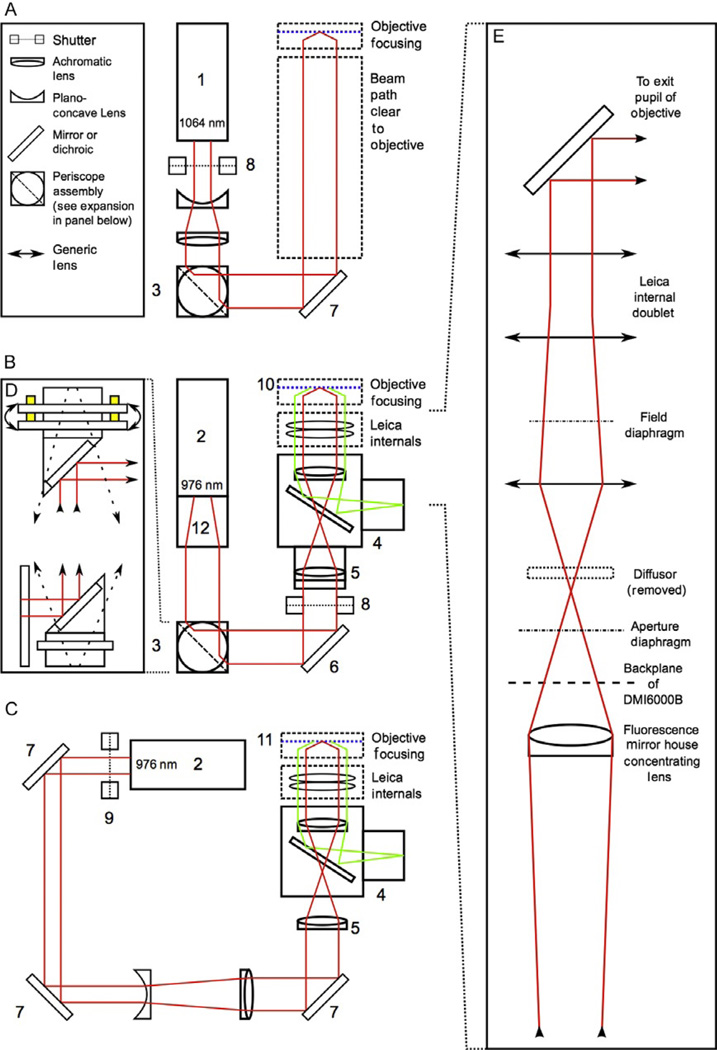
Figure 4.3.
Simplified schematic diagrams showing an idealized light path for an inverted microscope with a minimum of internal components (schematic A), and two functional designs based around the Leica DMI6000B microscope stand (schematics B and C). The suggested design in schematic A includes a 1064-nm, 3-W laser source (1) chosen for its high power and ready availability, as well as its wavelength, which is suitable for biological materials. The idealized design contains a beam expander consisting of a set of matched lenses preceded by a shutter (8; a Thorlabs part no. SH05 with a SC10 shutter controller was used in the configuration shown in B) placed at the head of the laser to enable control the beam without cycling the laser. Beam positioning and alignment is accomplished with a periscope consisting of two rotating mirrors mounted at 90 °C angles. An expanded view of the periscope used in the second configuration is presented in inset D (3; Thorlabs part no. RS99, equipped with dielectric broadband mirrors, Thorlabs part no. BB1-E03). By placing the periscope mirrors at right angles to each other, rotation of the mirrors can be used to coarsely position the beam horizontally and vertically on the third mirror, eliminating the need for three independent kinematic mounts. The periscope design used includes kinematic controls on the top mirror mount, allowing for the fine pitch-and-yaw adjustments needed for positioning at the second mirror. A third kinematic mount (6; Thorlabs part no. KM100 kinematic mount with BB1-E03 mirror) is used to adjust beam angle. The two designs utilizing the Leica DMI6000B microscope stand are somewhat more complex, largely due to the number of internal optics in the scope stand. Both of these setups utilized a 1.0–1.1-W, 976-nm laser (Crystalaser model DL-980-1W-OX/DL980-1W; 2), the wavelength chosen because of biological compatibility. The laser in the schematic B was modified at the factory with a spatial filter (12) to improve beam shape and power distribution. This spatial filter had the added effect of expanding the beam to ~8 mm, eliminating the need for additional beam expander optics. This design uses a periscope for two of the mirrors as described above. A third kinematic mount provides correction for beam angle (6). In this design, the shutter (8) is mounted outside of a customized mirror house (4) that includes a dichroic mirror to allow for simultaneous trapping and fluorescence. Due to the mirror house optics designed to bring the fluorescence excitation source into wide illumination field at the focal plane, a corrective achromatic lens (5) was installed in the mirror house at the factory to correct expansion of the trapping beam so that the beam would remain collimated upon entering the exit pupil of the trapping objective. This design utilized a 100 × magnification, 1.4 numerical aperture (NA) apochromatic oil immersion objective as the trapping objective (10; Leica part no. 11504107). An alternative design shown in schematic C was also used. This design uses an independent corrective lens in the free space laser setup to offset the fluorescence optics and also includes a free space Galilean beam expander made from two matching lenses located between the second and third kinematic mounts. In this setup, the laser was mounted at a height and three individual generic mirrors and kinematic mounts (7) were used to align the laser. As in the suggested setup, a shutter (9; Uniblitz) was placed at the head of the laser. The trapping objective used here was 100 × magnification, 1.3 NA apochromatic oil objective (Leica part no. 11506197; 11). A simplified diagram of the Leica DMI6000B internal components (inset E) is shown in the expansion on the right of the figure. Because the optical design inside the microscope is Leica-proprietary, the exact specifications are not indicated here. The important feature to note is that expansion and focusing optics placed external to the scope stand must be positioned so as to produce a collimated beam at the end of the light path prior to entering the exit pupil of the trapping objective. An additional, important modification is removal of the standard diffuser included in the Leica excitation light path (see inset E). The diffuser is located on a circuit board inside the scope at the position marked in inset E. Modifications to the scope internals are exacting and more accurate diagrams of the light path and scope internals can be requested from the manufacturer.
We specified the Leica DMI6000B stand due to its full automation and compatibility with particular Leica objectives known at the time to be available with high numerical aperture (NA), phase contrast, and acceptable infrared (IR) transmittance (Neuman & Block, 2004). However, all major microscope manufacturers today (Leica, Nikon, Olympus, Zeiss) are optimizing objectives for improved IR transmittance (transmittance should not be confused with correction for IR chromatic aberration), and useful features like external phase contrast are also now available. An additional consideration when choosing a microscope “stand” (the main body of the microscope) is competitive pricing as well as adaptability to your application and compatibility with particular software packages for automation. Prior to making a final decision on a microscope, be sure to demand IR transmission curves for the trapping objective lens and internal optics, as well as a comprehensive light path schematic indicating all internal optics the trap beam will encounter. One final consideration in choosing a microscope stand is selection of an appropriate automated stage. For our purposes, we used Leica DMI6000B stands equipped with either the standard Marhauser-Wetzlar stage available from Leica or the Applied Scientific Instrumentation MS-2000 (Eugene, OR). The stage must be able to be controlled both manually (e.g., via a joystick) and programmatically through a software interface (often drivers for RS-232, Labview or a .dll file is provided). Ensure the stage has a high level of movement precision and can be fluidly translated at speeds in the range 1–1000 µm/s. A linear encoder is also useful, as it improves the accuracy of repeated positioning.
3.2. Choosing an objective lens
By far, the single most important factors to consider when designing any optical trapping platform are IR transmission and NA of the objective lens that will be used for trapping. Higher NA provides oblique rays on which trapping in the focal dimension (z-coordinate) depends and dramatically improves overall trapping performance. A minimum of 1.0 NA is required for practical trapping, and NA of 1.3–1.5 is recommended. The objective also needs to be able to transmit IR wavelengths efficiently; however, priority should be given to maximizing the NA as low transmission can be mitigated by increased laser input power. If the trapping performance is too weak, the cell will easily escape the trap along the direction of the beam propagation. In a microfluidic channel, the upper channel surface may restrain the cell, allowing trapping of the cell in two dimensions against the upper surface of the microchannel. Even so, a better-performing trap able to hold the cell in three dimensions is strongly preferred because the cell can be moved more quickly when suspended in the center of the microchannel cross-section, and the possibility that the cell sticks to the channel surface is eliminated. Our applications utilized a 100 × magnification 1.3 or 1.4 NA apochromatic oil immersion objective (Leica part nos. 506211 and 11506197).
3.3. Alignment procedure and safety
IR light at the powers used for optical trapping is capable of quickly and permanently damaging the retina in certain exposure scenarios, and the upmost care needs to be taken in terms of safety. Alignment of laser optics should only be attempted with appropriate safety precautions. A laser safety class is recommended and is required at many research institutions. IR laser beams, invisible to the naked eye, merit extra caution because emissions are not visually apparent, and the IR light does not trigger a blink reflex. When working with lasers, appropriate shielding should be in place to ensure that laser light will not impinge on untrained or unaware individuals. Laser goggles blocking the appropriate wavelengths should be worn at all times when there is a possibility of laser emission. In general, the highly divergent laser light emitted from the microscope objective during trapping falls below a permissible exposure level after a short distance. A laser safety class should train users to calculate this distance for their particular configuration. Even so, it is good practice to place shielding above the stage to block user exposure to this divergent emission. All work should be done standing with the plane of the laser lines below the user’s eye level since stray beams are most likely to occur within this plane. Care should be taken to never look directly at the laser source or at reflective surfaces (e.g., mirrors) in the path of the laser. Reflective surfaces in the work area and on the users’ body (rings, watches, etc.) should be kept to a minimum. Importantly, the eyepieces on a microscope used for optical trapping with IR light must be removed or permanently disabled to eliminate the possibility that a user could directly view the laser beam in the case of user error or microscope malfunction.
The end goal of alignment is to position an expanded and collimated laser beam on the optical axis of the objective lens. To take advantage of the full NA of the objective lens, the beam should fully fill the back aperture of the objective, and thus prior to entry into the scope should be expanded to slightly overfill the objective’s back aperture. A convenient way to build a simple Galilean beam expander is from two lenses, one plano-concave and one convex (a Keplerian beam expander constructed from two positive lenses will also work). The two lenses should be selected to achieve the desired degree of beam expansion according to the equation:
where r is the radius of initial beam, f1 is the focal length of plano-concave lens, f2 is the focal length of convex lens, E is the expanded beam diameter, and the space between the two lenses is the sum of the two focal lengths.
When aligning the laser setup, at least three mirrors and adjustable kinematic mounts (Thorlabs part no. KM100 kinematic mount with BB1-E03 mirror were used in our application) should be used (alignment can be achieved with two kinematic mounts, but is easier with three). Dielectric broadband mirrors (Thorlabs part no. BB1-E03) are recommended due to their very high IR reflectance (hence low attenuation).The simplest setup positions the laser aperture at the same height above the table as the entry of the beam into the microscope. Alternatively, two of the mirrors may be configured as a periscope (we used Thorlabs part no. RS99 for this purpose) to raise the path of the beam (Fig. 4.3B). If the laser lacks a modulation/shuttering capability, a shutter is best placed between the laser source and the first mirror so that laser light can be excluded from the work area without turning the laser on and off. For our purposes, we mounted the Leica DMI6000B microscope stand in an amenable fixed position on a 1 m × 1 m × 50 cm optics table prior to placement of mirror optics. Some microscopes come with accessories to directly mount the stand to a standard optical table (drilled and tapped 1/4–20 on 1 in. centers or M6 on 25 mm centers). The laser was mounted in a fixed position appropriate for our setup. Initially, all mirrors should be positioned based on measurements such that the path of the laser will follow right angles as closely as possible at each mirror. It is important to use the reflective face of the mirrors as your reference for where the beam will be redirected, as the face of the mirror may project proud of the mount. When initially placing the mirror mounts and mount stands, it is good practice to physically verify their location and angles, and not simply “eyeball” their initial position (there will be plenty to adjust even with the most exacting initial placement of optics).Good practice demands that all optics on the table are fixed in position (directly bolted or clamped with “dogs”) when the laser is active to avoid unexpected redirection of the beam if an optic shifts or topples.
After the initial placement of the optics, alignment can begin. After ensuring all shielding is in place and everyone present is alert, the laser can be powered on. To align the laser, a clear way to visualize the end point of the beam and assess alignment needs to be in place. A useful tool consists of a short piece of optical tube (threaded aluminum tubing) 1–3 in. long with an adapter that allows mounting on the objective turret. The tool should have an iris next to the thread adapter, the length of the tube, then a second iris, and a cap consisting of a phosphorescent plate (allowing visualization of the invisible beam) with the center marked. For our purposes, we used Thorlabs part no. SM1A11 (adapter), SM1D12C (iris), SM1L30C (3 in. slotted lens tube), SM1D12C (iris), SM1L05 (0.5 in. lens tube), with VRC4D1 (phosphorescent disc) mounted using two retaining rings (SM1RR). The configuration of our alignment tool is detailed in supplementary figure 1 (http://dx.doi.org/10.1016/B978-0-12-407863-5.00004-6). Phosphorescent cards can be inserted into the laser path at multiple points in the beam to track the position and angle of the laser.
Initially, rotating the positions of the mirror mounts should allow for coarse positioning of the beam. When the laser is roughly aligned with the optomechanical hardware in fixed positions, adjustment using the finely threaded thumbscrews on the kinematic mounts can begin. The first two mirrors should be used to adjust the position of the beam on the third mirror. Adjustment of the third mirror should be reserved to correct the angle of the beam. Between proper positioning of the optomechanical hardware, coarse adjustment of mount position and fine alignment using the pitch-and-yaw screws of the kinematic mounts, some portion of the beam can be made to appear on the phosphorescent plate of the objective tool when it is positioned in place of the trapping objective. By continuing to use the first two mirrors for positioning, the beam can be centered. By adjusting the pitch-and-yaw of the third mirror, the angle of the beam can be corrected.
By placing an iris in between the final mirror and the entry point of the scope (centered on the optical axis of the microscope), the beam can be straightened as much as possible by ensuring that both this iris and the lower iris on the objective alignment tool close evenly around the beam (visualizing the projected beam on the alignment tool phosphorescent plate). When this is achieved, the beam is roughly aligned. The beam alignment can be fine-tuned by bringing the trapping objective into place, mounting a microscope coverslip or coverslip-mounted microfluidic device on the stage of the microscope, and visualizing the light reflected when the trap is focused on the upper surface of the coverslip with a CCD camera. By adjusting the mirrors in the beam path, the trap can be moved to the appropriate position within the field of view and the angle of the beam fine-tuned by optimizing the symmetry and concentricity of rings in the out-of-focus image of the trap reflection. The trap can be made coincident with the image plane of the microscope by adjusting the divergence of the trapping beam that enters the microscope by adjusting the distance between the lenses making up the beam expander. A diverging beam will form a trap above the imaging plane of the microscope, while a converging beam will form a trap below the imaging plane of the microscope.
4. MICROFLUIDICS CONFIGURATION AND SETUP
4.1. Microdevice design
The most current single-genome amplification device is the 48x_v4 device illustrated in Fig. 4.4. This chip can be ordered directly from the Stanford Microfluidics Foundry, and mold sets can also be obtained for in-house production of microfluidic devices. Supplement S4 (http://dx.doi.org/10.1016/B978-0-12-407863-5.00004-6) contains data files with the design of the 48x_v4 microfluidic device, Supplement S5 (http://dx.doi.org/10.1016/B978-0-12-407863-5.00004-6) describes the standard operating procedure for fabricating molds for the 48x_v4 device, and Supplement S7 (http://dx.doi.org/10.1016/B978-0-12-407863-5.00004-6) is a sample order sheet for obtaining molds and/or devices from the Stanford Microfluidics Foundry.
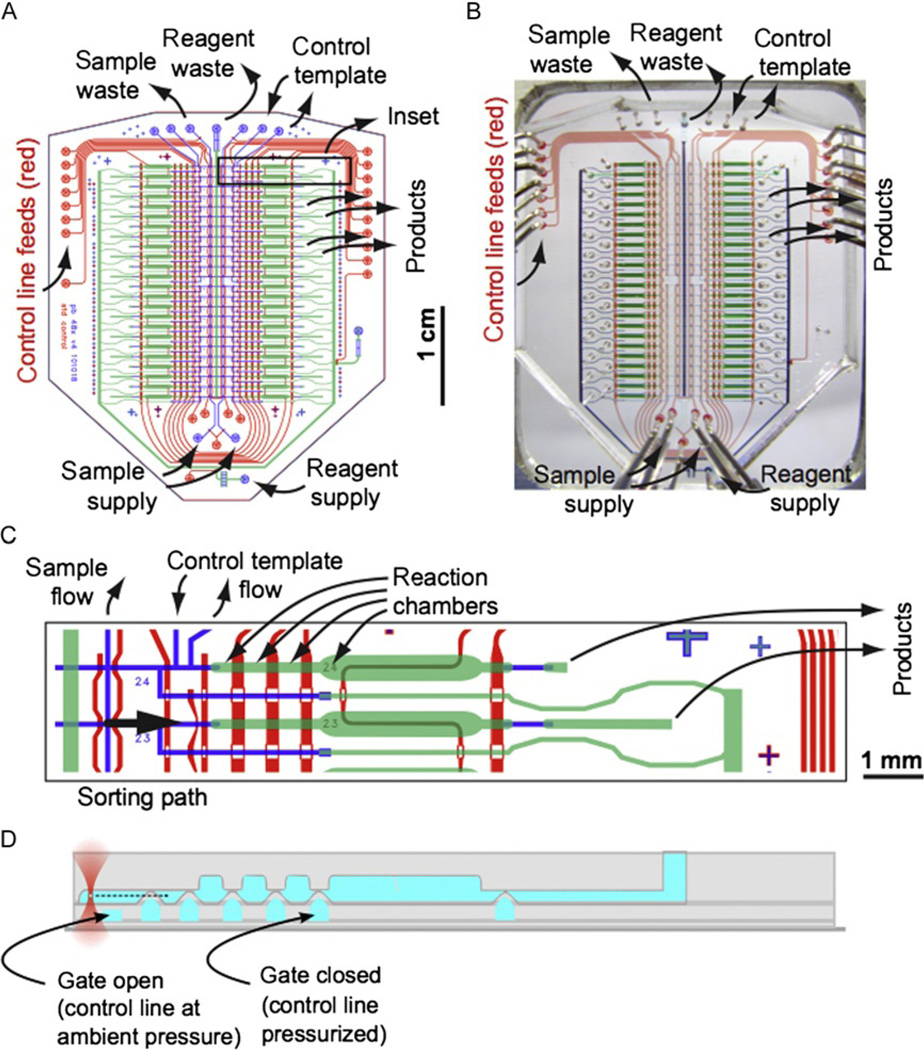
Figure 4.4.
48-chamber microfluidic chip for single cell sorting and whole-genome amplification. (A) Design of 48x_v4 device with key ports marked. (B) Photograph of 48-chamber device with corresponding ports marked. (C) Inset showing detail of two reaction chambers in the 48x_v4 device. (D) Cross-sectional schematic corresponding to part (C) indicating the trapping of a cell in the (upper) flow layer and the actuation of valves by pressurization of channels in the (lower) control layer.
The 48x_v4 device includes 48-arrayed independent reactor systems for cell selection, lysis, and whole-genome amplification. The device consists of two molded layers of PDMS mounted on a number one microscope coverslip. The transparent nature of the PDMS and mounting on the thin coverslip allows the application of most microscopy techniques and optical trapping. We have designed a custom mount for the 48x_v4 device that can be produced on a 3D printer and fits into the standard 110 × 160 mm stage insert opening. The holder securely grips the device, flattening any curling of the coverslip, protects the device from peeling forces, and prevents damage to the coverslip. The design files/renderings of the chip holder are provided in Supplement S3 (http://dx.doi.org/10.1016/B978-0-12-407863-5.00004-6).
The microfluidic device utilizes a “push-up” approach to the actuation of microfabricated valves. Control channels in the thin PDMS layer adjacent to the coverslip are filled with water and pressurized to actuate valves (also referred to as “gates”) on the device. Where the pressurized control channels cross flow channels in the top device layer, the thin membrane separating the two stretches to pinch off the microchannel in the flow layer (Fig. 4.4D). Control lines are typically configured as dead-end channels and are filled completely with water, taking advantage of the high gas permeability of PDMS, which allows air to rapidly escape microchannels as fluid is pushed in. Under normal use, the lines are under atmospheric pressure (valves open), and 10–20 psi of air when actuated to close valves.
Each control line is coupled to an individual fluid reservoir with an over-head pressure controlled by an electronic solenoid. The microfluidic controller used to control the solenoids consists of a USB microcontroller with 24 amplified digital output bits/channels, each of which powers a single solenoid to select either atmospheric (valves on chip open) or “control” (valves on chip closed) pressures. The solenoids are mounted on three 8-port manifolds supplied with the desired “control” pressure by a gas regulator. By sending a computer signal to control the solenoid states, the valves on the chip can be opened or closed at the behest of a user or in automated routines. The Stanford Microfluidics Foundry hosts information on the capabilities, assembly, use, limitations, and alternatives to the USB chip controller, as well as in-depth workshops on the design, fabrication, and use of microfluidic devices (http://www.stanford.edu/group/foundry/).
The 48x_v4 chip is designed using two nominal control channel dimensions (both square profile produced in SU8 resist): 100 µm wide and 25 µm high, and 200 µmwide and 25 µmhigh. Two nominal flow channel dimensions are also specified: small channels (round profile, produced in SPR resist, valved by the narrow control channels) 100 µm wide and 7–13 µm high (taller to accommodate larger cells) and large channels (round profile, produced in AZ50 resist, valved by the wide control channels) 200 µm wide and 50 µm high.
The 48x_v4 chip has been designed for robust operation, with valves tuned (by varying control channel width from the nominal dimensions) to operate at uniform pressure, extra space between components (and component redundancy) to allow proper function in the face of fabrication defects, and a large sealing area around the device perimeter for robust lamination. We produce three 48x_v4 devices on a 10-cm wafer, which allows ample space between devices for handling in the course of manufacture.
The 48_v4 variant was in fact designed for reuse, with extra features like tuned outlet resistances that facilitate clean-out. PDMS devices are mechanically robust, with valves able to cycle many thousands of times without showing wear (Unger, 2000). The most challenging aspect of reuse is preventing the DNA products of the previous cycle from contaminating reactions in the current cycle. We validated a clean-out protocol utilizing sodium hydroxide and 0.6% hypochlorite washes, plus a UV decontamination step (PDMS is transparent in the UV). This clean-out procedure was found to be sufficient in eliminating a PCR-traceable test template from the device (data not shown). With appropriate negative controls, reuse of the 48x_v4 device is certainly a feasible option.
5. PROCEDURE FOR SORTING AND AMPLIFYING SINGLE CELLS
Each reactor on the device is intended for the processing of an individual cell, and consists of a “template chamber,” two “lysis chambers,” and a “reaction chamber,” connected in series. Separating the template chamber from the “sample lines” is an antechamber filled with clean buffer and tapped for the addition of reagents from the “reagent line.” This configuration provides a “privileged” flow path for reagents to each reactor that does not trace any channel where the bulk sample flows. A full protocol for using the 48x_v4 device is provided in Supplement S6 (http://dx.doi.org/10.1016/B978-0-12-407863-5.00004-6).
Two independent “sample lines” run the length of the device, each intersecting half of the reactors. Cells identified for selection under the microscope are sorted by moving the targeted cell from the sample line to the template chamber using the optical trap. The antechamber buffer is kept clean by slightly pressurizing the antechamber (from a 2 psi feed via the reagent line) prior to opening the gate (valve) separating it from the sample selection line, preventing cells from inadvertently crossing into the antechamber. After a cell is selected and moved into the antechamber, this gate can be closed again and the cell can be taken through the antechamber to the template chamber by transiently opening a second valve, which is closed after the cell is placed in the template chamber. Using this “air-lock” setup (the cell must pass through two valves on the chip), cells can be individually passed to the template chambers while the bulk sample solution is completely excluded from the template chambers. This works despite the design necessity that the air-lock valves operate in blocks of 12 template chambers (four blocks in all). Cells in each of the four device blocks can be treated with different lysis or reaction conditions, since the second air-lock valve controls the flow of reagents into each cell’s reactor section. The ability to run different samples independently on the two halves of the chip, and to apply as many as four sets of reaction conditions, makes the device adaptable to different project configurations and allows efficient optimization of conditions for a given sample type. At the top of the device, provisions are made to inject positive control solutions (typically cells or nucleic acids) directly into reactors 24 and 48. Properly selected controls can be very useful to distinguish failures of lysis from failures of amplification.
The large surface area to volume ratios associated with microfluidic arrays present issues of “sticking” where hydrophobic or electrostatic interactions between the cells (when not held away from the walls using the optical trap) or reagent components and the walls of the device necessitate the use of surface treatments or surfactants to mediate the interaction between solution components and the walls of the device. We have found that 0.04–0.08% Pluronic F127 in the sample dilution buffer is extremely effective in preventing the sticking of most cell types to the device. Bovine serum albumin (BSA) can be supplemented if necessary, but the level of contaminating DNA should be assayed (Blainey et al., 2011), and measures taken to source a low-contamination sample or degrade contaminants (UV treatment is recommended, but can cause aggregation). Provided that amplifiable DNA can be removed or destroyed, 8 µg/ml of BSA can be used in both the sample dilution buffer as well as the sterile diluent. Many groups have developed chemical and physical coatings that prevent adsorption of cells and biomolecules to PDMS, but we find the application of dynamic coatings (soluble surfactants such as Pluronic F127 and Tween-20) convenient and effective.
Potassium hydroxide treatment (0.4 M, optionally with EDTA, DTT, surfactants, and elevated temperature) is the most commonly applied lysis treatment in single-cell genomics, but is not sufficient for the lysis of many environmental microorganisms. Pretreatment (of the bulk sample or individual cells on chip) with enzymes and/or detergents can be helpful in many cases, but appropriate positive controls should be used to ensure that WGA is not inhibited by the lysis reagents. As a side note, a coverslip-mounted chip was observed to survive cycling to liquid nitrogen temperatures with no ill effects, indicating the possibility of lysis by ice crystal formation within the device. Two 3.5 nl lysis chambers are provided in each reactor to allow the flexible application of sequential lysis treatments. When one reagent is changed for another, the entire reagent delivery manifold is flushed out of the central waste line to guarantee that the new reagent is applied at its full concentration.
After lysis, the third 3.5 nl chamber can be used to chemically neutralize any lysis conditions applied, for example, adding sodium acetate to neutralize an alkaline lysis step. Subsequently, the WGA reaction buffer can be prepared and applied to flush the lysate into the 50 nl reaction chamber for a total reaction volume of 60 nl. For MDA, the device is incubated at 30–31 °C on a digital hot plate with the hydration line, which runs directly under the reaction chambers, pressurized. The degree of hydration is an important parameter in optimizing reactivity. Too little hydration and the volume or the reaction will shrink as a net loss of water vapor occurs; too much, and the chamber can swell, “burping” products from the outlet and diluting the reactants. The correct amount of hydration depends on the gas permeability of the PDMS and the ambient humidity. A humidity chamber is useful and can consist of a sealed Pyrex dish that the microfluidic device is placed into with a small amount of purified water. This is placed on a hot plate with a temperature probe inserted into the chamber to regulate the supply of heat from the hot plate. (The Thermo Scientific Super-Nuova model was used for this purpose; its advantages include probe-controlled temperature as well as logging through an RS232 port.) Only the MDA chemistry has been tested in the device, although other WGA chemistries dependent on short high-temperature steps would likely work if provided increased hydration pressure.
We add an intercalating dye (0.5 × EVA Green) to the reactions and monitor product accumulation in real time by taking fluorescence images of the reaction chambers (a less informative alternative approach is endpoint fluorescence monitoring with 1 × SYBR). When complete, products are recovered in parallel by applying Tris buffer to the reagent supply and flushing the products into pipette tips inserted into the outlet of each reactor. Individual recovery of the product from each reaction eliminates the possibility of cross-contamination, while simultaneous recovery of the products in parallel (the device has been optimized for a uniform flow rate across all the chambers) enables rapid product collection in the pipette tips, with 5 µl recovered from every reaction within a few minutes. Although the products are diluted about 100-fold during this recovery step, leftover primers and considerable phi29 DNAP activity exist in the product mixture. To avoid interference with downstream assays such as PCR, it is strongly recommended that the products be heat treated (65 °C, 10 min will abolish the polymerase and exonuclease activities of phi29 DNAP) and further diluted and/or purified prior to other analyses.
6. SOFTWARE
The LabView graphical programming language was used to design an integrated software interface for the scope, stage, and microfluidic controller. Matlab, Micro-Manager, and a variety of commercial imaging programs can also be used to coordinate hardware control and image acquisition. In our implementation, the user is presented with a graphical interface (Fig. 4.5A) that shows a live view of the chip through the microscope (with fiducial marking the position of the optical trap within the field of view) and a chip schematic indicating the status of control lines. The interface provides for manual control of the on-chip valves, laser, stage, and camera, as well as automatic functions such as setting preprogrammed valve states, moving to predefined locations on the chip, time-lapse imaging across multiple fields of view, and initialization routines for calibrating the stage. Although a previous design relying on pumped flow for sorting was automated for user-independent sorting (Marcy, Ishoey, et al., 2007), the current software runs in a semiautomated mode where the human user selects cells and initiates a programmed sorting routine. Applications where the choice of freely diffusing cells can be reduced to a machine-vision task (e.g., by a fluorescence intensity metric) may benefit from complete automation of the sorting process.
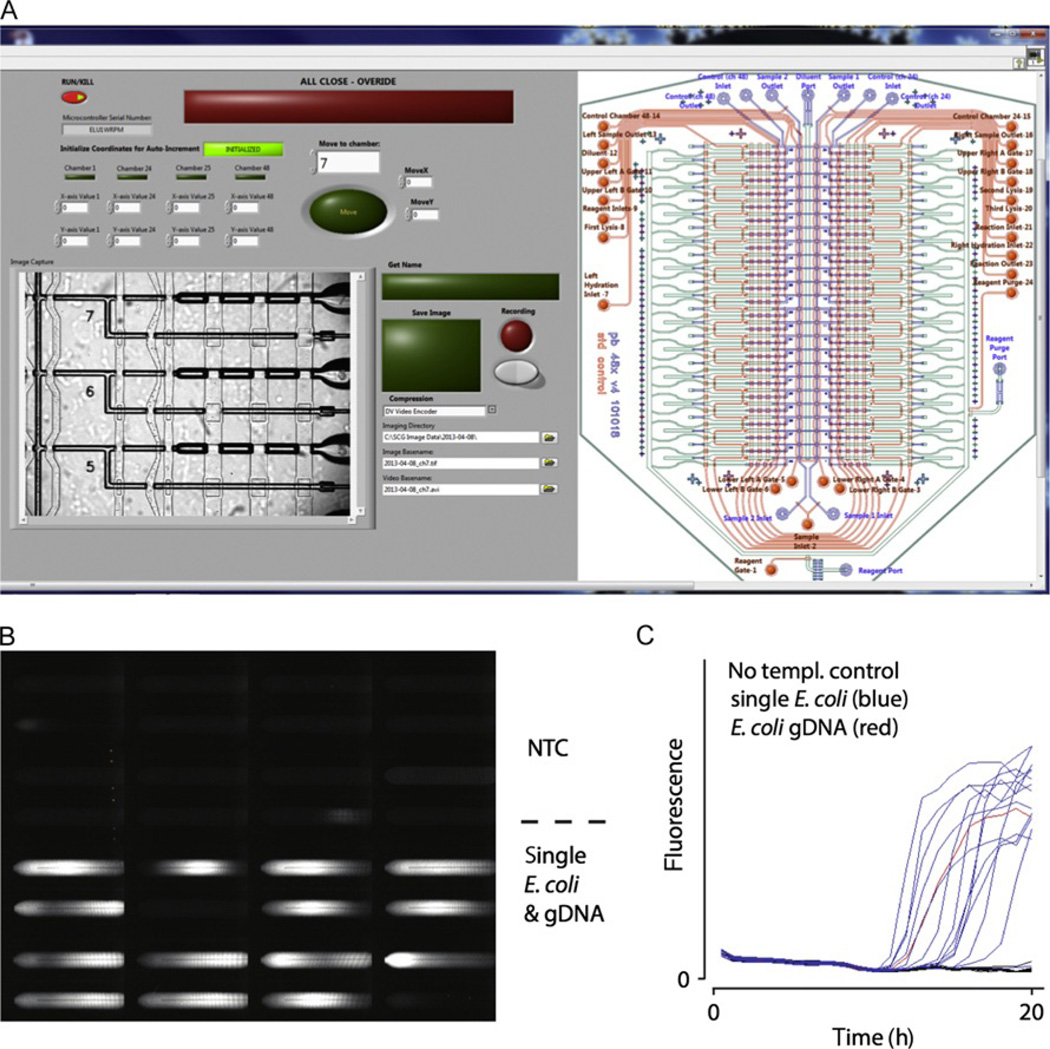
Figure 4.5.
Example of dual-screen graphical user interface with integrated microfluidic controls, stage automation, and video and image recording (A). Automated fluorescence data acquisition (B) and real-time kinetics curve of double-stranded DNA formation in individual microfluidic reaction chambers (C).
Valve control is integrated using the LabView drivers for the USB microcontroller card used in the Stanford valve manifold controller design. LabView drivers were used for control of the ASI and Marhauser-Wetzlar motorized stages. In both setups, a coordinate teaching system was developed to automatically calculate the location of each sorting intersection on the microfluidic device so as to improve the speed of cell sorting and help with navigation at higher magnifications. Our system measured focal positions as well, interpolating the z-position of the flow channel in between measurement points. For effective movement across large areas of the device, a chip-mounting system such as our 3D printed holder that flattens the coverslip and levels the device on the microscope stage is critical to prevent excessive focus run-out with high-power trapping objectives, which have a shallow depth of field. Alternatively, or in addition, an active autofocus system can be utilized to maintain the flow channels in sharp focus.
Computer-based control of the laser system is also highly recommended to improve safety and facilitate automatic control of sorting operations. The preferred solution is a laser that can be directly modulated by a digital or analog signal. Alternatively, a stand-alone electronic shutter is also suitable (e.g., Thorlabs part no. SH05 with SC10 shutter controller).
High-speed video imaging is critical to both manual and automatic manipulations of cells with the trap as well as documentation of the cells selected for analysis. A live-view frame rate of at least 25 frames per second is critical for eyestrain-free viewing over long periods of time. For optimally resolved and high-contrast fluorescence imaging of cells, the size of pixels on the detector should be between 0.5 × and 1 × the projected size of diffraction-limited objects in the field of view. For example, a 100 × NA 1.4 objective lens imaging green light would have a calculated resolution in the sample of ~ 0.6 × 525 nm/1.4 = 225 nm, and the optimal pixel size would be between 225 nm/2 × 100 = 11 µm and 225 nm × 100 = 22.5 µm. For this system, pixels smaller than 11 µm would compromise signal-to-noise ratio for little gain in spatial resolution, while pixels larger than 22.5 µm would compromise spatial resolution. Particularly useful is the integration of application-specific imaging modalities into the GUI. These may include fluorescence or interference-based imaging of cells and automated endpoint or time-lapse fluorescence of DNA stains in each MDA reaction chamber as illustrated in Fig. 4.5B and C. Additional facilities for video and image documentation of cells with automated file naming (by day, sample designation, and chamber number) during sorting may also be built into the GUI.
7. PERFORMANCE AND LIMITATIONS
In practice, the optofluidic laser-tweezing/microfluidic WGA approach provides a number of unique advantages as a method for highly targeted single-cell genomics studies. With a well-engineered system and medium-sized cells, the speed at which cells can be manipulated can reach ~200 µm/s or more; however, as trapping forces are dependent both on the size and shape of the cell, the maximum transport speed can vary. Optimal cell sizes for fast sorting range from 1 to 10 µmin diameter, but larger cells or groups of cells can be manipulated by trapping a portion of the structure and moving at slower rates. Smaller cells of 0.5 µmcan also be effectively manipulated by reducing the rate of movement; in fact, individual viruses (Ashkin et al., 1987), molecules (Katsura, Hirano, Matsuzawa, Mizuno, & Yoshikawa, 1998), and even atoms (Chu, Bjorkholm, Ashkin, & Cable, 1986) can also be confined in simple, single-beam optical traps. The limiting factor in trapping small cells is not usually the ability to achieve sufficient confinement of the cell, but rather the ability to visualize the trapped cell (highly sensitive approaches like fluorescence or scattering are typically employed for detection of small particles in optical traps). Even manual selection and sorting of cells can be performed quickly, with up to several cells per minute being placed into their respective reaction chambers. Full automation for amenable target cell types has a potential to increase this speed further, with the ultimate limit being placed by the time needed for image acquisition/processing and the maximum achievable speed of cell movement. The optical trap rejects contaminating DNA more effectively than methods based on subdivision of the input sample and does not require dilution of the sample. Laboratory experiments have shown that the trapping technique can remove cells from buffer containing 104 cp/µl of plasmid with no carry-over amplification of the plasmid DNA (Dodsworth et al., 2013).
The microfluidic technique produces nanogram quantities of DNA from each successful reaction (~30 ng) that are sufficient for many analyses including PCR and high-throughput sequencing (Blainey et al., 2011; Marcy, Ouverney, et al., 2007). The 48-chamber design of the microfluidic device provides ample opportunity for success. As an added benefit, the complexity of the system is not overwhelming. In the hands of an experienced user, the optical alignment is very repeatable and stable for months to years, and the relatively straightforward design can be broken down and set back up fairly easily, lending itself to travel or even field studies. As a demonstration, one of the systems used in our experiments was shipped from Oregon to Bermuda in 2012 for application as a teaching instrument in a course on microbial oceanography.
The most significant limitation of the current implementation of this optofluidic technology is throughput. The footprint of the 50 µm deep reactors and use of dedicated recovery ports limited the reactor density on the 48x_v4 device, although there are no fundamental barriers to increase the reactor density or size of the chip by reconfiguring the reactors and product recovery method to achieve batchwise improvements in throughput of several orders of magnitude. The genomic coverage achieved when sequencing single-cell WGA product is highly variable and is thought to be limited by incomplete postlysis accessibility of the genome and genome fragmentation. Coverage is limited further by amplification bias, or the uneven coverage of the genome common in single-cell WGA datasets. Single-cell WGA coverage of environmental microbial genomes has been reported at 13–41% with 16 × sequencing (Woyke et al., 2011), although much higher coverage has been observed in select cases. Since many reactions are unsuccessful or result in lower coverage than needed for desired analyses, it is advantageous to process many reactions to increase the number of high-quality reaction products obtained. Despite the current limitation of 48 reactions per chip in the optofluidic approach, applications with limited sample quantity, FACS-incompatible samples, selection criteria requiring high-resolution microscopy, or that benefit from a low-cost casual operating mode in the lab of an individual investigator may be suitable for the optical trapping approach.
Specific examples of where this approach would be particularly practical include:
-
–
separation of known or unknown components of an enrichment culture for WGA
-
–
separation of cells from the environment or enrichment culture for isolate culture
-
–
selection of bacteria from tissue or intracellular parasites from a cell lysate, where contamination from host DNA would be a serious concern
-
–
acquisition of cells with a defining or interesting morphology or visible phenotype
-
–
manipulation of cell aggregates
-
–
combinatorial approaches where high-quality microscopy documentation would be useful
-
–
single-cell genomics using extremely small volumes of sample (1–20 µl).
The reagent and personnel costs for this technology are likely to be significantly lower than those associated with a facility designed for very high-throughput single-cell genomics. In this vein, the approach is suitable for laboratories where it is desirable to have a single-cell genomics capability readily available, but where they are not necessarily the central focus of the research. It is also worth noting here, that while the microfluidic device described here was designed specifically for single-genome amplification, optical trapping is a microbiological technique that is applicable to a broad variety of studies and integration with other culture-based, imaging-based microfluidic and/or molecular approaches. We eagerly anticipate an expanding role for optical trapping in a variety of novel microbiological studies in the coming years.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the work of Geoffrey Schiebinger for software development and sequence data analysis, Nicholas Gobet in designing the first chip holder, and David McIntyre for assistance with optical trapping at OSU. P. C. B. was supported through the Burroughs Welcome Fund via a Career Award at the Scientific Interface. Z. C. L. and the educational laser-trapping system at OSU were supported through funding from the Gordon and Betty Moore Foundation.
REFERENCES
- Ashkin A. Forces of a single-beam gradient laser trap on a dielectric sphere in the ray optics regime. Biophysical Journal. 1992;61:569–582. doi: 10.1016/S0006-3495(92)81860-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A. Optical trapping and manipulation of neutral particles using lasers. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4853–4860. doi: 10.1073/pnas.94.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic JM. Internal cell manipulation using infrared laser traps. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7914–7918. doi: 10.1073/pnas.86.20.7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic JM, Yamane T. Optical trapping and manipulation of single cells using infrared laser beams. Nature. 1987;330:769–771. doi: 10.1038/330769a0. [DOI] [PubMed] [Google Scholar]
- Ashkin A, Gordon JP. Stability of radiation-pressure particle traps: An optical Earnshaw theorem. Optics Letters. 1983;8:511–513. doi: 10.1364/ol.8.000511. [DOI] [PubMed] [Google Scholar]
- Blainey PC. The future is now: Single-cell genomics of bacteria and archaea. FEMS Microbiology Reviews. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and meta-genomic analysis. PLoS One. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- Chu S, Bjorkholm JE, Ashkin A, Cable A. Experimental observation of optically trapped atoms. Physical Review Letters. 1986;57:314–317. doi: 10.1103/PhysRevLett.57.314. [DOI] [PubMed] [Google Scholar]
- Dean FB. Comprehensive human genome amplification using multiple displacement amplification. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Research. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG, et al. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nature Communications. 2013;4:1854. doi: 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Richter RA, Valas R, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. The ISME Journal. 2011;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson M, Hanstorp D, Hagberg P, Enger J, Nystrom T. Sorting out bacterial viability with optical tweezers. Journal of Bacteriology. 2000;182:5551–5555. doi: 10.1128/jb.182.19.5551-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nature Biotechnology. 2010;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumkin D, Wasserstrom A, Itzkovitz S, Stern T, Harmelin A, Eilam R, et al. Cell lineage analysis of a mouse tumor. Cancer Research. 2008;68:5924–5931. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- Huber R, Burggraf S, Mayer T, Barns SM, Rossnagel P, Stetter KO. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature. 1995;376:57–58. doi: 10.1038/376057a0. [DOI] [PubMed] [Google Scholar]
- Katsura S, Hirano K, Matsuzawa Y, Mizuno A, Yoshikawa K. Direct laser trapping of single DNA molecules in the globular state. Nucleic Acids Research. 1998;26:4943–4945. doi: 10.1093/nar/26.21.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist T, Ahring BK, Lasken RS, Westermann P. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Applied Microbiology and Biotechnology. 2006;74:926–935. doi: 10.1007/s00253-006-0725-7. [DOI] [PubMed] [Google Scholar]
- Liang H, Vu KT, Krishnan P, Trang TC, Shin D, Kimel S, et al. Wave-length dependence of cell cloning efficiency after optical trapping. Biophysical Journal. 1996;70:1529. doi: 10.1016/S0006-3495(96)79716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom RR, Rodrigue S, Huang KH, Kelly L, Kern SE, Thompson A, et al. Ecology of uncultured Prochlorococcus clades revealed through single-cell genomics and biogeographic analysis. The ISME Journal. 2013;7:184–198. doi: 10.1038/ismej.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Yann, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genetics. 2007;3:e155. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, et al. Inaugural article: Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Block SM. Optical trapping. The Review of Scientific Instruments. 2004;75:2787. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Chadd EH, Liou GF, Bergman K, Block SM. Characterization of photodamage to Escherichia coli in optical traps. Biophysical Journal. 1999;77:2856–2863. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: A ribosomal RNA approach. Annual Review of Microbiology. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Research. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annual Review of Microbiology. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Renesto P, Crapoulet N, Ogata H, La Scola B, Vestris G, Claverie J-M, et al. Genome-based design of a cell-free culture medium for Tropheryma whipplei. The Lancet. 2003;362:447–449. doi: 10.1016/S0140-6736(03)14071-8. [DOI] [PubMed] [Google Scholar]
- Rocha MS. Optical tweezers for undergraduates: Theoretical analysis and experiments. American Journal of Physics. 2009;77:704. [Google Scholar]
- Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Unger MA. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Digital Pcr. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Sczyrba A, Lee J, Rinke C, Tighe D, Clingenpeel S, et al. Decontamination of MDA reagents for single cell whole genome amplification. PLoS One. 2011;6:e26161. doi: 10.1371/journal.pone.0026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Xie G, Copeland A, González JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- Yilmaz S, Haroon MF, Rabkin BA, Tyson GW, Hugenholtz P. Fixationfree fluorescence in situ hybridization for targeted enrichment of microbial populations. The ISME Journal. 2010;4:1352–1356. doi: 10.1038/ismej.2010.73. [DOI] [PubMed] [Google Scholar]
- Youssef NH, Blainey PC, Quake SR, Elshahed MS. Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (zodletone spring, Oklahoma) Applied and Environmental Microbiology. 2011;77:7804–7814. doi: 10.1128/AEM.06059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov SD, Thanh NC. Combined absorption of light in the process of photoactivation of biosystems. Journal of Russian Laser Research. 2008;29:558–563. [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, et al. Sequencing genomes from single cells by polymerase cloning. Nature Biotechnology. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.