Abstract
Accumulation of foam cells in the neointima represents a key event in atherosclerosis. We previously demonstrated that Tanshinone IIA (Tan), a lipophilic bioactive compound extracted from Salvia miltiorrhiza Bunge, inhibits experimental atherogenesis, yet the detailed mechanisms are not fully understood. In this study, we sought to explore the potential effects of Tan on lipid accumulation in macrophage foam cells and the underlying molecular mechanisms. Our data indicate that Tan treatment reduced the content of macrophages, cholesterol accumulation, and the development of atherosclerotic plaque in apolipoprotein E-deficient mice. In human macrophages, Tan ameliorated oxidized low density lipoporotein (oxLDL)-elicited foam cell formation by inhibiting oxLDL uptake and promoting cholesterol efflux. Mechanistically, Tan markedly reduced the expression of scavenger receptor class A and increased the expression of ATP-binding cassette transporter A1 (ABCA1) and ABCG1 in lipid-laden macrophages via activation of the extracellular signal-regulated kinase (ERK)/nuclear factor-erythroid 2-related factor 2 (Nrf2)/heme oxygenase-1 (HO-1) pathway. Tan treatment induced the phosphorylation and nuclear translocation of Nrf2 and subsequently increased the expression of HO-1, and these effects were abolished by the specific ERK inhibitors, PD98059 and U0126. Moreover, HO-1 small interfering RNA or zinc protoporphyrin (a HO-1 inhibitor) abrogated Tan-mediated suppression of lipid accumulation in macrophages. Our current findings demonstrate that a novel HO-1-dependent mechanism is involved in the regulation of cholesterol balance by Tan.
Keywords: class A scavenger receptor, ATP-binding cassette transporter A1/G1, heme catabolism enzyme, cell signaling
Cardiovascular disease (CVD) is the leading cause of premature death worldwide. Atherosclerosis constitutes the single most important contributor to this growing burden of CVD. Lipid-laden macrophages, known as foam cells, which accumulate within the lesional area, represent the hallmark of early to mid-stage atherosclerotic lesion development. Uncontrolled uptake of modified low density lipoprotein (LDL) and/or impaired cholesterol efflux are major factors contributing to lipid overload and resultant foam cell formation of macrophages (1). Thus, maintaining the balanced flow of cholesterol into and out of the macrophage is the key to preventing lipid overload, and ultimately, the progression of atherosclerosis. In macrophages, the uptake of modified LDL is mainly mediated by a group of scavenger receptors, such as scavenger receptor class A (SR-A) and Cluster of Differentiation 36 (CD36) (2–4). On the contrary, SR-BI, ATP-binding cassette transporter A1 (ABCA1), and ABCG1 are responsible for the efflux of macrophage cholesterol (5–7). However, safe and efficacious therapeutic agents that prevent cholesterol uptake and/or promote cholesterol efflux are very limited.
Tanshinone IIA (Tan) (supplementary Fig. I), a lipophilic pharmacologically active compound derived from the Chinese herb Salvia miltiorrhiza Bunge (Danshen), exerts multiple cardioprotective effects (8). Tan has long been used clinically in Asian countries for the prevention and treatment of several CVDs, such as coronary heart disease, angina pectoris, and myocardial infarction (8–10). We and others have demonstrated that Tan attenuates the initiation and progression of atherosclerosis in experimental animals (11, 12). More recently, we have provided evidence that Tan can stabilize “vulnerable” atherosclerotic plaques in apolipoprotein E-deficient (ApoE−/−) mice fed a high-cholesterol diet (13). However, the precise molecular mechanisms by which Tan attenuates and stabilizes atherosclerotic plaques remain largely unknown.
Heme oxygenase-1 (HO-1), the key enzyme in heme catabolism, has been reported to display several beneficial effects in atherosclerosis-related CVDs (14–16). In addition, emerging evidence suggests that HO-1 participates in the protective effects of Tan in several cell types (17, 18). However, it remains unclear whether HO-1 is involved in the atheroprotective effect of Tan on foam cell formation. Therefore, the aim of the present study was to investigate whether Tan may suppress lipid accumulation in macrophage foam cells by activating HO-1.
MATERIALS AND METHODS
Chemicals and reagents
Tan (>98% purity assayed by HPLC) was a kind gift from Prof. Lianquan Gu (Department of Medicinal Chemistry, Sun Yat-sen University, Guangzhou, China). RPMI 1640 medium was purchased from Gibco BRL (Grand Island, NY). Fetal bovine serum (FBS), Alexa Fluor 594-conjugated anti-rabbit IgG (H+L) antibody, Lipofectamine 2000, nuclear factor-erythroid 2-related factor 2 (Nrf2) small interfering RNA (siRNA), HO-1 siRNA, and liver X receptor (LXR) siRNA were obtained from Invitrogen (Carlsbad, CA). Rabbit anti-CD36 and goat anti-SR-A polyclonal antibodies were purchased from R&D Systems (Minneapolis, MN). Rabbit anti-Nrf2, anti-c-Fos, and anti-c-Jun were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-ABCA1 antibody was obtained from Abcam (Cambridge, MA). Rabbit anti-SR-BI and anti-ABCG1 antibodies were obtained from Novus Biologicals (Littleton, CO). Rabbit anti-HO-1 and anti-phospho-Nrf2 (Ser40) antibodies were obtained from Epitomics (Burlingame, CA). Mouse anti-α-tubulin, rabbit anti-histone H1 antibody, phorbol 12-myristate 13-acetate (PMA), fluorescent dye DiI, Oil Red O, 4′,6-diamidino-2-phenylindole (DAPI), Harris hematoxylin, actinomycin D, zinc protoporphyrin (ZnPP), and human HDL were purchased from Sigma-Aldrich (St. Louis, MO). Mouse anti-Mac3 antibody was obtained from BD Transduction Laboratories (San Jose, CA). PD98059, U0126, SB203580, SP600125, and calphostin C were from Calbiochem (La Jolla, CA). Probes for activator protein (AP)-1 used in electrophoretic mobility shift assay (EMSA) were from Beyotime (Haimen, China). pRL-TK, AP-1 reporter plasmids, and a dual-luciferase reporter assay system were purchased from Promega (Madison, WI). All other chemicals were purchased from Sigma-Aldrich unless otherwise specified.
Cell culture
Human monocyte-derived THP-1 cells (CTCC, Shanghai, China) were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 μg/ml streptomycin, and 100 U/ml penicillin. The cells were incubated under a humidified atmosphere of 95% O2 and 5% CO2 at 37°C (Thermo) to a density of 106 cells per ml. Monocytic THP-1 cells were stimulated with 100 nM PMA for 48 h to differentiate into adherent macrophages. When the differentiated phenotype was achieved, the PMA-containing medium was removed and replaced with complete RPMI 1640 supplemented with 10% FBS. The macrophages were cultured for another 24 h before treatment.
Human peripheral blood monocytes were isolated from the blood of healthy donors by density gradient centrifugation on Ficoll/Hypaque. Monocytes were purified by adherence to plastic in RPMI 1640 supplemented with 10 % FBS and antibiotics for 2 14h. Nonadherent cells were removed by several washes with warm PBS. Freshly isolated monocytes were thereafter differentiated into macrophages using macrophage colony-stimulating factor (50 ng/ml) for 6 days in RPMI 1640 medium containing 10% FBS. The investigation conforms to the principles outlined in the Declaration of Helsinki. The study protocol was approved by the ethics committee of Sun Yat-sen University.
The isolation and culture of macrophages from the mouse peritoneal cavity were carried out as reported previously (19). Briefly, the mice were euthanized and ice-cold PBS was injected into the peritoneal cavity of each mouse. This fluid was carefully collected and centrifuged at 3,000 rpm. Then the supernatant was withdrawn and the cell pellet was resuspended in RPMI 1640 medium and allowed to adhere for 3 h, then was washed three times with prewarmed PBS to remove nonadherent cells. The medium was then replaced with fresh RPMI 1640 medium with 10% FBS. The adherent macrophages were used for further experiments.
Animals and procedures
ApoE−/− mice (C57BL/6J background) were obtained from the Jackson Laboratory (Bar Harbor, ME) at 6 weeks of age. Mice were fed a high-cholesterol diet (containing 1.25% cholesterol) and dosed daily via intragastric gavage with 30 mg/kg Tan dissolved in 0.5% carboxy-methyl-cellulose sodium (CMC-Na) or administered 0.5% CMC-Na alone (vehicle control). After 12 weeks, mice were euthanized and aortas were collected and subjected to Western blotting analysis. The aortic sinus cryosections were analyzed for atherosclerotic lesion size with Oil Red O staining and immunohistochemistry staining as previously described (13). The dosage of the drug used in the current study was evaluated according to a small pilot study by us (13) and reports by others (20). All animal experimental procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Ethical Committee for Animal Research at Sun Yat-sen University.
Oil Red O staining
Foam cell formation was revealed by Oil Red O staining according to our previous report (21). In brief, cells were fixed with 4% paraformaldehyde and then stained by filtered 0.5% Oil Red O, followed by counterstaining the nuclei with Harris hematoxylin. Photomicrographs were taken by phase-contrast microscopy (IX71, Olympus, Tokyo, Japan). Images were obtained from at least five randomly chosen fields for each condition.
LDL isolation, modification, and labeling
The use of human plasma in this study conforms to the principles outlined in the Declaration of Helsinki. LDL was isolated from fresh plasma of healthy subjects as previously described (21). LDL was either oxidized with 5 μM Cu2+ for 24 h to generate oxidized low density lipoprotein (oxLDL), or labeled with the fluorescent probe DiI and then oxidized to DiI-oxLDL followed by dialysis against PBS + 10 μM EDTA. Endotoxin levels were tested using the Limulus Amebocyte Lysate assay kit (Bio Whittaker, Walkersville, MD). Preparations containing less than 0.5 EU/mg (LDL protein) of endotoxin were used for the experiments. The oxidative extent of each lot was monitored by agarose gel electrophoresis and colorimetrically by the thiobarbituric acid reactive substances assay. The protein concentration was determined using a BCATM protein assay kit (Pierce, Rockford, IL). All lipoproteins were filter sterilized, stored at 4°C in the dark, and used within 3 weeks.
Cellular uptake of Dil-oxLDL
THP-1 monocytes were cultured on chamber slides (Warner Instruments) and stimulated with PMA (100 nM) for 48 h to differentiate them into adherent macrophages. THP-1-derived macrophages were incubated for 24 h with Tan (1–10 μM) or the vehicle (0.1% DMSO). After incubation, the cells were washed twice with PBS (to bar physical interference of Tan) and incubated with DiI-oxLDL (10 μg/ml) at 37°C for an additional 4 h. At the end of the incubation period, cells were washed, mounted on coverslips with glycerin jelly and examined by confocal microscopy (LSM 710, Carl Zeiss, Germany). Data were analyzed with Image-Pro Plus 6.0 software. The DiI-oxLDL uptake was determined and expressed as fold of the control.
Cholesterol contents measurement
Lipids in the thoraco-abdominal aortas of the mice and in macrophages were extracted by hexane/isopropanol [3/2 (v/v)], the solvents were evaporated, and the tissue or cell pellet was resuspended in aqueous solution with 1% Triton X-100-water. The level of cholesterol was measured using Amplex Red cholesterol assay kit (Invitrogen) according to the manufacturer's instructions (22). Protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit. Data are normalized to cellular protein content.
Cholesterol efflux analysis
Cholesterol efflux analysis was performed as previously described (23). Briefly, THP-1-derived macrophages were labeled in 0.2% BSA, serum-free RPMI 1640 medium with 50 μg/ml oxLDL (used as the carrier for free cholesterol labeling), and 1 μCi/ml [3H]labeled cholesterol for 24 h. After labeling, the cells were washed twice with PBS and incubated with Tan at the indicated concentrations for another 24 h. To equilibrate cholesterol pools, cells were washed twice with PBS and incubated for 24 h in RPMI 1640 containing 0.2% BSA with no lipoproteins. Cells were washed again and switched to serum-free medium containing 0.2% BSA or purified apoAI (10 μg/ml) (Calbiochem, Darmstadt, Germany) or native human HDL (50 μg/ml) (Sigma). After incubation for 12 h, the supernatant was collected and adherent cells were lysed with 1 mol/l NaOH. Radioactivity was quantified in both the supernatant and cell lysate by scintillation counting (Beckmann LS6000SC; Beckman Coulter, Somerset, NJ). Cholesterol efflux was expressed as the percentage of counts in the supernatant compared with the total [3H]cholesterol in the supernatant and cell lysate.
RNA isolation and real-time PCR
Total RNA of cells was extracted using Trizol reagent (Invitrogen) and was converted into cDNA with reverse transcriptase according to the manufacturer's instructions (Takara, Dalian, China). Quantitative real-time PCR was used to measure the relative levels of gene expression. All primers used were custom synthesized by Invitrogen and well-validated. Quantification of relative gene expression was calculated with the efficiency-corrected 2−ΔΔCT method using the housekeeping gene, GAPDH, for RNA input standardization, and data were presented as fold change over control group.
Western blotting analysis
Western blotting was conducted as previously described in detail (24). Nuclear protein, cytosol protein, or total cell protein was extracted using CelLytic nuclear/cytosol extraction kit (Sigma) with protease and phosphatase inhibitor according to the recommendations of the supplier. Anti-α-tubulin or anti-histone H1 antibodies were used for equal protein loading of cytosolic and nuclear proteins, respectively.
EMSA
EMSA was performed using the LightShiftTM chemiluminescent EMSA kit (Pierce) according to our established procedure (21). In brief, cells were washed with ice-cold PBS and then were scraped off the plates with a cell scraper. Nuclear extracts were prepared using the CelLytic nuclear extraction kit (Sigma) according to the manufacturer's instructions. Nuclear protein (2 μg) was then used to assess DNA binding activity using 3′ biotin-labeled oligonucleotide probes containing the AP-1 consensus sequence: sense, 5′-CGC TTG ATG ACT CAG CCG GAA-3′ antisense, 3′-GCG AAC TAC TGA GTC GGC CTT-5′ (Beyotime). Each sample was electrophoresed in a 6% nondenaturing polyacrylamide gel in 0.5× Tris/Borate/EDTA buffer at 100 V for 60 min. For cold competition experiments, 100-fold molar excess unlabeled duplex oligonucleotides containing AP-1 consensus sequence were added to the nuclear extracts before incubation with the biotin-labeled oligonucleotides.
Luciferase reporter assay
In the AP-1 promoter activation assay, cells were seeded at 5 × 104 cells/well in 96-well plates, and cotransfected with AP-1 reporter plasmid (100 ng/well) and pRL-TK (10 ng/well) as an internal control for 12 h using Lipofectamine LTX and PLUS reagents (Invitrogen) according to the manufacturer's instructions. After transfection, cells were treated with Tan (1, 3, or 10 μM) for 30 min and then stimulated with PMA (100 nM) for 48 h. The luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega), according to the manufacturer's protocol. Relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity. For LXR activation studies, LXR response element (LXRE)-driven luciferase reporter vector (LXRE-tk-Luc), which was kindly provided by Dr. In-Kyu Lee (Kyungpook National University School of Medicine, Korea), and β-galactosidase control vector (Promega) were used. Luciferase and β-galactosidase activities were determined in cell lysates. The amount of luciferase activity was normalized for β-galactosidase and reported as relative light units.
siRNA
Cells were transfected with negative control siRNA, HO-1 siRNA, Nrf2 siRNA, or LXR siRNA using Lipofectamine 2000 (Invitrogen) for 48 h according to the manufacturer's instructions. After transfection, the cells were incubated with Tan for another 24 h. The cells were then subjected to Western blotting analysis or cholesterol content measurement.
Immunofluorescent staining
THP-1 monocytes were cultured on 6-well chamber slides and stimulated with PMA (100 nM) for 48 h to differentiate them into adherent macrophages, followed by exposure to oxLDL (50 μg/ml) for 24 h to induce macrophage foam cells. Cells were then treated with 10 μM Tan or the vehicle (0.1% DMSO) for 4 h. At the end of incubation period, cells were fixed with cold 4% (w/v) paraformaldehyde for 30 min, washed with PBS for 5 min, and permeabilized in 0.1% (w/v) Triton X-100 at room temperature for 10 min. After being washed with PBS, the cells were incubated with primary antibody against Nrf2 at 4°C overnight followed by Alexa Fluor 594-conjugated anti-rabbit IgG (H+L) secondary antibody for 1 h at room temperature. The slides were counterstained with DAPI (5 mg/ml; Sigma) and mounted in glycerin jelly medium, then subjected to confocal microscopy (LSM 710; Carl Zeiss).
Statistical analysis
All values are expressed as means ± SEM unless otherwise specified. Data were analyzed by two-tailed unpaired Student's t-test between two groups and by one-way ANOVA followed by the Bonferroni post hoc test for multiple comparisons. The analyses were performed using GraphPad Prism 5.0 software (GraphPad Software Inc., La Jolla, CA). P < 0.05 was considered statistically significant.
RESULTS
Tan inhibits atherogenesis and regulates the expression of scavenger receptors and cholesterol transporters in ApoE−/− mice
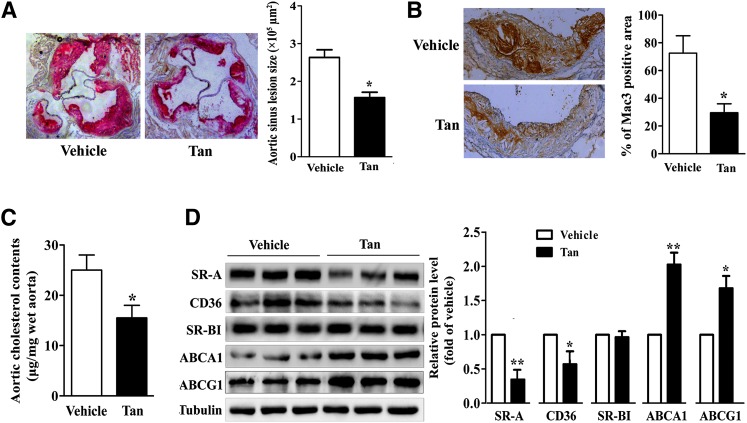
We first evaluated the impact of Tan treatment on the formation of atherosclerotic plaques. Six-week-old ApoE−/− mice fed a high-cholesterol diet were randomized to receive either Tan (30 mg/kg/day) or vehicle for 12 weeks. We then analyzed the development of atherosclerotic lesions in the aortic sinus by Oil Red O staining. As shown in Fig. 1A, Tan treatment markedly diminished atherosclerotic plaque size by 46.23 ± 10.68%, compared with the vehicle control group (n = 8, P < 0.05). However, Tan did not affect the serum lipid profile (supplementary Table I), which agrees with previous data (13). Accumulation of macrophage foam cells in the neointima plays a key role in the development of atherosclerosis. To further evaluate the impact of Tan on macrophage content in the atherosclerotic plaques, immunohistochemical staining of the aortic sinus was performed. Notably, compared with vehicle-treated ApoE−/− control mice, the macrophage-positive area in the atherosclerotic plaques of Tan-treated mice was markedly reduced by 61.64 ± 12.28% (Fig. 1B). Furthermore, cholesterol content was lower in Tan-treated aortas than it was in vehicle-treated ApoE−/− aortas (Fig. 1C). Scavenger receptors (such as SR-A and CD36) and cholesterol transporters (such as SR-BI, ABCA1, and ABCG1) play crucial roles in cholesterol metabolism (2, 3, 5–7). We therefore determined whether Tan modulates cholesterol homeostasis by altering the expression of these receptors and transporters. As indicated in Fig. 1D, treatment with Tan decreased the protein level of SR-A (1.00 in controls vs. 0.35 ± 0.06 in Tan-treated mice, P < 0.01) and CD36 (1.00 vs. 0.58 ± 0.16, P < 0.05), and increased the protein expression of ABCA1 (1.00 vs. 2.09 ± 0.13, P < 0.01) and ABCG1 (1.00 vs. 1.67 ± 0.15, P < 0.05) in the aortas of ApoE−/− mice. These findings imply that chronic treatment with Tan might affect lipid accumulation in macrophages in ApoE−/− mice through regulating the expression of scavenger receptors and cholesterol transporters.
Fig. 1.
Effect of Tan on atherogenesis in ApoE−/− mice. Six-week-old ApoE−/− mice fed a high-cholesterol diet were dosed daily with Tan (30 mg/kg/day, intragastrically.) or vehicle (CMC-Na) for 12 weeks. A: Determination of atherosclerotic lesion size in ApoE−/− mice. The aortic sinuses of the animals treated as described in the Materials and Methods section were analyzed for atherosclerotic lesion size with Oil Red O staining. Magnification ×400. The atherosclerotic lesion areas in the aortic sinus were quantified (n = 8). *P < 0.05. B: Percentage of Mac3-positive area in comparison to total plaque area in ApoE−/− mice treated with Tan (30 mg/kg/day) or vehicle. *P < 0.05. C: Cholesterol content in aortas was measured by the Amplex Red cholesterol assay kit (n = 8). *P < 0.05. D: Aortas were collected from ApoE−/− mice treated with Tan (30 mg/kg/day) or vehicle. Lysates of aortas from three mice were prepared and subjected to Western blotting to examine the expression of SR-A, CD36, SR-BI, ABCA1, ABCG1, and α-tubulin. *P < 0.05; **P < 0.01 versus vehicle-treated mice.
Tan regulates the expression of scavenger receptors and cholesterol transporters in human macrophages
We next investigated the effects of Tan on the expression of scavenger receptors and cholesterol transporters in human macrophages. As indicated in supplementary Fig. II, Tan treatment dose-dependently decreased mRNA expression of SR-A, but enhanced that of ABCA1 and ABCG1, without affecting CD36 and SR-BI in THP-1 macrophages. We then examined the effects of Tan on protein expression of these receptors and transporters, and the changes in protein expression paralleled with the changes in mRNA expression (Fig. 2A). Notably, treatment with Tan (1, 3, or 10 μM) for 24 h significantly reduced the protein expression of SR-A (10 μM Tan decreased the SR-A protein level to 12.82 ± 3.36% of the control, P < 0.05). Similar results were obtained in primary human monocyte-derived macrophages (Fig. 2B).
Fig. 2.
Effect of Tan on the protein expression of scavenger receptors and cholesterol transporters in human macrophages. THP-1 macrophages (A) or primary human macrophages (B) were treated with the indicated concentrations (1, 3, or 10 μM) of Tan or vehicle (0.1% DMSO) for 24 h in the presence of oxLDL (50 μg/ml) and subjected to Western blotting to determine the protein level of SR-A, CD36, SR-BI, ABCA1, ABCG1, or α-tubulin. *P < 0.05 versus untreated group.
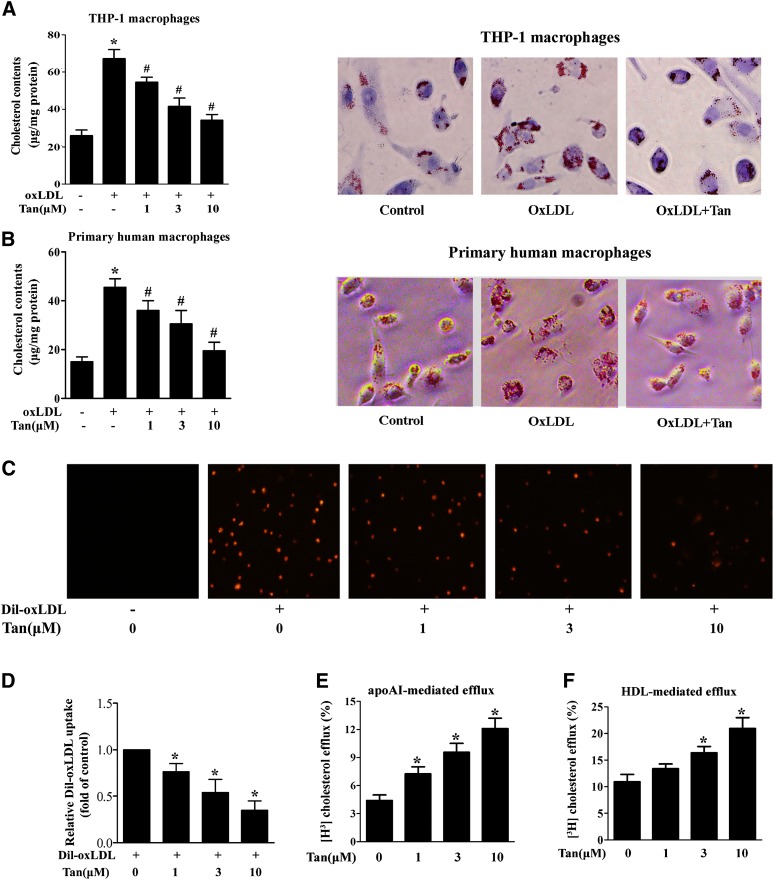
Tan attenuates foam cell formation by inhibiting oxLDL uptake and promoting cholesterol efflux in lipid-laden macrophages
Based on the fact that Tan regulates the expression of scavenger receptors and cholesterol transporters, we surmised that Tan might play a role in macrophage foam cell formation. To further explore the effect of Tan on foam cell formation, differentiated THP-1 and primary human macrophages were loaded with oxLDL (50 μg/ml) for 24 h in the absence or presence of Tan. Tan significantly ameliorated intracellular lipid accumulation as determined by the measurement of cellular cholesterol content and by Oil Red O staining (Fig. 3A, B). As the regulation of cholesterol homeostasis is a finely-tuned process determined by net outcome of cholesterol uptake and efflux, we next examined the effect of Tan on cholesterol uptake and efflux. We found that Tan treatment (1, 3, or 10 μM) markedly decreased DiI-oxLDL uptake (Fig. 3C, D), but significantly promoted apoAI- and HDL-mediated cholesterol efflux (Fig. 3E, F). However, Tan did not affect the mRNA expression of cholesterol synthesis-related genes (supplementary Fig. III); implying that de novo lipid synthesis was not involved in Tan-mediated reduction of foam cell formation. Taken together, these data suggest that Tan protects against the formation of macrophage foam cells by decreasing SR-A-mediated oxLDL uptake and increasing ABCA1/G1-mediated cholesterol efflux.
Fig. 3.
Effect of Tan on oxLDL uptake and cholesterol efflux in human macrophages. A, B: THP-1-derived macrophages (A) or primary human macrophages (B) were coincubated with Tan (10 μM) and oxLDL (50 μg/ml) for 24 h. After incubation, intracellular cholesterol was extracted and determined by the Amplex Red cholesterol assay kit or cells were fixed and stained with Oil Red O. Cellular nuclei were stained with hematoxylin. The magnification of each panel is ×400. *P < 0.05 versus vehicle-treated group; #P < 0.05 versus oxLDL-treated alone group. C, D: Tan suppresses the uptake of DiI-oxLDL by THP-1 macrophages. THP-1 macrophages were incubated with Tan (1, 3, or 10 μM) or vehicle (0.1% DMSO) for 24 h. The cells were then washed twice with PBS and incubated with DiI-oxLDL (10 μg/ml) at 37°C for 4 h. As a negative control, cells were incubated without DiI-oxLDL. DiI-oxLDL uptake was assessed by confocal microscopy (C) and was quantified by Image-Pro Plus software (D). The experiments were repeated three times independently. Data are expressed as means ± SEM (n = 3). *P < 0.05 versus Dil-oxLDL-treated alone group. E, F: THP-1-derived macrophages were incubated with 50 μg/ml oxLDL and 1 μCi/ml [3H]labeled cholesterol for 24 h. After labeling, the cells were incubated with Tan at the indicated concentrations for another 24 h. ApoAI- (E) or HDL-dependent (F) cholesterol efflux was measured as described in the Materials and Methods. *P < 0.05 versus vehicle-treated group.
Tan inhibits AP-1 expression, DNA binding activity, and transcriptional activity
AP-1 is a crucial transcription factor that regulates SR-A expression (25, 26). Therefore, we examined the effects of Tan on the expression of c-Fos and c-Jun (two key subunits of AP-1) in cell nuclei. As shown in Fig. 4A, a concentration-dependent suppression occurred on c-Fos expression when the cells were treated with Tan (1–10 μM) for 6 h. However, Tan did not affect c-Jun expression in nuclei. Additionally, EMSA and luciferase reporter assays demonstrated that Tan significantly inhibited AP-1 DNA binding activity and transcriptional activity (Fig. 4B, C). These findings indicate that the inhibitory effects of Tan on SR-A expression are partly associated with the suppression of AP-1 activity.
Fig. 4.
Effect of Tan on AP-1 expression, DNA binding activity, and transcriptional activity. A: THP-1-derived macrophages were treated with Tan (1, 3, or 10 μM) or vehicle (0.1% DMSO) for 30 min followed by stimulation with oxLDL (50 μg/ml) for 6 h and the nuclear protein level of c-Fos, c-Jun, or histone H1 were determined by Western blotting. B: Nuclear extracts (2 μg aliquots) were complexed with biotin-labeled AP-1 probe and assayed for AP-1 DNA binding activity by EMSA. Specificity of binding was confirmed by cold competition experiments with a 100-fold molar excess of unlabeled AP-1 duplex oligonucleotide (n = 3). C: Dual luciferase reporter assays were performed to evaluate the influence of Tan treatment on AP-1-dependent transcriptional activity. Data are expressed as the means ± SEM (n = 3). *P < 0.05 versus vehicle-treated group.
Tan upregulates ABCA1/G1 expression independent of LXRα activation
To address whether the LXRα/retinoid X receptor (RXR) is involved in Tan-induced upregulation of ABCA1/G1, we determined the protein expression of LXRα and RXR in the nuclei of Tan-treated THP-1 macrophages. Intriguingly, treatment with Tan did not affect the nuclear expression of LXRα and RXR (supplementary Fig. IV). Furthermore, Tan exerted no effect on LXRα transactivation, as determined by LXRE-mediated luciferase activity (supplementary Fig. IV). While, LXRα knockdown by siRNA transfection or pharmacological inhibition by GGPP failed to abrogate the effect of Tan-mediated upregulation of ABCA1/G1 expression (supplementary Fig. IV), implying that LXRα activation was not involved in Tan-induced ABCA1/G1 upregulation.
To further explore the mechanisms by which Tan increased ABCA1/G1 mRNA levels in macrophages, actinomycin D was added to both vehicle- and Tan-treated macrophages for specified periods, and the half-life of ABCA1/G1 mRNA was determined by quantitative RT-PCR. Based on the observed decay curve, we found that Tan prolonged the turnover rate of ABCG1 transcripts by approximately 3-fold (t1/2, 125 vs. 352 min; supplementary Fig. V), whereas Tan did not affect the turnover rate of ABCA1 transcripts (supplementary Fig. V). These data indicate that Tan promotes ABCG1 expression through inhibition of rapid mRNA decay.
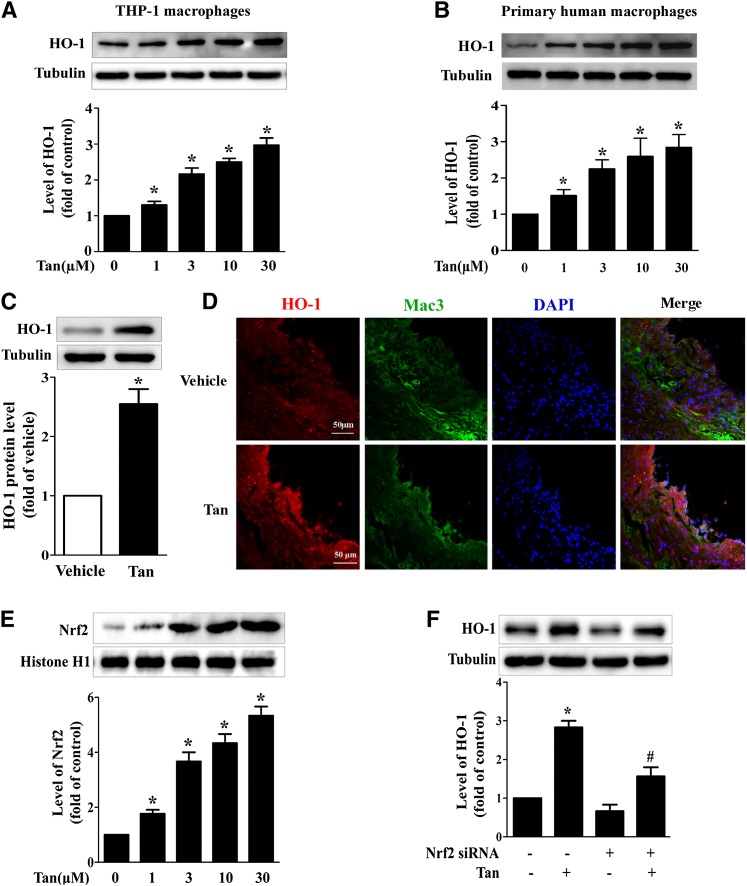
Tan modulates cholesterol metabolism through HO-1 activation
To explore the potential role of HO-1 in Tan-mediated upregulation of ABCA1 expression and suppression of cholesterol accumulation in macrophage foam cells, first, we demonstrated that treatment with Tan significantly upregulated protein expression of HO-1 in a dose-dependent manner in THP-1 macrophages and human primary macrophages (Fig. 5A, B). The expression of HO-1 was increased in mouse peritoneal macrophages (Fig. 5C) and in lesional macrophages in atherosclerotic plaques (Fig. 5D) from Tan-treated ApoE−/− mice compared with vehicle-treated mice. As HO-1 was transcriptionally regulated by Nrf2, we next determined the effect of Tan on Nrf2 expression in the nuclei. As shown in Fig. 5E, Tan significantly increased Nrf2 expression dose-dependently. Moreover, transfection of siRNA targeting Nrf2 completely abrogated the effects of Tan on HO-1 expression (Fig. 5F).
Fig. 5.
Effect of Tan on the expression of HO-1 in macrophages in vitro and in vivo. THP-1-derived macrophages (A) or primary human macrophages (B) were incubated with the indicated concentrations of Tan for 12 h and whole cell lysates were subjected to Western blotting to determine the protein expression of HO-1, with α-tubulin as the loading control. *P < 0.05 versus vehicle-treated group. C: Six-week-old ApoE−/− mice fed a high-cholesterol diet were dosed daily with Tan (30 mg/kg/day, i.g.) or vehicle (CMC-Na) for 12 weeks. The peritoneal macrophages were isolated, lysed, and subjected to Western blotting to evaluate the protein levels of HO-1 and α-tubulin. *P < 0.05 versus. vehicle-treated group. D: HO-1 immunohistochemical staining (red) of the aortic sinuses from ApoE−/− mice receiving Tan or vehicle treatment. Macrophages were stained with Mac3 (green) and nuclei were stained with DAPI (blue). E: THP-1 macrophages were incubated with the indicated concentrations of Tan for 4 h, then the nuclear extracts were subjected to Western blotting to determine the protein expression of Nrf2 and histone H1. *P < 0.05 versus vehicle-treated group. F: THP-1 macrophages were transfected with Nrf2 siRNA for 24 h, followed by Tan (10 μM) incubation for an additional 12 h. The expression of HO-1 or α-tubulin was examined by Western blotting. *P < 0.05 versus vehicle-treated group, #P < 0.05 versus Tan-treated alone group.
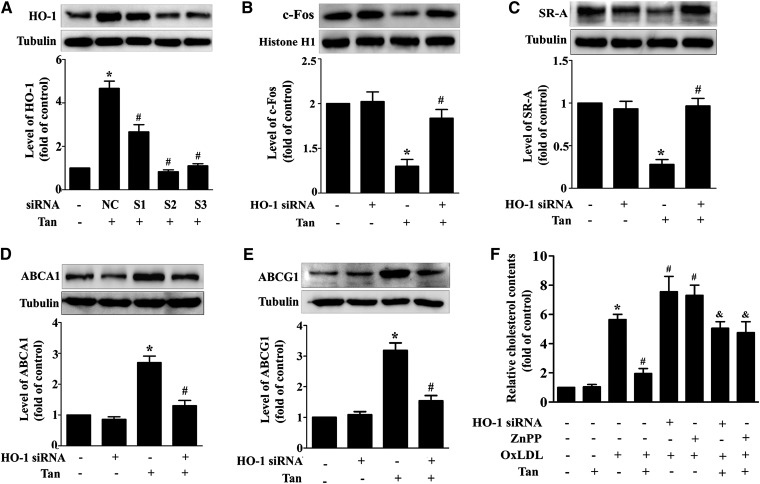
To further determine whether HO-1 was involved in Tan-mediated inhibitory effects on foam cell formation, the basal expression of HO-1 was knocked down in THP-1 macrophages by using three independent siRNAs, marked S1, S2, and S3, respectively. We observed that S2 exhibited the best efficacy for HO-1 knockdown (supplementary Fig. VI). In addition, Tan-induced HO-1 upregulation was completely abolished by HO-1 siRNA (S2) transfection (Fig. 6A). HO-1 siRNA (S2) also attenuated the effect of Tan on the downregulation of c-Fos (Fig. 6B) and SR-A protein expression (Fig. 6C), and upregulation of ABCA1 and ABCG1 protein expression (Fig. 6D, E). To verify these findings, ZnPP, a specific HO inhibitor, was used to inhibit HO activity (supplementary Fig. VII). Likewise, the effect of Tan on c-Fos, SR-A, ABCA1, and ABCG1 was blocked by ZnPP. Accordingly, both ZnPP and HO-1 siRNA reversed Tan's suppressive effects on oxLDL uptake and promotion of cholesterol efflux (data not shown). Moreover, Tan-mediated diminution of lipid accumulation was also attenuated by HO-1 siRNA or ZnPP (Fig. 6F). These data indicate that HO-1 functions as an upstream regulator of cholesterol uptake and efflux in macrophages.
Fig. 6.
Tan regulates cholesterol metabolism through HO-1 activation. A: THP-1 macrophages were transfected with three different siRNAs against HO-1 (marked as S1, S2, and S3), as well as the negative control (NC) for 24 h, followed by Tan (10 μM) treatment for an additional 12 h. Protein expression of HO-1 and α-tubulin was measured by Western blotting. *P < 0.05 versus vehicle-treated group, #P < 0.05 versus NC group. B–E: Macrophages were pretreated with HO-1 siRNA (S2) for 24 h, followed by Tan treatment for an additional 24 h. Protein expression of c-Fos (B), SR-A (C), ABCA1 (D), ABCG1 (E), α-tubulin and histone H1 were determined by Western blotting. *P < 0.05 versus vehicle-treated group; #P < 0.05 versus Tan-treated alone group. F: Macrophages were pretreated with HO-1 siRNA (S2) for 24 h, or preincubated with ZnPP (10 μM) for 6 h, followed by Tan for an additional 24 h in the presence or absence of oxLDL (50 μg/ml); cholesterol content was detected by the Amplex Red cholesterol assay kit. *P < 0.05 versus vehicle-treated group; #P < 0.05 versus oxLDL-treated alone group; and &P < 0.05 versus Tan/oxLDL-treated group.
Tan enhances Nrf2-mediated HO-1 expression via activating extracellular signal regulated kinases
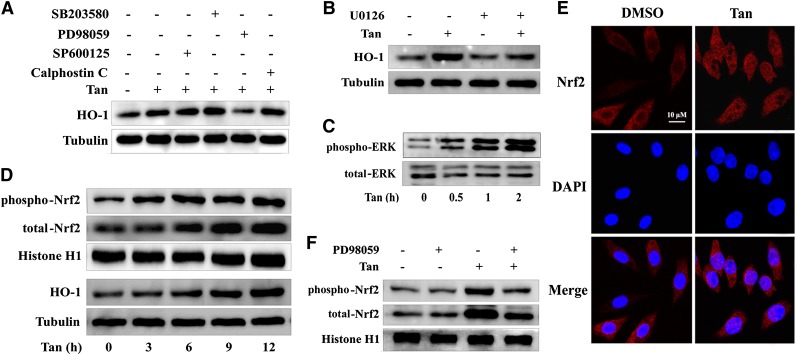
To elucidate the upstream signaling events leading to induction of HO-1 expression in Tan-treated macrophages, different kinase inhibitors were used, including PD98059 [extracellular signal regulated kinase (ERK) inhibitor], SB203580 (p38 MAPK inhibitor), SP600125 [c-Jun-N-terminal kinase (JNK) inhibitor], and calphostin C [protein kinase C (PKC) inhibitor]. We observed that Tan-mediated upregulation of HO-1 protein expression was almost completely abolished by incubation with PD98059 (Fig. 7A). This effect was also confirmed by another ERK inhibitor U0126 (Fig. 7B). However, SB203580, SP600125, and calphostin C had no effect on HO-1 protein expression (Fig. 7A).
Fig. 7.
Upregulation of HO-1 expression by Tan is dependent on ERK activation. A, B: THP-1 macrophages were pretreated with different kinase inhibitors, including SP 600125 (20 μM), SB 203580 (5 μM), PD 98059 (10 μM), calphostin C (0.5 μM), and U0126 (0.5 μM) for 30 min before treatment with Tan (10 μM) for 12 h. Cell lysates were collected and probed with anti-HO-1 and α-tubulin antibodies. C, D: Macrophages were treated with or without Tan (10 μM) for different time points as indicated. Whole cell lysates were subjected to Western blotting using antibodies specific for the phosphorylated forms of ERK or total ERK (C), and HO-1 or α-tubulin (D). The nuclear extracts were subjected to Western blotting to determine the protein expression of phospho-Nrf2 (S40), total Nrf2, or histone H1 (D). E: Macrophages were treated with 10 μM Tan for 4 h and then fixed and labeled with rabbit anti-Nrf2 antibody and Alexa Fluor 594-conjugated anti-rabbit IgG (H+L) secondary antibody. Cells were counterstained with DAPI for visualization of the nuclei. Slides were viewed using confocal microscopy. F: Macrophages were preincubated with PD98059 for 30 min and then exposed to 10 μM Tan for an additional 4 h before preparation of nuclear extracts for Western blotting analysis. Phospho-Nrf2 (S40), total Nrf2, or histone H1 were detected using their specific antibodies (n = 3).
To further determine whether Tan induced HO-1 expression by activating ERK, we treated THP-1 macrophages with Tan at different time points and assessed the levels of phosphorylated ERK and HO-1 in vehicle- and Tan-treated macrophages. Tan rapidly induced ERK phosphorylation at 30 min and the kinetics of ERK activation preceded the upregulation of HO-1 expression by Tan (Fig. 7C, D), suggesting that ERK activation functions as an upstream event in Tan-mediated HO-1 upregulation.
We have demonstrated that Tan profoundly elicited the induction of HO-1 protein in an Nrf2-dependent manner (Fig. 5E, F). Previous studies have described the phosphorylation of Nrf2 (on Ser40) by several kinases as a critical process for the nuclear translocation of Nrf2, and the subsequent transactivation of various targeted genes, such as HO-1 (27). To further explore the mechanisms of ERK-mediated increased HO-1 expression by Tan, we investigated whether ERK was involved in Tan-induced Nrf2 activation. We first examined the phosphorylation and subcellular localization of Nrf2 in THP-1 macrophages after Tan treatment. As shown in Fig. 7D, Western blotting analysis of the nuclear fraction demonstrated significant augmentation of Nrf2 phosphorylation and translocation after Tan treatment in a time-dependent manner. Consistent with the Western blotting analysis, immunofluorescent analysis showed augmented Nrf2 translocation into cell nuclei upon Tan treatment (Fig. 7E). We then examined the effect of ERK inhibition on Tan-induced Nrf2 activation. As shown in Fig. 7F, nuclear Nrf2 phosphorylation and accumulation increased after treatment with Tan (10 μM). Inhibition of the ERK pathway by PD98059 acutely reduced the capacity of Tan to increase nuclear Nrf2 phosphorylation and accumulation. Another ERK inhibitor, U0126, showed a similar effect (data not shown). These results demonstrate the crucial role of ERK in Nrf2-dependent activation of HO-1 and suggest that Nrf2 is a downstream effector of ERK in response to Tan treatment.
DISCUSSION
Tan, one of the most bioactive constituents isolated from Salvia miltiorrhiza Bunge, has long been clinically used in Asian countries for the prevention and treatment of CVDs (8–10). Accumulating evidence suggests that Tan prevents the development of atherosclerosis by inhibiting LDL oxidation (28), scavenger receptor expression (21), matrix metalloproteinase expression and activity (11), pro-inflammatory cytokine expression (29), and smooth muscle cell proliferation (30) and migration (31). Although the protective effects of Tan against atherosclerosis have been widely investigated, the molecular mechanisms by which Tan prevents atherosclerosis remain elusive, especially because the direct target of Tan is unknown. In this study, we investigated the efficacy and the possible molecular mechanisms of Tan involved in cholesterol metabolism of macrophage-derived foam cells. The salient findings of this study are summarized as: i) Tan markedly decreases SR-A expression and oxLDL uptake in lipid-laden macrophages via inhibition of AP-1; ii) Tan induces ABCA1 and ABCG1 expression and promotes cholesterol efflux from macrophages in an LXR-independent fashion; and iii) Tan regulates the expression of SR-A, ABCA1, and ABCG1 and suppresses cholesterol accumulation in human macrophage foam cells via activation of the ERK/Nrf2/HO-1 axis. These observations shed a new light on the potential anti-atherogenic properties of Tan in addition to anti-oxidative/anti-inflammatory effects.
In Fig. 1, we have shown that Tan attenuates atherosclerotic plaque formation in ApoE−/− mice. This is consistent with previous studies showing that Tan can inhibit atherogenesis (13, 32). Macrophages play a critical role in the pathogenesis of atherosclerosis. The neointimal retention of macrophage-derived foam cells is a central pathological event of atherosclerotic lesion formation (1). Our data demonstrate that Tan treatment markedly decreases the macrophage-positive area in the aortic sinuses of ApoE−/− mice. The decreased macrophage-positive area in atherosclerotic plaques could result in a decreased number of macrophage foam cells in atherosclerotic lesions and decreased size of lesional macrophages because of decreased lipid loading. Furthermore, we found that Tan inhibited cholesterol accumulation and regulated the expression of scavenger receptors and cholesterol transporters in the aorta. Collectively, these data imply that Tan treatment may have a significant impact on macrophage foam cells.
A critical step in foam cell formation is the recognition and uptake of oxLDL by multiple macrophage scavenger receptors. SR-A and CD36 are the principal scavenger receptors responsible for the binding and uptake of modified LDL in macrophages (33). Combined inhibition of these two receptors blocks human and murine foam cell formation in vitro (33), and genetic deletion of either SR-A or CD36 retards lesion development in ApoE−/− mice (34, 35). In the present study, we showed that Tan decreases the expression of SR-A and CD36 in the atherosclerotic plaques of ApoE−/− mice (supplementary Fig. VIII). In accordance with this result, Tan markedly decreased mRNA and protein expression of SR-A in human macrophages. Intriguingly, the expression of CD36 was not altered in human macrophages, which is inconsistent with the in vivo data in ApoE−/− mice (Fig. 1D) and the in vitro data in cultured mouse peritoneal macrophages (supplementary Fig. IX). Thus, it is highly plausible that the effect of Tan on CD36 expression may be different in humans and mice. This discrepancy also reminds us that we have to be cautious with extrapolation of the mouse data to humans. Therefore, it will be of interest to examine the effect of Tan on patients with atherosclerosis in future clinical studies.
Another aspect in maintaining cholesterol homeostasis and foam cell formation is cholesterol efflux, mainly mediated by ABCA1, ABCG1, and SR-BI. In animals with deficiency in these three transporters, foam cell accumulation and atherosclerotic lesions were increased (36–38). We observed that Tan enhanced ABCA1/G1 expression in a dose-dependent manner, without altering the expression of SR-BI in human macrophages and in the aortas of ApoE−/− mice, indicating that upregulation of ABCA1/G1 expression by Tan is mainly responsible for the increase in cholesterol efflux and the alleviation of foam cell formation. It has been demonstrated that the expression of ABCA1/G1 is mediated through LXRα, a key nuclear receptor involved in the regulation of intracellular lipid metabolism (39, 40). However, our results indicated that Tan-mediated upregulation of ABCA1/G1 expression was independent of classic LXRα activation. Therefore, the posttranscriptional regulation might be a potential mechanism underlying the effects of Tan on ABCA1/G1 expression. When in the presence of actinomycin D, a general inhibitor of gene transcription, we observed that the mRNA half-life of ABCG1 was considerably extended in Tan-treated macrophages versus vehicle-treated cells, suggesting that Tan increased the steady-state expression of ABCG1. However, we did not observe significant alterations in the mRNA half-life of ABCA1. Previous studies reported that ABCA1 was transcriptionally regulated via LXR-independent pathways such as upstream stimulatory factor (USF) (41), AP-2 (42), and specificity protein 1 (Sp1) (43). Whether these pathways contribute to the induction of the ABCA1 gene by Tan warrants further investigation.
To gain further insight into the mechanism of Tan on ABCA1 expression and lipid accumulation, we next examined the effect of Tan on HO-1 expression. HO-1, which is an important cytoprotective, anti-inflammatory, and anti-oxidant enzyme, has been considered as a novel therapeutic target for the treatment of atherosclerotic diseases (14, 15). Genetic deletion of HO-1 in ApoE−/− mice exhibited accelerated atherogenesis with more extensive and complex atherosclerotic plaques. Macrophages from HO-1−/− mice demonstrated increased foam cell formation in response to oxLDL stimulation (16). Conversely, overexpression of HO-1 by adenoviral vectors significantly protected against atherosclerotic plaque formation in ApoE−/− mice (44). In this study, we demonstrated that Tan profoundly induces HO-1 protein expression in human macrophages. This observation sparked our interest to investigate the potential role of HO-1 in the Tan-mediated protective effect against foam cell formation. Our results showed that both the pharmacological inhibitor (ZnPP) and gene knockdown of HO-1 (siRNA) abolished the beneficial effects of Tan on SR-A, ABCA1, and ABCG1 expression, and on cholesterol uptake and efflux, and finally, attenuated the inhibitory effect of Tan on lipid accumulation in macrophages. These data suggest that Tan protects against foam cell formation, at least in part, through HO-1 activation.
HO-1 gene promoter regions contain anti-oxidant response elements (AREs), and the Nrf2 family of transcription factors can translocate and bind to AREs in response to ARE activation signals (i.e., protein kinase pathway and redox-active components), initiating HO-1 gene transcription (45, 46). In the present study, we revealed that Tan induces HO-1 protein expression through an Nrf2-dependent mechanism. Activation of the Nrf2/HO-1 signaling pathway is regulated by several upstream protein kinases, such as MAPK and PKC (45, 46). The MAPK cascade, including ERK, JNK1/2, and p38 MAPK, as well as the PKC pathway, have been reported to regulate HO-1 activation by different stimuli (45). For example, isothiocyanates mediate HO-1 gene upregulation via the ERK/JNK/Nrf2 axis (47); curcumin induces HO-1 and GCLM expression through the PKC/p38 pathway (48); and diallyl sulfide activates the ERK and p38 MAPK pathways to stimulate HO-1 gene expression (49). To identify which signal cascade controlled the activation of HO-1 by Tan, we examined the effects of MAPK and PKC inhibitors on Tan-mediated upregulation of HO-1 expression, and the results suggest that Tan-induced HO-1 expression is dependent on the activation of ERK, rather than JNK1/2, p38 MAPK, and PKC.
Nrf2 phosphorylation is a critical process for nuclear translocation and transcriptional activity (27, 50). Given that Tan rapidly activated ERK, which is involved in Tan-induced HO-1 expression, we hypothesized that ERK might represent an upstream effector in regulating Nrf2 phosphorylation and the subsequent transcriptional response of ARE-driven HO-1 gene expression. In the present study, we showed that Tan treatment significantly enhanced the phosphorylation and nuclear translocation of Nrf2, and inhibition of ERK signaling by PD98059 or U0126 led to decreased Tan-induced Nrf2 activation. Collectively, these results suggest that Tan activates the ERK pathway, and subsequently induces Nrf2 phosphorylation and nuclear translocation, and ultimately, leads to the upregulation of HO-1 expression.
The previous studies by Sussan et al. (51) and Barajas et al. (52) have consistently shown that Nrf2 expression promotes atherosclerotic lesion formation in ApoE−/− mice, which is associated with increased expression of scavenger receptor CD36. It is therefore paradoxical that Tan activates Nrf2 and inhibits atherogenesis. Nevertheless, our results show that Tan activates Nrf2 without enhancing the expression of CD36. Thus, it seems that Tan can achieve anti-oxidant and anti-inflammatory protection mediated by Nrf2 activation and avoid enhanced oxLDL uptake mediated by increased expression of CD36. Another potential explanation is that Tan-mediated HO-1 induction may not be entirely Nrf2-dependent. In this way, increased HO-1 expression by other pathways could have overshadowed the proatherogenic effects that could potentially be derived from Nrf2 activation.
One study limitation is that the present studies were mainly performed in human macrophages in vitro. Thus, the interpretation of the present data should be cautious, and further studies using in vivo approaches are necessary to verify the present results in future experiments. Systemic HO-1−/−ApoE−/− mice or macrophage-specific HO-1−/− mice are feasible approaches to demonstrate the direct role of HO-1 in the Tan-mediated suppressive effects on foam cells in vivo. Importantly, Tan has been widely used in Asian countries to prevent and treat CVDs. Therefore, further experiments in patients with atherosclerosis are required to confirm the anti-atherogenic effect of Tan.
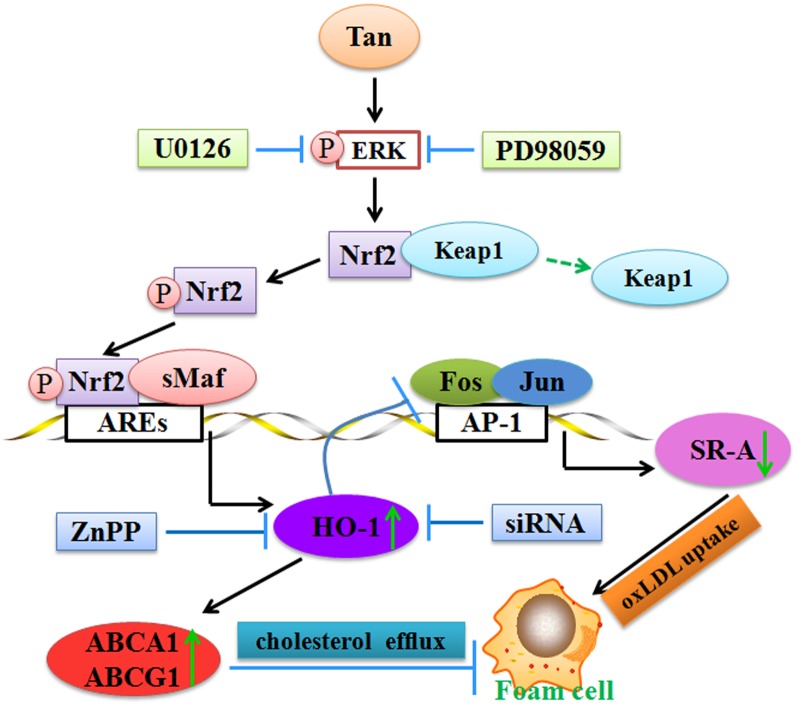
In summary, our study demonstrates that Tan decreases SR-A-mediated oxLDL uptake via inhibition of AP-1 and increases ABCA1/G1-mediated cholesterol efflux via the ERK/Nrf2/HO-1 pathway, ultimately resulting in reduced cholesterol accumulation in foam cells and atherosclerotic plaques (Fig. 8). These findings indicate a critical role of HO-1 in Tan-mediated suppressive effects on foam cell formation. Our present study sheds a novel light on the understanding of the molecular mechanisms of Tan and the clinical application of Tan to treat patients with atherosclerotic CVDs.
Fig. 8.
Schematic illustration of the molecular mechanisms of Tan. Under normal conditions, Nrf2 is sequestered by Keap1 in the cytoplasm. Tan induces ERK activation as an ARE activation signal to disrupt the Nrf2/Keap1 complex and subsequently leads to the phosphorylation and nuclear translocation of Nrf2. Nrf2 then heterodimerizes with small Maf (sMaf) and binds to AREs, eventually resulting in transcriptional activation of ARE-mediated HO-1 expression. HO-1 is involved in the beneficial effects of Tan on lipid-laden macrophages by downregulation of SR-A-mediated oxLDL uptake through inhibition of AP-1, and upregulation of ABCA1 and ABCG1-mediated cholesterol efflux eventually alleviates the formation of foam cells and atherosclerotic plaques.
Supplementary Material
Footnotes
Abbreviations:
- AP
- activator protein
- ApoE−/−
- apolipoprotein E-deficient
- ARE
- anti-oxidant response element
- CD36
- Cluster of Differentiation 36
- CMC-Na
- carboxy-methyl-cellulose sodium
- DAPI
- 4′,6-diamidino-2-phenylindole
- EMSA
- electrophoretic mobility shift assay
- HO-1
- heme oxygenase-1
- JNK
- c-Jun-N-terminal kinase
- LXR
- liver X receptor
- LXRE
- liver X receptor response element
- Nrf2
- nuclear factor-erythroid 2-related factor 2
- oxLDL
- oxidized low density lipoprotein
- PKC
- protein kinase C
- RXR
- retinoid X receptor
- Sp1
- specificity protein 1
- SR-A
- scavenger receptor class A
- Tan
- Tanshinone IIA
- USP
- upstream stimulatory factor
- ZnPP
- zinc protoporphyrin
This work was supported by research grants from the National Natural Science Foundation of China (81072641, 81273499), the National Science and Technology Major Project of China “Key New Drug Creation and Manufacturing Program” (2011ZX09401-307), Team Item of Natural Science Foundation of Guangdong Province (S2011030003190), and Major Project of Guangdong Province (2008A030201013, 2012A080201007). The authors have no conflicts of interest to disclose.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of text, nine figures, and one table.
REFERENCES
- 1.Li A. C., Glass C. K. 2002. The macrophage foam cell as a target for therapeutic intervention. Nat. Med. 8: 1235–1242 [DOI] [PubMed] [Google Scholar]
- 2.Platt N., Gordon S. 2001. Is the class A macrophage scavenger receptor (SR-A) multifunctional? The mouse's tale. J. Clin. Invest. 108: 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park Y. M., Febbraio M., Silverstein R. L. 2009. CD36 modulates migration of mouse and human macrophages in response to oxidized LDL and may contribute to macrophage trapping in the arterial intima. J. Clin. Invest. 119: 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greaves D. R., Gordon S. 2005. The immune system and atherogenesis. Recent insights into the biology of macrophage scavenger receptors. J. Lipid Res. 46: 11–20 [DOI] [PubMed] [Google Scholar]
- 5.Van Eck M., Pennings M., Hoekstra M., Out R., Van Berkel T. J. 2005. Scavenger receptor BI and ATP-binding cassette transporter A1 in reverse cholesterol transport and atherosclerosis. Curr. Opin. Lipidol. 16: 307–315 [DOI] [PubMed] [Google Scholar]
- 6.Attie A. D., Kastelein J. P., Hayden M. R. 2001. Pivotal role of ABCA1 in reverse cholesterol transport influencing HDL levels and susceptibility to atherosclerosis. J. Lipid Res. 42: 1717–1726 [PubMed] [Google Scholar]
- 7.Schmitz G., Langmann T., Heimerl S. 2001. Role of ABCG1 and other ABCG family members in lipid metabolism. J. Lipid Res. 42: 1513–1520 [PubMed] [Google Scholar]
- 8.Gao S., Liu Z., Li H., Little P. J., Liu P., Xu S. 2012. Cardiovascular actions and therapeutic potential of tanshinone IIA. Atherosclerosis. 220: 3–10 [DOI] [PubMed] [Google Scholar]
- 9.Han J. Y., Fan J. Y., Horie Y., Miura S., Cui D. H., Ishii H., Hibi T., Tsuneki H., Kimura I. 2008. Ameliorating effects of compounds derived from Salvia miltiorrhiza root extract on microcirculatory disturbance and target organ injury by ischemia and reperfusion. Pharmacol. Ther. 117: 280–295 [DOI] [PubMed] [Google Scholar]
- 10.Xu S., Liu P. 2013. Tanshinone II-A: new perspectives for old remedies. Expert Opin. Ther. Pat. 23: 149–153 [DOI] [PubMed] [Google Scholar]
- 11.Fang Z. Y., Lin R., Yuan B. X., Yang G. D., Liu Y., Zhang H. 2008. Tanshinone IIA downregulates the CD40 expression and decreases MMP-2 activity on atherosclerosis induced by high fatty diet in rabbit. J. Ethnopharmacol. 115: 217–222 [DOI] [PubMed] [Google Scholar]
- 12.Chen W., Tang F., Xie B., Chen S., Huang H., Liu P. 2012. Amelioration of atherosclerosis by tanshinone IIA in hyperlipidemic rabbits through attenuation of oxidative stress. Eur. J. Pharmacol. 674: 359–364 [DOI] [PubMed] [Google Scholar]
- 13.Xu S., Little P. J., Lan T., Huang Y., Le K., Wu X., Shen X., Huang H., Cai Y., Tang F., et al. 2011. Tanshinone II-A attenuates and stabilizes atherosclerotic plaques in apolipoprotein-E knockout mice fed a high cholesterol diet. Arch. Biochem. Biophys. 515: 72–79 [DOI] [PubMed] [Google Scholar]
- 14.Morita T. 2005. Heme oxygenase and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 25: 1786–1795 [DOI] [PubMed] [Google Scholar]
- 15.Stocker R., Perrella M. A. 2006. Heme oxygenase-1: a novel drug target for atherosclerotic diseases? Circulation. 114: 2178–2189 [DOI] [PubMed] [Google Scholar]
- 16.Orozco L. D., Kapturczak M. H., Barajas B., Wang X., Weinstein M. M., Wong J., Deshane J., Bolisetty S., Shaposhnik Z., Shih D. M., et al. 2007. Heme oxygenase-1 expression in macrophages plays a beneficial role in atherosclerosis. Circ. Res. 100: 1703–1711 [DOI] [PubMed] [Google Scholar]
- 17.Chen T. H., Hsu Y. T., Chen C. H., Kao S. H., Lee H. M. 2007. Tanshinone IIA from Salvia miltiorrhiza induces heme oxygenase-1 expression and inhibits lipopolysaccharide-induced nitric oxide expression in RAW 264.7 cells. Mitochondrion. 7: 101–105 [DOI] [PubMed] [Google Scholar]
- 18.Qi Y. Y., Xiao L., Zhang L. D., Song S. H., Mei Y., Chen T., Tang J. M., Liu F., Ding G. S., Shi Y. Z., et al. 2012. Tanshinone IIA pretreatment attenuates hepatic ischemia-reperfusion. Front. Biosci. (Elite Ed.). 4: 1303–1313 [DOI] [PubMed] [Google Scholar]
- 19.Gu X., Trigatti B., Xu S., Acton S., Babitt J., Krieger M. 1998. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J. Biol. Chem. 273: 26338–26348 [DOI] [PubMed] [Google Scholar]
- 20.Gong Z., Huang C., Sheng X., Zhang Y., Li Q., Wang M. W., Peng L., Zang Y. Q. 2009. The role of tanshinone IIA in the treatment of obesity through peroxisome proliferator-activated receptor gamma antagonism. Endocrinology. 150: 104–113 [DOI] [PubMed] [Google Scholar]
- 21.Xu S., Liu Z., Huang Y., Le K., Tang F., Huang H., Ogura S., Little P. J., Shen X., Liu P. 2012. Tanshinone II-A inhibits oxidized LDL-induced LOX-1 expression in macrophages by reducing intracellular superoxide radical generation and NF-kappaB activation. Transl. Res. 160: 114–124 [DOI] [PubMed] [Google Scholar]
- 22.Robinet P., Wang Z., Hazen S. L., Smith J. D. 2010. A simple and sensitive enzymatic method for cholesterol quantification in macrophages and foam cells. J. Lipid Res. 51: 3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin S., Zhou C., Neufeld E., Wang Y. H., Xu S. W., Lu L., Wang Y., Liu Z. P., Li D., Li C., et al. 2013. BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein modulates ATP-binding cassette transporter A-1 trafficking and function. Arterioscler. Thromb. Vasc. Biol. 33: e31–e38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou S. G., Zhou S. F., Huang H. Q., Chen J. W., Huang M., Liu P. Q. 2006. Proteomic analysis of hypertrophied myocardial protein patterns in renovascularly hypertensive and spontaneously hypertensive rats. J. Proteome Res. 5: 2901–2908 [DOI] [PubMed] [Google Scholar]
- 25.Horvai A., Palinski W., Wu H., Moulton K. S., Kalla K., Glass C. K. 1995. Scavenger receptor A gene regulatory elements target gene expression to macrophages and to foam cells of atherosclerotic lesions. Proc. Natl. Acad. Sci. USA. 92: 5391–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moulton K. S., Semple K., Wu H., Glass C. K. 1994. Cell-specific expression of the macrophage scavenger receptor gene is dependent on PU.1 and a composite AP-1/ets motif. Mol. Cell. Biol. 14: 4408–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Autréaux B., Toledano M. B. 2007. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8: 813–824 [DOI] [PubMed] [Google Scholar]
- 28.Niu X. L., Ichimori K., Yang X., Hirota Y., Hoshiai K., Li M., Nakazawa H. 2000. Tanshinone II-A inhibits low density lipoprotein oxidation in vitro. Free Radic. Res. 33: 305–312 [DOI] [PubMed] [Google Scholar]
- 29.Jang S. I., Kim H. J., Kim Y. J., Jeong S. I., You Y. O. 2006. Tanshinone IIA inhibits LPS-induced NF-kappaB activation in RAW 264.7 cells: possible involvement of the NIK-IKK, ERK, p38 and JNK pathways. Eur. J. Pharmacol. 542: 1–7 [DOI] [PubMed] [Google Scholar]
- 30.Li X., Du J. R., Yu Y., Bai B., Zheng X. Y. 2010. Tanshinone IIA inhibits smooth muscle proliferation and intimal hyperplasia in the rat carotid balloon-injured model through inhibition of MAPK signaling pathway. J. Ethnopharmacol. 129: 273–279 [DOI] [PubMed] [Google Scholar]
- 31.Jin U. H., Suh S. J., Chang H. W., Son J. K., Lee S. H., Son K. H., Chang Y. C., Kim C. H. 2008. Tanshinone IIA from Salvia miltiorrhiza BUNGE inhibits human aortic smooth muscle cell migration and MMP-9 activity through AKT signaling pathway. J. Cell. Biochem. 104: 15–26 [DOI] [PubMed] [Google Scholar]
- 32.Tang F., Wu X., Wang T., Wang P., Li R., Zhang H., Gao J., Chen S., Bao L., Huang H., et al. 2007. Tanshinone II A attenuates atherosclerotic calcification in rat model by inhibition of oxidative stress. Vascul. Pharmacol. 46: 427–438 [DOI] [PubMed] [Google Scholar]
- 33.Kunjathoor V. V., Febbraio M., Podrez E. A., Moore K. J., Andersson L., Koehn S., Rhee J. S., Silverstein R., Hoff H. F., Freeman M. W. 2002. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J. Biol. Chem. 277: 49982–49988 [DOI] [PubMed] [Google Scholar]
- 34.Febbraio M., Podrez E. A., Smith J. D., Hajjar D. P., Hazen S. L., Hoff H. F., Sharma K., Silverstein R. L. 2000. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J. Clin. Invest. 105: 1049–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki H., Kurihara Y., Takeya M., Kamada N., Kataoka M., Jishage K., Ueda O., Sakaguchi H., Higashi T., Suzuki T., et al. 1997. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 386: 292–296 [DOI] [PubMed] [Google Scholar]
- 36.McNeish J., Aiello R. J., Guyot D., Turi T., Gabel C., Aldinger C., Hoppe K. L., Roach M. L., Royer L. J., de Wet J., et al. 2000. High density lipoprotein deficiency and foam cell accumulation in mice with targeted disruption of ATP-binding cassette transporter-1. Proc. Natl. Acad. Sci. USA. 97: 4245–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy M. A., Barrera G. C., Nakamura K., Baldan A., Tarr P., Fishbein M. C., Frank J., Francone O. L., Edwards P. A. 2005. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 1: 121–131 [DOI] [PubMed] [Google Scholar]
- 38.Trigatti B., Rayburn H., Vinals M., Braun A., Miettinen H., Penman M., Hertz M., Schrenzel M., Amigo L., Rigotti A., et al. 1999. Influence of the high density lipoprotein receptor SR-BI on reproductive and cardiovascular pathophysiology. Proc. Natl. Acad. Sci. USA. 96: 9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li A. C., Glass C. K. 2004. PPAR- and LXR-dependent pathways controlling lipid metabolism and the development of atherosclerosis. J. Lipid Res. 45: 2161–2173 [DOI] [PubMed] [Google Scholar]
- 40.Calkin A. C., Tontonoz P. 2010. Liver x receptor signaling pathways and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 30: 1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X. P., Freeman L. A., Knapper C. L., Amar M. J., Remaley A., Brewer H. B., Jr, Santamarina-Fojo S. 2002. The E-box motif in the proximal ABCA1 promoter mediates transcriptional repression of the ABCA1 gene. J. Lipid Res. 43: 297–306 [PubMed] [Google Scholar]
- 42.Iwamoto N., Abe-Dohmae S., Ayaori M., Tanaka N., Kusuhara M., Ohsuzu F., Yokoyama S. 2007. ATP-binding cassette transporter A1 gene transcription is downregulated by activator protein 2alpha. Doxazosin inhibits activator protein 2alpha and increases high-density lipoprotein biogenesis independent of alpha1-adrenoceptor blockade. Circ. Res. 101: 156–165 [DOI] [PubMed] [Google Scholar]
- 43.Chen X., Zhao Y., Guo Z., Zhou L., Okoro E. U., Yang H. 2011. Transcriptional regulation of ATP-binding cassette transporter A1 expression by a novel signaling pathway. J. Biol. Chem. 286: 8917–8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juan S. H., Lee T. S., Tseng K. W., Liou J. Y., Shyue S. K., Wu K. K., Chau L. Y. 2001. Adenovirus-mediated heme oxygenase-1 gene transfer inhibits the development of atherosclerosis in apolipoprotein E-deficient mice. Circulation. 104: 1519–1525 [DOI] [PubMed] [Google Scholar]
- 45.Kang K. W., Lee S. J., Kim S. G. 2005. Molecular mechanism of nrf2 activation by oxidative stress. Antioxid. Redox Signal. 7: 1664–1673 [DOI] [PubMed] [Google Scholar]
- 46.Kim Y. M., Pae H. O., Park J. E., Lee Y. C., Woo J. M., Kim N. H., Choi Y. K., Lee B. S., Kim S. R., Chung H. T. 2011. Heme oxygenase in the regulation of vascular biology: from molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 14: 137–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu C., Yuan X., Pan Z., Shen G., Kim J. H., Yu S., Khor T. O., Li W., Ma J., Kong A. N. 2006. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol. Cancer Ther. 5: 1918–1926 [DOI] [PubMed] [Google Scholar]
- 48.Rushworth S. A., Ogborne R. M., Charalambos C. A., O'Connell M. A. 2006. Role of protein kinase C delta in curcumin-induced antioxidant response element-mediated gene expression in human monocytes. Biochem. Biophys. Res. Commun. 341: 1007–1016 [DOI] [PubMed] [Google Scholar]
- 49.Gong P., Hu B., Cederbaum A. I. 2004. Diallyl sulfide induces heme oxygenase-1 through MAPK pathway. Arch. Biochem. Biophys. 432: 252–260 [DOI] [PubMed] [Google Scholar]
- 50.Nguyen T., Sherratt P. J., Huang H. C., Yang C. S., Pickett C. B. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 278: 4536–4541 [DOI] [PubMed] [Google Scholar]
- 51.Sussan T. E., Jun J., Thimmulappa R., Bedja D., Antero M., Gabrielson K. L., Polotsky V. Y., Biswal S. 2008. Disruption of Nrf2, a key inducer of antioxidant defenses, attenuates ApoE-mediated atherosclerosis in mice. PLoS ONE. 3: e3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barajas B., Che N., Yin F., Rowshanrad A., Orozco L. D., Gong K. W., Wang X., Castellani L. W., Reue K., Lusis A. J., et al. 2011. NF-E2-related factor 2 promotes atherosclerosis by effects on plasma lipoproteins and cholesterol transport that overshadow antioxidant protection. Arterioscler. Thromb. Vasc. Biol. 31: 58–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.