Abstract
The first step in reverse cholesterol transport is a process by which lipid-free or lipid-poor apoA-1 removes cholesterol from cells through the action of ATP binding cassette transporter A1 at the plasma membrane. However the structure and composition of lipid-free or -poor apoA-1 in plasma remains obscure. We previously obtained a monoclonal antibody (MAb) that specifically recognizes apoA-1 in preβ1-HDL, the smallest apoA-1-containing particle in plasma, which we used to establish a preβ1-HDL ELISA. Here, we purified preβ1-HDL from fresh normal plasma using said antibody, and analyzed the composition and structure. ApoA-1 was detected, but neither phospholipid nor cholesterol were detected in the purified preβ1-HDL. Only globular, not discoidal, particles were observed by electron microscopy. In nondenaturing PAGE, no difference in the mobility was observed between the purified preβ1-HDL and original plasma preβ1-HDL, or between the preβ1-HDL and lipid-free apoA-1 prepared by delipidating HDL. In sandwich ELISA using two anti-preβ1-HDL MAbs, reactivity with intact plasma preβ1-HDL was observed in ELISA using two MAbs with distinct epitopes but no reactivity was observed in ELISA using a single MAb, and the same phenomenon was observed with monomolecular lipid-free apoA-1. These results suggest that plasma preβ1-HDL is lipid-free monomolecular apoA-1.
Keywords: pre-beta-high density lipoprotein, lipid-free apolipoprotein A-1, reverse cholesterol transport
HDL plays a central role in reverse cholesterol transport of excessively accumulated cholesterol from peripheral tissues to the liver (1–3). The first and most important step in reverse cholesterol transport is a reaction in which lipid-free apoA-1, composed of only a single apoA-1, or lipid-poor apoA-1, composed of a single apoA-1 and a small amount of phospholipid, accepts unesterified cholesterol from cells through ATP binding cassette transporter A1 (ABCA1) on the plasma membrane to form a discoidal nascent HDL composed of two molecules of apoA-1, phospholipid, and unesterified cholesterol (4, 5). However, the presence or absence of lipid-free- or -poor apoA-1 in plasma and its composition and structure remain obscure (5).
Preβ1-HDL is an apoA-1-containing particle known as the initial plasma acceptor of cellular cholesterol (1, 6–8). Although preβ1-HDL accounts for only 1–5% of total apoA-1 in plasma, it is considered to play an important role in cholesterol efflux because efflux capacity is remarkably reduced by adding anti-preβ1-HDL-specific antibody to plasma or depleting preβ1-HDL from plasma in cholesterol efflux experiments where plasma is added to culture cells (9, 10). Plasma preβ1-HDL can be separated by nondenaturing two-dimensional gel electrophoresis (2D electrophoresis). Plasma preβ1-HDL migrates to the preβ position in agarose gel electrophoresis for the first dimension, and to the position of the smallest particle among plasma apoA-1-containing particles in nondenaturing PAGE for the second dimension (6, 11). Preβ1-HDL is known to be present in the plasma of Tangier disease patients as the only apoA-1-containing particle (12, 13). Tangier disease is a disorder in which cholesterol efflux to apoA-1 is rendered defective by mutations in the ABCA1 gene (14–16). In fibroblasts obtained from Tangier patients, cholesterol and phospholipid efflux to lipid-free apoA-1 is completely defective, and HDL is not formed (17). Hence plasma preβ1-HDL may be lipid-free or -poor apoA-1, the substrate (precursor) for ABCA1-mediated cholesterol efflux. However some reports claim that preβ1-HDL contains cholesterol (8, 18, 19), is composed of two molecules of apoA-1 (5), and is a discoidal particle (2, 3).
The reason the composition and structure of preβ1-HDL are still obscure may be that adequate preβ1-HDL for analysis cannot be easily purified from plasma. The concentration of plasma preβ1-HDL is much lower than that of other HDL subfractions, and preβ1-HDL is very unstable in plasma (20). We previously obtained a monoclonal antibody (MAb) 55201 (MAb55201) that specifically recognizes apoA-1 in preβ1-HDL and developed an ELISA for easily measuring plasma preβ1-HDL concentrations using the MAb (21). Recently a variety of research results on preβ1-HDL using the ELISA were reported, in particular the relationships between plasma preβ1-HDL concentrations and coronary artery disease and medications have been noted (22–26).
The aim of the present study is to clarify the composition and structure of plasma preβ1-HDL. The MAb55201 we obtained reacts with apoA-1 in plasma preβ1-HDL and delipidated apoA-1 (lipid-free apoA-1), and does not react with apoA-1 in other HDL sub-fractions (21). We first purified preβ1-HDL from fresh plasma of healthy subjects with affinity chromatography using MAb55201, and with gel filtration chromatography to analyze the protein and lipid composition and to examine its shape under electron microscopy. We then analyzed intact plasma preβ1-HDL and lipid-free apoA-1 by nondenaturing PAGE, nondenaturing 2D electrophoresis, and size-exclusion chromatography, and examined whether the concentration of lipid-free apoA-1 added to preβ1-HDL-depleted plasma decreases in an LCAT-dependent conversion into α-HDL, which is a phenomenon unique to plasma preβ1-HDL, for comparison of intact plasma preβ1-HDL and lipid-free apoA-1. In addition, we obtained another anti-preβ1-HDL MAb (MAb55205), which recognizes a different epitope on apoA-1 from that of MAb55201, and investigated whether the apoA-1 in preβ1-HDL is a monomer or dimer by an immunochemical procedure using the anti-preβ1-HDL antibodies.
MATERIALS AND METHODS
Materials
Blood was obtained on the day experiments were performed from healthy volunteers (employees of Sekisui Medical Co., Ltd.) having a normal plasma lipid profile (total cholesterol, <220 mg/dl; triglyceride, <150 mg/dl; and HDL-cholesterol, >40 mg/dl). Written informed consent was obtained from all volunteers. The subjects fasted overnight, and venous blood was drawn into vacuum blood collection tubes containing EDTA-2Na (Terumo Corporation, Tokyo, Japan). The blood samples were immediately chilled in ice water for 30 min and centrifuged at 2°C, 3,000 rpm for 30 min to separate the plasma. Lipid-free apoA-1 was prepared by delipidating the HDL fraction (1.063 g/ml < d < 1.21 g/ml) and subsequent purification as described previously (21). Lipid-free apoA-1 is known to self-associate to form dimers and larger multimers, depending on various conditions that include apoA-1 concentration and ionic strength in the solution (27). The purified lipid-free apoA-1 was dialyzed against PBS [0.15 mol/l NaCl, 20 mmol/l phosphate buffer (pH 7.2)] to a concentration of 0.1 mg/ml or lower, and stored at −80°C to avoid self-association before use. Twenty-five peptides of apoA-1 were purchased from Protein Purity Ltd. (Gunma, Japan). The molecular mass of each peptide was confirmed to be the same as the theoretical molecular mass by MALDI-MS, and the purity was confirmed to be more than 95% by HPLC. Cyanogen bromide (CNBr) fragments of apoA-1 were prepared using the procedure reported by Morrison et al. (28) as follows. Lipid-free apoA-1 (0.1 mg) dialyzed with purified water was lyophilized and dissolved in 0.2 ml of 70% trifluoroacetic acid (TFA). CNBr dissolved in 0.2 ml of 70% TFA to a concentration of 1 mg/ml was added to the apoA-1 solution; the bottle was suffused with nitrogen gas and sealed, followed by incubation at room temperature for 24 h in the dark. After incubation, 3 ml of purified water was added and lyophilized. Each fragment was identified by molecular mass using SDS-PAGE and N-terminal amino acid analysis. MAb55201 was obtained by our group as described previously (21). The mouse anti-influenza A MAb obtained by our group was used as the control IgG. MAb55201 or the control IgG-bound sepharose column was prepared by using CNBr-activated Sepharose 4B (GE Healthcare) in accordance with the procedure recommended by the manufacturer. Polyacrylamide slab gels used for nondenaturing PAGE or SDS-PAGE, and the protein transfer kits used for Western blotting were purchased from Cosmo Bio Co., Ltd. (Tokyo). DTNB used for the LCAT-dependent conversion experiment was purchased from Sigma-Aldrich, and dissolved with the phosphate buffer [0.1 mol/l KH2PO4-Na2HPO4 (pH 7.4)] just prior to use, followed by addition to plasma or preβ1-HDL-depleted plasma to a final concentration of 2 mmol/l. Goat anti-apoA-1 polyclonal antibody (PAb) and goat anti-apoA-2 PAb were prepared as described previously (21). Biotin-labeled antibodies were prepared with EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific, Inc.).
Purification of plasma preβ1-HDL
Plasma (40 ml) was passed through the control IgG-bound Sepharose column (5 ml) to remove fractions nonspecifically binding to mouse IgG or the Sepharose beads. The eluent (40 ml) was then passed through the MAb55201-bound Sepharose column (2 ml) to bind preβ1-HDL. The unbound fraction was used as the preβ1-HDL-depleted plasma for the experiments described later. After washing the MAb55201-bound Sepharose column with 30 ml of PBS, the adsorbed fraction was eluted with 10 ml of 0.1 mol/l glycine-HCl buffer (pH 2.0) containing 0.15 mol/l NaCl, and collected in 1 ml fractions. The absorbance at 280 nm of each fraction was measured, and the fractions (Fr. 4–6) having absorbance of more than 0.4 were immediately pooled and applied to a Superdex200 16/60 column (GE Healthcare) for further separation by gel filtration chromatography. The chromatography was run with 0.15 mol/l NaCl solution at a flow rate of 1.0 ml/min and the eluent was collected in 1.0 ml fractions by a fraction collector. The preβ1-HDL concentration of each fraction was determined by ELISA using MAb55201 (preβ1-HDL-ELISA) (21). Those fractions containing a high concentration of preβ1-HDL were then collected. The purification process was performed four times, once each for the plasma samples obtained from the four healthy subjects, and all operations were performed at 4°C. The purity of preβ1-HDL in the purified sample was confirmed by nondenaturing 2D electrophoresis, and the concentration of preβ1-HDL was determined by preβ1-HDL-ELISA. In order to examine the possibility that MAb55201 bound to Sepharose beads may strip apoA-1 from HDL, we carried out affinity chromatography under the same conditions at 1/10 scale of the above using HDL (1.063 g/ml < d < 1.21 g/ml) prepared by ultracentrifugation instead of plasma. Two milliliters of HDL (140 μg/ml of cholesterol) was applied to a MAb55201-bound Sepharose column (0.2 ml). After washing the column with 3 ml of PBS, the adsorbed fraction was eluted with 1 ml of glycine-HCl buffer (pH 2.0). The eluted fraction was neutralized by adding 0.05 ml of 2 mol/l Tris-HCl buffer (pH 8.0), followed by analysis by nondenaturing PAGE.
Nondenaturing 2D electrophoresis
Purified preβ1-HDL (4 μl) or plasma (2 μl) was added to a 0.75% agarose gel in 50 mmol/l Barbital buffer (pH 8.6) prepared on gel-bond film with a thickness of 1 mm, and run at 540 V for 60 min at 4°C. Strips of agarose gel were cut out, set on 10–20% polyacrylamide gradient slab gels, and run at 75 V for 24 h at 4°C in 90 mmol/l Tris, 80 mmol/l boric acid, and 3 mmol/l EDTA (pH 8.3). The separated HDL subfractions in the polyacrylamide gel were transferred onto poly vinylidene difluoride (PVDF) membranes using a semidry electroblotter, and then analyzed by Western blot using goat anti-apoA-1 PAb, HRP-conjugated rabbit anti-goat IgG antibody (DAKO), and diaminobenzidine.
Electron microscopy
Purified preβ1-HDL solution was dropped on a 400 mesh grid coated with carbon film for dispersion. The adsorbed specimen was soaked with 2% uranyl acetate at 4°C for 10 s for negative staining and examined in a transmission electron microscope, JEM-1200EX (JEOL Ltd., Tokyo, Japan).
Protein analysis
The protein content was determined with a Micro BCA kit (Thermo). The protein composition was investigated by SDS-PAGE and mass spectrometric analysis as follows. The purified fraction (0.22 μg) diluted in a sample buffer containing SDS and 2-mercaptoethanol was boiled and electrophoresed on a 15% polyacrylamide gel. Mass spectrometric analysis and the pretreatment were performed as described previously (29). Briefly, protein in the gel was detected by MS-compatible silver staining. The stained band was cut out and put into a 1.5 ml tube. Cysteine disulfide bonds were reduced with DTT and alkylated with iodoacetamide. In-gel digestion with trypsin was performed at 37°C overnight, followed by extracting the peptides with 50% acetonitrile containing 5% TFA. The extracts were dried and redissolved in 0.1% formic acid, followed by LC/MS/MS analysis. LC/MS/MS analysis was carried out in a LCQ Deca XP ion trap mass spectrometer (ThermoFinnigan, USA) equipped with a nano-LC electrospray ionization source (AMR, Japan), interfaced on-line with a capillary HPLC system (Paradigm MS4; Michrom BioResources, USA). MS/MS data were analyzed using SEQUEST, a computer program that allows the correlation of experimental data with theoretical spectra generated from known protein sequences, to be compared against the latest version of the public nonredundant protein database of the National Center for Biotechnology Information.
Lipid analysis
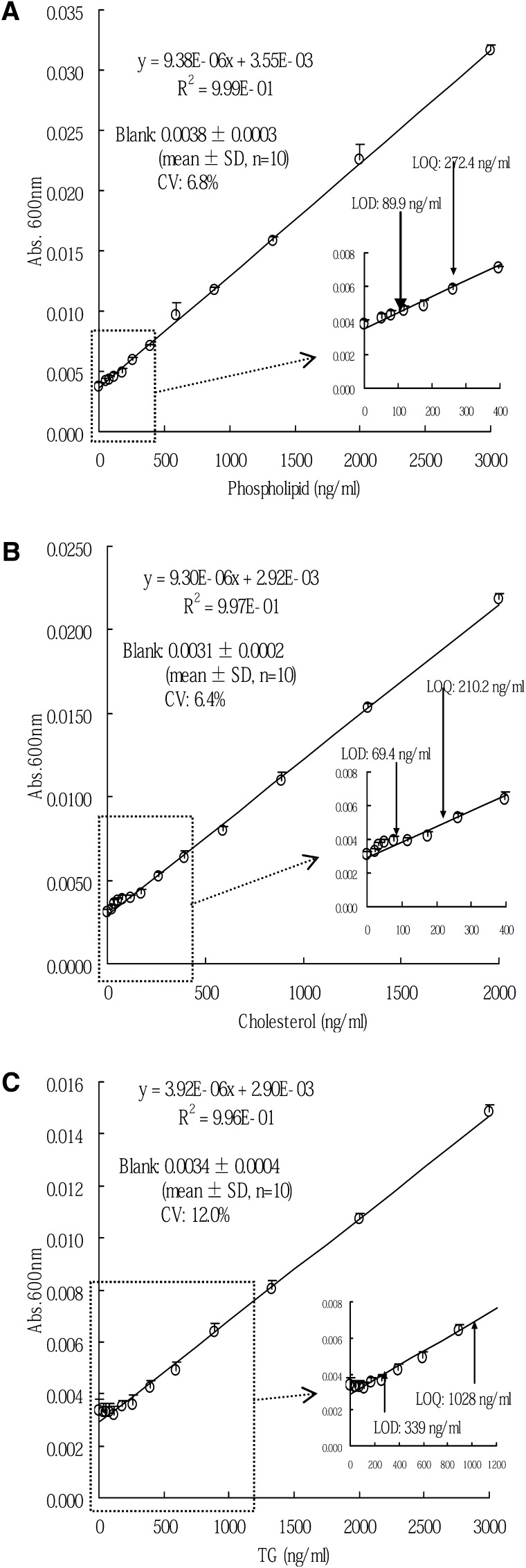
The amounts of choline-containing phospholipids, cholesterol, and triglyceride were determined by enzymatic methods using commercial reagents (Wako Pure Chemical Industries, Ltd., Osaka, Japan) as follows. Each standard, diluted with saline (0.15 mol/l NaCl) and purified preβ1-HDL (0.2 ml), were added to the appropriate enzyme solution (0.2 ml) in glass tubes and incubated at 37°C for 5 min. Absorbance at 600 nm was measured by a spectrophotometer, UV-2400PC (Shimadzu Corporation, Kyoto, Japan). In order to validate each lipid assay, limit of detection (LOD) and limit of quantification (LOQ) were calculated using the following equations, in accordance with the appropriate ICH guideline (30).
where σ and S represent standard deviations of blank absorbance (n = 10) and slopes of calibration curves, respectively.
Nondenaturing PAGE
Plasma and preβ1-HDL-depleted plasma were diluted 10-fold with sample buffer containing 31% sucrose, 0.06% EDTA, and 0.01% Bromophenol blue; and purified preβ1-HDL and lipid-free apoA-1 were diluted with the sample buffer to a concentration of 10 μg/ml. The unbound fraction and the eluted fraction in affinity chromatography using HDL were each diluted 2-fold with the sample buffer. Each sample was applied to a 15–25% polyacrylamide gradient slab gel with 5 μl/well and electrophoresed at 75 V for 24 h at 4°C. Then Western blot analysis was performed in the same manner as described above for nondenaturing 2D electrophoresis. A high molecular mass calibration kit for native electrophoresis (GE Healthcare) was used as the molecular size marker.
Size-exclusion chromatography
Plasma, preβ1-HDL-depleted plasma, and lipid-free apoA-1 added to preβ1-HDL-depleted plasma were individually separated using size-exclusion chromatography in accordance with the methods of Nanjee and Brinton (31). Fifty microliters of each sample was applied to a Superdex 200 HR 10/30 column connected in series to a Superdex 75 HR 10/30 column and separated in a 50 mmol/l Tris-HCl buffer (pH 7.4), containing 150 mmol/l NaCl, 1 g/l sodium EDTA, and 1 g/l NaN3 at a flow rate of 0.25 ml/min. After discarding 12.5 ml of the eluent, 40 fractions of 0.5 ml each were collected. The concentrations of preβ1-HDL and lipid-free apoA-1 in all fractions were determined by preβ1-HDL-ELISA (21). The apoA-1 concentrations of fractions separated from plasma were determined by sandwich ELISA using goat anti-apoA-1 PAb as described previously (21).
LCAT-dependent conversion experiment
Plasma and lipid-free apoA-1 added to preβ1-HDL-depleted plasma (0.5 ml) were individually incubated at 37°C with or without DTNB. A portion of each sample was taken at intervals and diluted 11-fold with a stabilization buffer (20) containing 50% sucrose. The diluted samples were immediately stored at −80°C, and later the concentrations of preβ1-HDL and lipid-free apoA-1 were determined by preβ1-HDL-ELISA (21).
Preparation of new anti-apoA-1 MAbs
A hybridoma cell line producing MAb55205 was developed in the same manner as described previously for MAb55201 (21). Briefly, after immunization of a Balb/c mouse with lipid-free apoA-1, the mouse was euthanized, and the spleen cells were fused with murine myeloma cells according to the method of Kohler and Milstein (32). Screening of hybridoma cells was performed using two kinds of ELISA with biotin-labeled goat anti-apoA-1 or apoA-2 PAb. Hybridomas were selected that produced antibodies showing reactivity in ELISA using anti-apoA-1 PAb, but not in ELISA using anti-apoA-2 PAb. A hybridoma cell line producing anti-apoA-1 MAb 14208 (MAb14208) was prepared as follows. After immunization and cell fusion were performed as described above, a hybridoma cell producing an antibody that reacted strongly with apoA-1 in apoA-1-immobilized ELISA was selected. The selected hybridomas were cloned by limited dilution and then injected into the peritoneal region of mice to harvest ascites, followed by purification of IgG from the ascites using protein A-Sepharose.
Specificity of anti-apoA-1 MAbs
The specificity of the newly developed antibodies (MAb55205 and MAb14208) was assessed based on their reactivity to plasma proteins, LpA-1 (lipoprotein containing apoA-1 but not apoA-2), LpA-1:A-2 (lipoprotein containing both apoA-1 and apoA-2), and plasma gel filtration fractions, as described previously (21). Reactivity to plasma proteins was investigated by Western blotting using plasma proteins separated by SDS-PAGE. Reactivity to LpA-1 and LpA-1:A-2 was investigated by sandwich ELISA using plasma diluted 2,121-fold as the antigen, and goat anti-apoA-1 or anti-apoA-2 PAb as the biotin-labeled antibody in each plate. Reactivity to plasma gel filtration fractions was investigated by sandwich ELISA using HRP-labeled goat anti-apoA-1 PAb with plasma fractions separated on a fast protein liquid chromatography system (GE Healthcare) composed of two TSK gel G3000SW columns (7.5 mm × 60 cm), a TSK gel G3000SW column (7.5 mm × 30 cm) (Tosoh, Japan), and a Superdex 200HR1030 column (10 mm × 30 cm).
Epitope of anti-apoA-1 MAbs
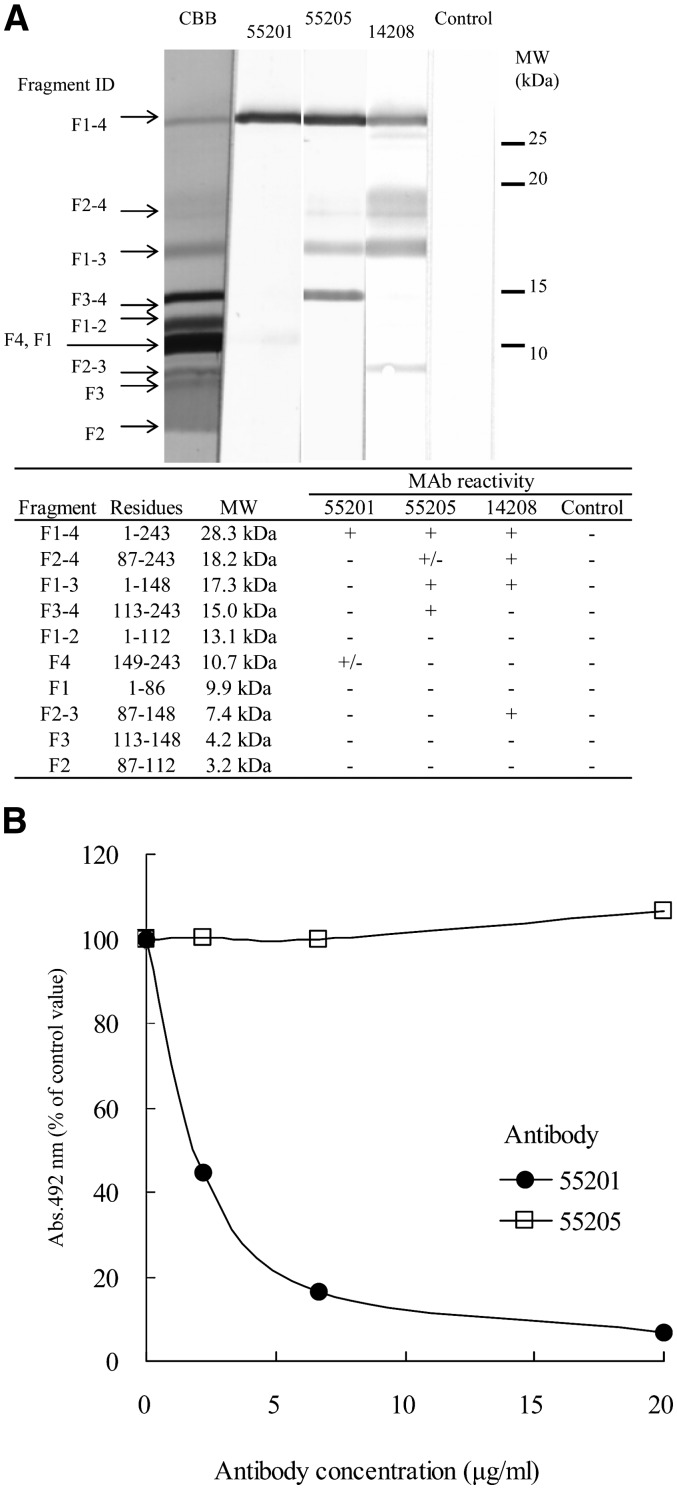
To determine the epitope of each antibody, first, reactivity to 25 kinds of apo-A1 peptide was investigated in the following peptide-competitive ELISA. Lipid-free apoA-1 (50 μl/well), diluted to 0.5 μg/ml with PBS, was added to an ELISA plate, followed by incubation at 4°C overnight. After washing and blocking the plate, various concentrations of each peptide were added (25 μl/well). Then each MAb (25 μl/well), diluted to 200 ng/ml, was added, followed by incubation at room temperature for 1 h. After washing, HRP-labeled goat anti-mouse IgG antibody (50 μl/well) was added, followed by incubation at room temperature for 1 h. After washing, the substrate solution (50 μl/well) containing o-phenylenediamine and H2O2 was added, followed by incubation at room temperature for 10 min. After stopping the enzymatic reaction by adding 1.5N H2SO4 (50 μl/well), the absorbance at 492 nm was measured by a plate reader. Next, reactivity of each MAb to CNBr fragments of apoA-1 was investigated by Western blot analysis. After separating apoA-1 CNBr fragments (5 μg) by SDS-PAGE, the separated fragments in the polyacrylamide gel were transferred onto a PVDF membrane. The fragments on the membrane were first reacted with each MAb and then reacted with HRP-conjugated goat anti-mouse IgG antibody, followed by detection of bands by diaminobenzidine. We then carried out an antibody-competitive ELISA to examine whether the epitope of MAb55205 is different from that of MAb55201. Various concentrations of each MAb (25 μl/well) were added (25 μl/well) to a plate coated with lipid-free apoA-1, as described above in the peptide-competitive ELISA test. Then biotin-labeled MAb55201 (1 μg/ml) was added (25 μl/well), followed by incubation at room temperature for 1 h. After washing, HRP-labeled streptavidin was added (50 μl/well), followed by incubation at room temperature for 0.5 h. After a further washing, the enzymatic reaction was carried out as in the peptide-competitive ELISA.
Sandwich ELISA using anti-apoA-1 MAbs
To investigate the number of apoA-1 molecules per particle of plasma preβ1-HDL, sandwich ELISA using MAb55201 and MAb55205 was performed, with sandwich ELISA using only MAb14208 serving as control. Each MAb (50 μl/well), diluted to 5 μg/ml with PBS, was added to an ELISA plate, followed by incubation at 4°C overnight. After washing the plate with PBS, blocking solution was added, followed by incubation at room temperature for 1 h. This was then removed and various concentrations of lipid-free apoA-1 or plasma (50 μl/well) were added to the plate, followed by incubation at room temperature for 1 h. After washing, each biotin-labeled MAb (50 μl/well), diluted to 1 μg/ml, was added and incubated for 1 h. After washing, HRP-labeled streptavidin (50 μl/well) was added and incubated for 30 min. After washing, the HRP reaction was performed as described above in the peptide-competitive ELISA test. The blocking and dilution steps in the sandwich ELISA were performed using 1% BSA-PBS containing no detergent in order to preserve the three-dimensional structure of the lipoprotein, and the washing steps after the antigen reaction were performed using 0.1% BSA-PBS.
RESULTS
Purification and analysis of plasma preβ1-HDL
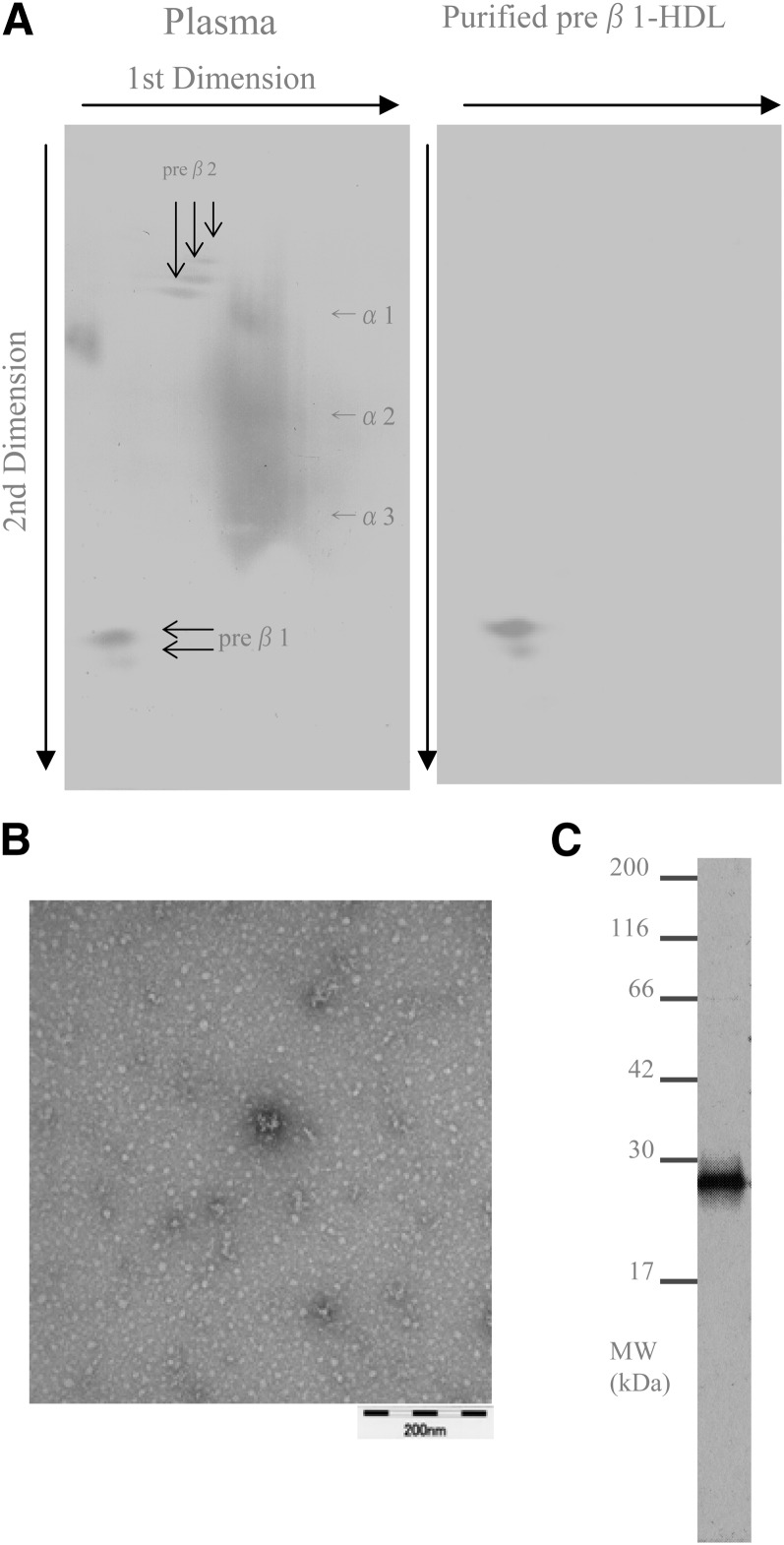
We purified preβ1-HDL from each plasma sample of the four healthy volunteers. The concentration and the amount of preβ1-HDL in the purified fractions were 19.0 ± 1.7 μg/ml (mean ± SD, n = 4) and 152 ± 13.4 μg (mean ± SD, n = 4), respectively. In nondenaturing 2D electrophoresis, only preβ1-HDL spots were detected in the purified fraction (Fig. 1A). In electron microscopy, only globular, not discoidal, particles were observed (Fig. 1B). In SDS-PAGE, only a band with a molecular mass of 28 kDa was detected (Fig. 1C). In LC/MS/MS analysis of the band, apoA-1 was detected, but other proteins, including apoM, were not detected (supplementary Table I). The protein concentration in the purified fraction was 22.5 ± 3.5 μg/ml (mean ± SD, n = 4), while any lipids, including phospholipids, cholesterol, and triglyceride, were not detected at all (Table 1). The LOD of the lipid assays for phospholipids, cholesterol, and triglyceride were 89.9 ng/ml, 69.4 ng/ml, and 339 ng/ml, respectively (Fig. 2).
Fig. 1.
Analysis of purified preβ1-HDL. A: Nondenaturing 2D electrophoresis; plasma (left panel), purified preβ1-HDL (right panel). B: Electron microscopy. C: SDS-PAGE.
TABLE 1.
Composition of purified preβ1-HDL
| Concentration (μg/ml) | Mass Ratio (protein:lipid) | Molar Ratio (apoA-1:lipid)e | |
| Protein | 22.5 ± 3.5a | — | — |
| Phospholipid | <0.0899b | 100:<0.40 | 1:<0.11 |
| Cholesterol | <0.0694c | 100:<0.31 | 1:<0.23 |
| Triglyceride | <0.339d | 100:<1.5 | 1:<0.48 |
Mean ± SD (n = 4).
The absorbance of each sample was 0.0031, 0.0033, 0.0043, and 0.0033, respectively (mean of duplicates). All values were less than LOD (0.0899 μg/ml).
The absorbance of each sample was 0.0022, 0.0020, 0.0024, and 0.0023, respectively (mean of duplicates). All values were less than LOD (0.0694 μg/ml).
The absorbance of each sample was 0.0023, 0.0044, 0.0022, and 0.0036, respectively (mean of duplicates). All values were less than LOD (0.339 μg/ml).
Values are calculated on the basis of the following molecular weights: apoA-1, 28,331 Da; phospholipid, 774 Da; cholesterol, 387 Da; and triglyceride, 886 Da.
Fig. 2.
Calibration curves of lipid assays for phospholipids (A), cholesterol (B), and triglyceride (C). The error bars represent one standard deviation of absorbance (n = 10).
Comparison of preβ1-HDL and lipid-free apoA-1
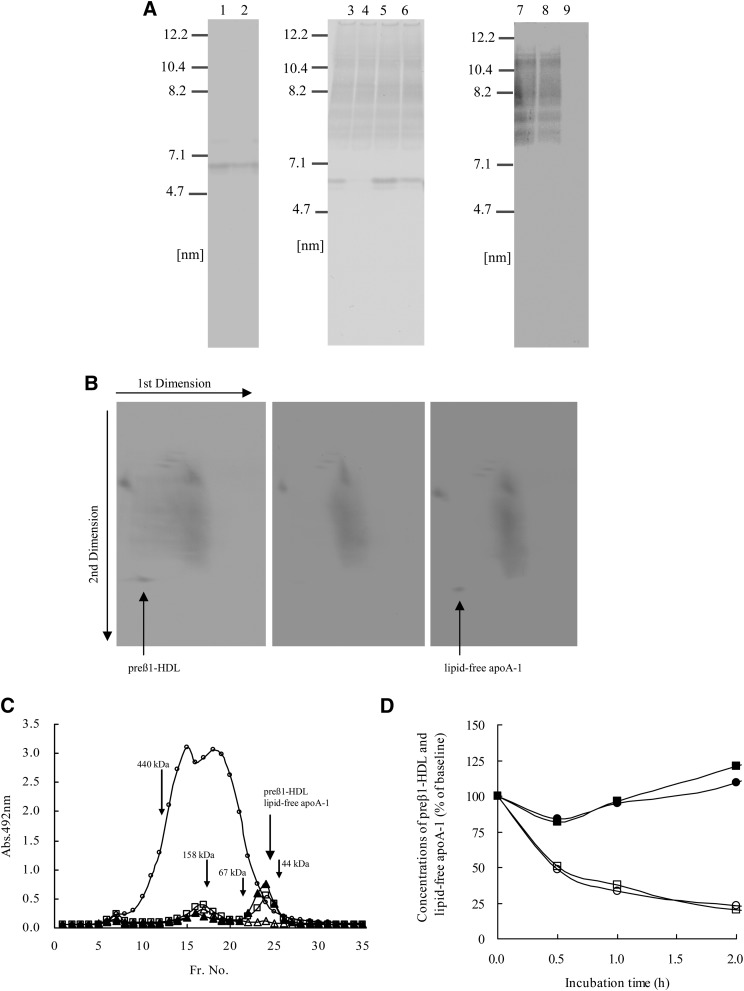
In order to compare preβ1-HDL with lipid-free apoA-1, nondenaturing PAGE, nondenaturing 2D-electrophoresis, size-exclusion chromatography, and an LCAT-dependent conversion experiment were carried out. In nondenaturing PAGE, the particle sizes (diameter) of purified preβ1-HDL, lipid-free apoA-1, intact plasma preβ1-HDL, and lipid-free apoA-1 added to preβ1-HDL-depleted plasma showed no significant variation, all being 6.0 ± 0.1 nm (mean ± SD, n = 3) (Fig. 3A). A 6.0 nm band was not detected in the fractions separated by the affinity chromatography using HDL prepared by ultracentrifugation (Fig. 3A, lanes 8 and 9). In nondenaturing 2D electrophoresis, intact plasma preβ1-HDL was detected at the same position as lipid-free apoA-1 added to preβ1-HDL-depleted plasma (Fig. 3B). In size-exclusion chromatography, intact plasma preβ1-HDL and lipid-free apoA-1 were both eluted in the same number fraction (Fr. 24) (Fig. 3C). In the LCAT-dependent conversion experiment, the concentration of lipid-free apoA-1, added to preβ1-HDL-depleted plasma, decreased in a time-dependent manner when incubated at 37°C, while this decrease was not observed in the presence of DTNB. Moreover, the shift in the lipid-free apoA-1 concentration approximately corresponded to that in the preβ1-HDL concentration when the plasma was incubated (Fig. 3D).
Fig. 3.
Comparison of preβ1-HDL with lipid-free apoA-1. A: Nondenaturing PAGE: lane 1, purified preβ1-HDL; lane 2, lipid-free apoA-1; lane 3, plasma; lane 4, preβ1-HDL-depleted plasma; lane 5, preβ1-HDL-depleted plasma + purified preβ1-HDL; lane 6, preβ1-HDL-depleted plasma + lipid-free apoA-1; lane 7, HDL prepared by ultracentrifugation; lane 8, unbound fraction of affinity chromatography using HDL; lane 9, fraction eluted with glycine-HCl buffer (pH 2.0) in affinity chromatography using HDL. B: Nondenaturing 2D electrophoresis: plasma (left panel), preβ1-HDL-depleted plasma (middle panel), and preβ1-HDL-depleted plasma + lipid-free apoA-1 (right panel). C: Size-exclusion chromatography. A portion of each fraction was diluted 625-fold and 11-fold with each dilution buffer in order to measure apoA-1 and preβ1-HDL concentrations, respectively. The y-axis represents apoA-1 or preβ1-HDL absorbance levels at 492 nm measured by apoA-1-ELISA using goat anti-apoA-1 PAb or preβ1-HDL-ELISA. Open circles represent apoA-1 levels of each fraction (Fr.) separated from plasma. Open squares, open triangles, and closed triangles represent preβ1-HDL levels of each fraction separated from plasma, preβ1-HDL-depleted plasma, and preβ1-HDL-depleted plasma + lipid-free apoA-1, respectively. Molecular markers: ferritin (440 kDa), aldorase (158 kDa), albumin (67 kDa), and ovalbumin (44 kDa) were eluted in fraction numbers 12, 17, 22, and 25, respectively. D: Variations in preβ1-HDL and lipid-free apoA-1 levels in plasma during incubation at 37°C. ○, plasma (plasma 0.4 ml, phosphate buffer 0.05 ml, PBS 0.05 ml); ●, plasma + DTNB (plasma 0.4 ml, DTNB solution 0.05 ml, PBS 0.05 ml); □, preβ1-HDL-depleted plasma + lipid-free apoA-1 [preβ1-HDL-depleted plasma, 0.4 ml; phosphate buffer, 0.05 ml; lipid-free apoA-1 (0.1 mg/ml), 0.05 ml]; ■, preβ1-HDL-depleted plasma + lipid-free apoA-1 + DTNB [preβ1-HDL-depleted plasma, 0.4 ml; DTNB solution, 0.05 ml; lipid-free apoA-1 (0.1 mg/ml), 0.05 ml]. Values are expressed as percent of the baseline value (before incubation). Each point represents the average of duplicate measurements.
Specificity of anti-apoA-1 MAbs
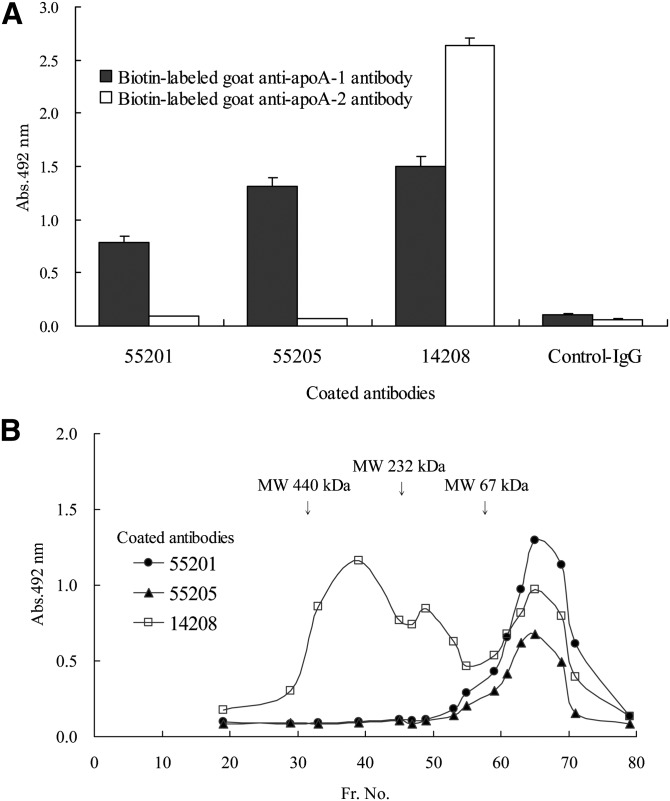
In Western blotting analysis, MAb55205 and MAb14208 specifically reacted with apoA-1 having a molecular mass of 28 kDa (data not shown). In sandwich ELISA using biotin-labeled goat anti-apoA-1 or anti-apoA-2 PAb, MAb55205, like MAb55201, showed reactivity in ELISA using anti-apoA-1 PAb but not anti-apoA-2 PAb; whereas MAb14208 showed reactivity in ELISA using either anti-apoA-1 PAb or anti-apoA-2 PAb (Fig. 4A). With regard to the plasma gel filtration fractions, MAb55205, like MAb55201, showed reactivity only to those fractions with a molecular mass of less than 67 kDa containing preβ1-HDL; whereas MAb14208 showed reactivity not only for that fraction but also for other fractions containing HDL of higher molecular mass (Fig. 4B). These findings indicate that MAb55205, like MAb55201, specifically recognizes apoA-1 associated with preβ1-HDL; whereas MAb14208 reacts with apoA-1 associated with all plasma HDL fractions.
Fig. 4.
Specificity of anti-apoA-1 MAbs. A: Reactivity with LpA-1 and LpA-1:A-2. Error bars represent the SD of three measurements. B: Reactivity with plasma lipoprotein fractions (Fr.) separated by gel filtration chromatography. Each point represents the average of duplicate measurements.
Epitope of anti-apoA-1 MAbs
In peptide-competitive ELISA, MAb14208 reacted with only a peptide of aa97-116 in 25 peptides of apoA-1, whereas MAb55201 and MAb55205 did not react with any peptides (Table 2). In Western blot analysis using apoA-1 CNBr fragments, MAb55201 reacted slightly with F4, but did not react with F2-4 and F3-4. MAb55205 reacted weakly with F2-4 and clearly with F1-3 and F3-4, but did not react with F3 (Fig. 5A). In antibody-competitive ELISA, MAb55205 did not inhibit the reactivity of biotin-labeled MAb55201 for apoA-1, while unlabeled MAb55201 did (Fig. 5B). These findings indicate that the epitope of MAb14208 is included in aa97-116 and that MAb55205 and MAb55201 recognize different epitopes on apoA-1, though each epitope was not identified.
TABLE 2.
Reactivity of each MAb to apoA-1 peptides defined by competitive ELISA
| Peptide | Sequence | IC50 (μmol/l) | |||||||||||||||||||||
| 55201 | 55205 | 14208 | |||||||||||||||||||||
| aa1-20 | D | E | P | P | Q | S | P | W | D | R | V | K | D | L | A | T | V | Y | V | D | >100 | >100 | >100 |
| aa13-32 | D | L | A | T | V | Y | V | D | V | L | K | D | S | G | R | D | Y | V | S | Q | >100 | >100 | >100 |
| aa25-44 | S | G | R | D | Y | V | S | Q | F | E | G | S | A | L | G | K | Q | L | N | L | >100 | >100 | >100 |
| aa37-56 | A | L | G | K | Q | L | N | L | K | L | L | D | N | W | D | S | V | T | S | T | >100 | >100 | >100 |
| aa49-68 | N | W | D | S | V | T | S | T | F | S | K | L | R | E | Q | L | G | P | V | T | >100 | >100 | >100 |
| aa61-80 | R | E | Q | L | G | P | V | T | Q | E | F | W | D | N | L | E | K | E | T | E | >100 | >100 | >100 |
| aa73-92 | D | N | L | E | K | E | T | E | G | L | R | Q | E | M | S | K | D | L | E | E | >100 | >100 | >100 |
| aa85-104 | E | M | S | K | D | L | E | E | V | K | A | K | V | Q | P | Y | I | D | D | F | >100 | >100 | >100 |
| aa97-116 | V | Q | P | Y | I | D | D | F | Q | K | K | W | Q | E | E | M | E | L | Y | R | >100 | >100 | 7.9 |
| aa109-128 | Q | E | E | M | E | L | Y | R | Q | K | V | E | P | L | R | A | E | L | Q | E | >100 | >100 | >100 |
| aa121-140 | P | L | R | A | E | L | Q | E | G | A | R | Q | K | L | H | E | L | Q | E | K | >100 | >100 | >100 |
| aa129-143 | G | A | R | Q | K | L | H | E | L | Q | E | K | I | S | P | >100 | >100 | >100 | |||||
| aa134-153 | L | H | E | L | Q | E | K | I | S | P | L | G | E | E | M | R | D | R | A | R | >100 | >100 | >100 |
| aa144-158 | L | G | E | E | M | R | D | R | A | R | A | H | V | D | A | >100 | >100 | >100 | |||||
| aa149-168 | R | D | R | A | R | A | H | V | D | A | L | R | T | H | L | A | P | Y | S | D | >100 | >100 | >100 |
| aa159-173 | L | R | T | H | L | A | P | Y | S | D | E | L | R | Q | R | >100 | >100 | >100 | |||||
| aa164-183 | A | P | Y | S | D | E | L | R | Q | R | L | A | A | R | L | E | A | L | K | E | >100 | >100 | >100 |
| aa174-188 | L | A | A | R | L | E | A | L | K | E | N | G | G | A | R | >100 | >100 | >100 | |||||
| aa179-198 | E | A | L | K | E | N | G | G | A | R | L | A | E | Y | H | A | K | A | T | E | >100 | >100 | >100 |
| aa189-203 | L | A | E | Y | H | A | K | A | T | E | H | L | S | T | L | >100 | >100 | >100 | |||||
| aa194-213 | A | K | A | T | E | H | L | S | T | L | S | E | K | A | K | P | A | L | E | D | >100 | >100 | >100 |
| aa204-218 | S | E | K | A | K | P | A | L | E | D | L | R | Q | G | L | >100 | >100 | >100 | |||||
| aa209-228 | P | A | L | E | D | L | R | Q | G | L | L | P | V | L | E | S | F | K | V | S | >100 | >100 | >100 |
| aa219-235 | L | P | V | L | E | S | F | K | V | S | F | L | S | A | L | E | E | >100 | >100 | >100 | |||
| aa224-243 | S | F | K | V | S | F | L | S | A | L | E | E | Y | T | K | K | L | N | T | Q | >100 | >100 | >100 |
IC50, Concentration of competitive peptide required to cause 50% inhibition; >100, inhibition was not detected even at peptide concentrations of 100 μmol/l.
Fig. 5.
Epitope of anti-apoA-1 MAbs. A: Western blot analysis for reactivity with apoA-1 CNBr-fragments. B: Antibody-competitive ELISA using biotin-labeled MAb55201. Values are expressed as percent of the control value (without competitive antibody). Each point represents the average of duplicate measurements.
Sandwich ELISA using anti-apoA-1 MAbs
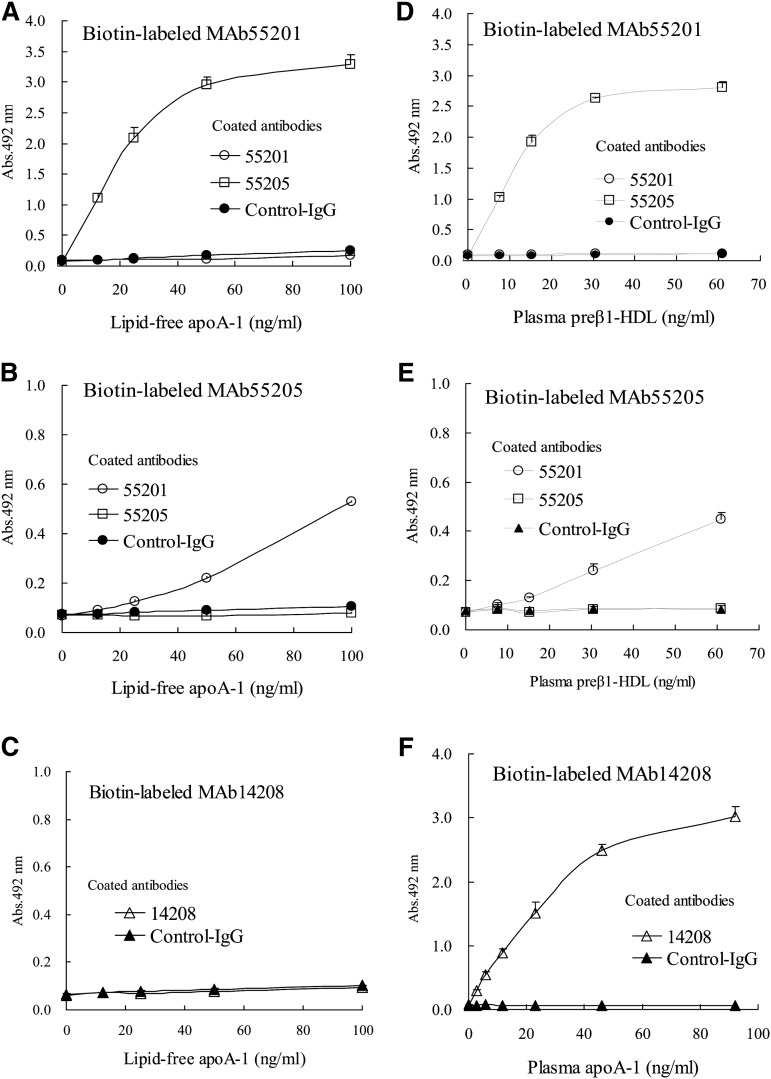
Reactivity was not observed for either lipid-free apoA-1 or plasma preβ1-HDL in sandwich ELISA using only MAb55201 or only MAb55205 as the immobilized and labeled antibody, although it was observed for both lipid-free apoA-1 and plasma preβ1-HDL in sandwich ELISA using a combination of MAb55201 and MAb55205 (Fig. 6A, B, D, E). On the other hand, in sandwich ELISA using MAb14208 as both the immobilized and labeled antibody, reactivity was not observed for lipid-free apoA-1, but was seen for plasma HDL (Fig. 6C, F). We performed this experiment two more times using plasma obtained from two other healthy volunteers and obtained the same results (data not shown). These results suggest that there is only one epitope recognized by each MAb in a particle of plasma preβ1-HDL, i.e., plasma preβ1-HDL is composed of one molecule of apoA-1, as is lipid-free apoA-1.
Fig. 6.
Sandwich ELISA using anti-apoA-1 MAbs. A–C: Reactivity with lipid-free apoA-1 in each sandwich ELISA using biotin-labeled MAb55201 (A), biotin-labeled MAb55205 (B), and biotin-labeled MAb14208 (C). D, E: Reactivity with plasma preβ1-HDL in each sandwich ELISA using biotin-labeled MAb55201 (D) and biotin-labeled MAb55205 (E). Plasma with a preβ1-HDL concentration of 26.9 μg/ml was used after diluting 441- to 3,580-fold. The horizontal axis indicates the plasma preβ1-HDL concentration. F: Reactivity with plasma HDL in sandwich ELISA using biotin-labeled MAb14208. Plasma with an apoA-1 concentration of 1.3 mg/ml was used after diluting 14,112- to 451,584-fold. The horizontal axis indicates the plasma apoA-1 concentration. Values are presented as the mean value ± SD of triplicate measurements.
DISCUSSION
In the present study, we investigated the composition and structure of plasma preβ1-HDL. With respect to the composition of preβ1-HDL, several different results have been reported (8, 18, 19, 31). Castro and Fielding (8) analyzed a small amount of preβ1-HDL extracted from polyacrylamide gel after separating plasma by nondenaturing 2D electrophoresis, and reported that preβ1-HDL was composed of apoA-1, phospholipids, and unesterified cholesterol. Their report is the basis of the view that preβ1-HDL is composed of apoA-1, phospholipids, and unesterified cholesterol. Nanjee and Brinton (31) separated a very small apoA-1-containing particle having a size of 5.8–6.3 nm (diameter) from plasma by using two kinds of analytical columns for size-exclusion chromatography. The particle is speculated to be the same as preβ1-HDL because its content in plasma is 2–15% of total apoA-1, its particle size is the smallest among plasma apoA-1-containing particles, it has preβ mobility in agarose gel electrophoresis, and its concentration markedly falls when plasma is incubated at 37°C for 2 h. It has been shown that the particle is composed only of apoA-1 and phospholipids in a molar ratio of one to two. In the present study, we tried to obtain a large amount of pure preβ1-HDL, sufficient to analyze its composition by affinity chromatography using a MAb55201-bound column and subsequent preparative gel filtration chromatography using 40 ml of fresh normal plasma. As a result, we succeeded in purifying 139–169 μg of preβ1-HDL from each plasma sample. We first investigated the protein composition of the purified preβ1-HDL, and found that only apoA-1 was detected. Wolfrum, Poy, and Stoffel (33) have reported that apoM is required for preβ-HDL formation and is a component of preβ-HDL. However, apoM was not detected in the purified preβ1-HDL in the present study (supplementary Table I). The apoM-containing preβ-HDL previously reported might have included the larger preβ-HDLs, preβ2-HDL, or preβ3-HDL, which could account for the different results in the two studies. In line with our results, low or undetectable levels of apoM in preβ-HDL have been reported in other studies (34, 35). We next investigated the lipid composition of the purified preβ1-HDL, and found that no lipid was detected (Table 1). We determined the LOD of each lipid assay to examine the possibility that sensitivity may have been insufficient. The results showed that the LOD values are, relative to the amount of protein, 0.40, 0.31, and 1.5% for phospholipids, cholesterol, and triglyceride, respectively. In molar ratios, these are 0.11 mol of phospholipids, 0.23 mol of cholesterol, and 0.48 mol of triglyceride to 1 mol of apoA-1. In other words, even if lipid is contained in the purified preβ1-HDL, the amount is less than one molecule to one molecule of apoA-1. The enzymatic method we used to measure phospholipids cannot detect choline-free phospholipids such as phosphatidyl ethanolamine, although it can detect choline-containing phospholipids including phosphatidyl choline, sphingomyelin, and lysophosphatidyl choline, comprising about 95% of plasma phospholipids (36). Therefore, although we cannot assert that no phospholipids are present at all in the purified preβ1-HDL, we can affirm that at least phosphatidyl choline and sphingomyelin, previously reported (1) to be constituents of preβ1-HDL, are not present. We compared the purified preβ1-HDL with intact plasma preβ1-HDL by nondenaturing PAGE, to examine the possibility that lipids were dissociated from apoA-1 in preβ1-HDL during the purification. Our results showed no difference in mobility in nondenaturing PAGE between the purified preβ1-HDL and intact plasma preβ1-HDL (Fig. 3A). It has been reported that the particle size in nondenaturing PAGE remarkably increases when a small amount of lipid binds to lipid-free apoA-1 (37, 38). Conversely, the possibility that lipids were dissociated from apoA-1 in preβ1-HDL during the purification process is extremely low because the particle size of preβ1-HDL remained unchanged. We also performed the affinity chromatography under the same conditions using HDL prepared by ultracentrifugation, which does not contain lipid-free apoA-1 and/or preβ1-HDL, and confirmed that neither lipid-free apoA-1 nor preβ1-HDL were stripped from HDL by the action of MAb55201 bound to Sepharose beads during the chromatography process (Fig. 3A). These findings suggest that plasma preβ1-HDL does not contain lipids at all or, if it does, the amount is extremely small.
With respect to the structure, preβ1-HDL is postulated to be discoidal in many articles (2, 3, 39, 40). However there is no report to directly prove that plasma preβ1-HDL has a discoidal shape (6). Discoidal HDL, having a preβ electrophoretic mobility, is known to be synthesized in vitro as a model particle of plasma preβ-HDL. The particle forms a structure which is called the double belt model that characteristically contains two molecules of apoA-1 per particle (39, 41). In the present study, we did not find any discoidal particle, but found only globular particles in the purified preβ1-HDL through electron microscopy (Fig. 1B). In addition, we found that apoA-1 in preβ1-HDL is one molecule per particle through an immunochemical procedure using anti-preβ1-HDL MAbs (Fig. 6). The view that preβ1-HDL has a discoidal shape may be based on the fact that preβ1-HDL exists in lymph at a relatively high concentration (42) and some HDL in lymph has a discoidal shape (43). However the density of discoidal HDL in lymph is reported to be 1.063–1.21 g/ml (43), whereas the density of plasma preβ1-HDL is reported to be higher than 1.21 g/ml (11). This fact suggests that the discoidal HDL in lymph is different from plasma preβ1-HDL. Regarding the number of apoA-1 molecules per particle of preβ1-HDL, there are two hypotheses, one suggesting that there are two molecules and the other proposing one molecule (7). The hypothesis of two molecules is speculation based on the molecular mass determined by nondenaturing PAGE and size-exclusion chromatography (5, 31). In the present study, the molecular masses of plasma preβ1-HDL determined by nondenaturing PAGE and size-exclusion chromatography are 60 kDa (Fig. 3A) and 50 kDa (Fig. 3C), respectively. It is well known that molecular mass determined by nondenaturing PAGE and size-exclusion chromatography is affected by the electrical charge and shape of the particle. Formisano, Brewer, and Osborne (27) reported the molecular mass of the apoA-1 monomer, determined to be 28 kDa by sedimentation equilibrium, to be 50 kDa in size-exclusion chromatography. Because the frictional ratio of apoA-1 has been reported to be 1.4 (44), it may be difficult to determine the exact molecular mass of the apoA-1 monomer or dimer by size exclusion chromatography or nondenaturing PAGE. On the other hand, the hypothesis of one molecule is based on the results of a cross-linking study (37, 45, 46). Nakamura et al. (45) reported that only the apoA-1 monomer was detected when plasma preβ1-HDL separated by nondenaturing PAGE was cross-linked in polyacrylamide gel, followed by SDS-PAGE. Our result, based on immunochemical analysis, is consistent with their report (Fig. 6).
We compared intact plasma preβ1-HDL with monomolecular lipid-free apoA-1 obtained by delipidating plasma HDL and subsequent purification, due to the fact that plasma preβ1-HDL appeared to be monomolecular lipid-free apoA-1. We demonstrated that no difference is to be found in their mobility in nondenaturing PAGE and nondenaturing 2D electrophoresis (Fig. 3A, B) and in their elution volume in size-exclusion chromatography (Fig. 3C). We also compared lipid-free apoA-1 with plasma preβ1-HDL in a LCAT-dependent conversion experiment. It is known that when plasma is incubated at 37°C, the concentration of plasma preβ1-HDL decreases and this decrease is blocked by the presence of DTNB (10, 21, 45, 47). Although the mechanism has not yet been clarified, the phenomenon seems to be a LCAT-dependent effect because it has been reported that the same effect is shown when anti-LCAT antibody is used instead of DTNB (47), and that plasma LCAT activity is positively correlated with the decreased level of plasma preβ1-HDL in the experiment (25). In the present study, lipid-free apoA-1 added to preβ1-HDL-depleted plasma showed the same variation in concentration as plasma preβ1-HDL in a LCAT-dependent manner (Fig. 3-D). These results suggest that plasma preβ1-HDL is monomolecular lipid-free apoA-1.
Plasma preβ1-HDL is known to play an important role in cholesterol efflux, but it is still controversial whether preβ1-HDL is the substrate or the first product of the ABCA1-mediated cholesterol efflux (1, 2, 5–7). Castro and Fielding (8) reported that cell-derived cholesterol is first detected at the position of preβ1-HDL in nondenaturing 2D electrophoresis when fibroblasts loaded with 3H-labeled cholesterol are incubated with plasma. The result seems to demonstrate that preβ1-HDL is the first product of cholesterol efflux by apoA-1. In recent years it has been reported that “preβ1-HDL-like” lipid-poor apoA-1 or nascent HDL is formed when lipid-free apoA-1 retrieves phospholipids and cholesterol from cells mediated by ABCA1 in experiments using various culture cells (38, 46, 48, 49). However, the particle sizes of those preβ1-HDL-like particles determined by nondenaturing PAGE are all larger than albumin with a diameter of 7.1 nm, and are clearly different from plasma preβ1-HDL, which is a particle smaller than albumin. On the other hand, it is known that the cholesterol efflux from fibroblasts to plasma markedly declines when preβ1-HDL is depleted from plasma (10), or when antibodies specifically recognizing preβ1-HDL are added to plasma (9). The results seem to demonstrate preβ1-HDL itself retrieves cholesterol, i.e., preβ1-HDL is the substrate of the cholesterol efflux. It is also reported that preβ1-HDL-specific cholesterol efflux remarkably declines when fibroblasts are pretreated with protease, while it is not influenced by protease when blood cells not expressing ABCA1 are used instead of fibroblasts (10). The result implies that preβ1-HDL retrieves cellular cholesterol mediated by some protein on cell membranes, probably ABCA1. Moreover, it is known that preβ1-HDL exists in the plasma of patients with Tangier disease in which ABCA1-mediated cholesterol efflux is defective (12, 13). This fact strongly suggests that preβ1-HDL is the substrate, not the product, of the ABCA1-mediated cholesterol efflux. Our present data, suggesting plasma preβ1-HDL is lipid-free apoA-1, seems to make the view clearer.
With reference to the source of the lipid-free apoA-1 in blood, three mechanisms are considered. One is direct secretion from the liver and intestine. ApoA-1, produced from liver and intestine cells, accepts phospholipids and cholesterol from cells in these organs through ABCA1 on the plasma membrane to form nascent HDL, and the nascent HDL is then released to the circulating blood (2, 7, 38). However a portion of the apoA-1 may be directly released into the circulating blood without lipidation mediated by ABCA1. The other mechanisms are dissociation from mature spherical HDL and dissociation from triglyceride-rich lipoproteins. It is known that a preβ1-HDL-like apoA-1-containing small particle dissociates from mature spherical HDL during HDL remodeling by phospholipid transfer protein, cholesteryl ester transfer protein, hepatic triglyceride lipase, secretory phospholipase A2, and serum amyloid A (3, 5). It is also reported that preβ1-HDL dissociates from triglyceride-rich lipoproteins, including VLDL, during hydrolysis by lipoprotein lipase (50). It may prove useful in the future to investigate which of these is the main mechanism by which lipid-free apoA-1 in the blood is generated and how the amount of lipid-free apoA-1 generated by each mechanism changes in a variety of diseases and conditions.
Lipid-free apoA-1 is stated to be the substrate of the ABCA1-mediated cholesterol efflux in many review articles (2–4, 7, 51). However, clear evidence that lipid-free apoA-1 exists in plasma has not been reported (5). In the present study, we analyzed preβ1-HDL purified from plasma and intact plasma preβ1-HDL, and defined plasma preβ1-HDL as monomolecular lipid-free apoA-1. Our findings indicate that lipid-free apoA-1 circulates in blood where it serves to promote reverse cholesterol transport by mediating cholesterol efflux from cells.
Supplementary Material
Footnotes
Abbreviations:
- CNBr
- cyanogen bromide
- 2D electrophoresis
- two-dimensional gel electrophoresis
- LOD
- limit of detection
- LOQ
- limit of quantification
- MAb
- monoclonal antibody
- PAb
- polyclonal antibody
- PVDF
- poly vinylidene difluoride
- TFA
- trifluoroacetic acid
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table.
REFERENCES
- 1.Fielding C. J., Fielding P. E. 1995. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 36: 211–228 [PubMed] [Google Scholar]
- 2.Sviridov D., Nestel P. 2002. Dynamics of reverse cholesterol transport: protection against atherosclerosis. Atherosclerosis. 161: 245–254 [DOI] [PubMed] [Google Scholar]
- 3.von Eckardstein A., Nofer J. R., Assmann G. 2001. High density lipoproteins and arteriosclerosis. Role of cholesterol efflux and reverse cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 21: 13–27 [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama S. 2006. Assembly of high-density lipoprotein. Arterioscler. Thromb. Vasc. Biol. 26: 20–27 [DOI] [PubMed] [Google Scholar]
- 5.Rye K. A., Barter P. J. 2004. Formation and metabolism of prebeta-migrating, lipid-poor apolipoprotein A-I. Arterioscler. Thromb. Vasc. Biol. 24: 421–428 [DOI] [PubMed] [Google Scholar]
- 6.Barrans A., Jaspard B., Barbaras R., Chap H., Perret B., Collet X. 1996. Pre-beta HDL: structure and metabolism. Biochim. Biophys. Acta. 1300: 73–85 [DOI] [PubMed] [Google Scholar]
- 7.Wróblewska M. 2011. The origin and metabolism of a nascent pre-β high density lipoprotein involved in cellular cholesterol efflux. Acta Biochim. Pol. 58: 275–285 [PubMed] [Google Scholar]
- 8.Castro G. R., Fielding C. J. 1988. Early incorporation of cell-derived cholesterol into pre-beta-migrating high density lipoprotein. Biochemistry. 27: 25–29 [DOI] [PubMed] [Google Scholar]
- 9.Fielding P. E., Kawano M., Catapano A. L., Zoppo A., Marcovina S., Fielding C. J. 1994. Unique epitope of apolipoprotein A-I expressed in pre-beta-1 high-density lipoprotein and its role in the catalyzed efflux of cellular cholesterol. Biochemistry. 33: 6981–6985 [DOI] [PubMed] [Google Scholar]
- 10.Kawano M., Miida T., Fielding C. J., Fielding P. E. 1993. Quantitation of pre beta-HDL-dependent and nonspecific components of the total efflux of cellular cholesterol and phospholipid. Biochemistry. 32: 5025–5028 [DOI] [PubMed] [Google Scholar]
- 11.Asztalos B. F., Sloop C. H., Wong L., Roheim P. S. 1993. Two-dimensional electrophoresis of plasma lipoproteins: recognition of new apo A-I-containing subpopulations. Biochim. Biophys. Acta. 1169: 291–300 [DOI] [PubMed] [Google Scholar]
- 12.Huang Y., von Eckardstein A., Shili W., Langer C., Assmann G. 1995. Generation of pre-beta 1-HDL and conversion into alpha-HDL. Evidence for disturbed HDL conversion in Tangier disease. Arterioscler. Thromb. Vasc. Biol. 15: 1746–1754 [DOI] [PubMed] [Google Scholar]
- 13.Asztalos B. F., Brousseau M. E., McNamara J. R., Horvath K. V., Roheim P. S., Schaefer E. J. 2001. Subpopulations of high density lipoproteins in homozygous and heterozygous Tangier disease. Atherosclerosis. 156: 217–225 [DOI] [PubMed] [Google Scholar]
- 14.Brooks-Wilson A., Marcil M., Clee S. M., Zhang L. H., Roomp K., van Dam M., Yu L., Brewer C., Collins J. A., Molhuizen H. O., et al. 1999. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 22: 336–345 [DOI] [PubMed] [Google Scholar]
- 15.Bodzioch M., Orsó E., Klucken J., Langmann T., Böttcher A., Diederich W., Drobnik W., Barlage S., Büchler C., Porsch-Ozcürümez M., et al. 1999. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22: 347–351 [DOI] [PubMed] [Google Scholar]
- 16.Rust S., Rosier M., Funke H., Real J., Amoura Z., Piette J. C., Deleuze J. F., Brewer H. B., Duverger N., Denèfle P., et al. 1999. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 22: 352–355 [DOI] [PubMed] [Google Scholar]
- 17.Francis G. A., Knopp R. H., Oram J. F. 1995. Defective removal of cellular cholesterol and phospholipids by apolipoprotein A-I in Tangier disease. J. Clin. Invest. 96: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guendouzi K., Jaspard B., Barbaras R., Motta C., Vieu C., Marcel Y., Chap H., Perret B., Collet X. 1999. Biochemical and physical properties of remnant-HDL2 and of pre beta 1-HDL produced by hepatic lipase. Biochemistry. 38: 2762–2768 [DOI] [PubMed] [Google Scholar]
- 19.Jaspard B., Collet X., Barbaras R., Manent J., Vieu C., Parinaud J., Chap H., Perret B. 1996. Biochemical characterization of pre-beta 1 high-density lipoprotein from human ovarian follicular fluid: evidence for the presence of a lipid core. Biochemistry. 35: 1352–1357 [DOI] [PubMed] [Google Scholar]
- 20.Miida T., Miyazaki O., Nakamura Y., Hirayama S., Hanyu O., Fukamachi I., Okada M. 2003. Analytical performance of a sandwich enzyme immunoassay for pre beta 1-HDL in stabilized plasma. J. Lipid Res. 44: 645–650 [DOI] [PubMed] [Google Scholar]
- 21.Miyazaki O., Kobayashi J., Fukamachi I., Miida T., Bujo H., Saito Y. 2000. A new sandwich enzyme immunoassay for measurement of plasma pre-beta1-HDL levels. J. Lipid Res. 41: 2083–2088 [PubMed] [Google Scholar]
- 22.Sethi A. A., Sampson M., Warnick R., Muniz N., Vaisman B., Nordestgaad B. G., Tybjaerg-Hansen A., Remaley A. T. 2010. High pre-beta1 HDL concentrations and low lecithin:cholesterol acyltransferase activities are strong positive risk markers for ischemic heart disease and independent of HDL-cholesterol. Clin. Chem. 56: 1128–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tashiro J., Miyazaki O., Nakamura Y., Miyazaki A., Fukamachi I., Bujo H., Saito Y. 2009. Plasma pre beta1-HDL level is elevated in unstable angina pectoris. Atherosclerosis. 204: 595–600 [DOI] [PubMed] [Google Scholar]
- 24.Troutt J. S., Alborn W. E., Mosior M. K., Dai J., Murphy A. T., Beyer T. P., Zhang Y., Cao G., Konrad R. J. 2008. An apolipoprotein A-I mimetic dose-dependently increases the formation of prebeta1 HDL in human plasma. J. Lipid Res. 49: 581–587 [DOI] [PubMed] [Google Scholar]
- 25.Kawano M., Nagasaka S., Yagyu H., Ishibashi S. 2008. Pitavastatin decreases plasma prebeta1-HDL concentration and might promote its disappearance rate in hypercholesterolemic patients. J. Atheroscler. Thromb. 15: 41–46 [DOI] [PubMed] [Google Scholar]
- 26.Miida T., Seino U., Miyazaki O., Hanyu O., Hirayama S., Saito T., Ishikawa Y., Akamatsu S., Nakano T., Nakajima K., et al. 2008. Probucol markedly reduces HDL phospholipids and elevated prebeta1-HDL without delayed conversion into alpha-migrating HDL: putative role of angiopoietin-like protein 3 in probucol-induced HDL remodeling. Atherosclerosis. 200: 329–335 [DOI] [PubMed] [Google Scholar]
- 27.Formisano S., Brewer H. B., Jr, Osborne J. C., Jr 1978. Effect of pressure and ionic strength on the self-association of apo-A-I from the human high density lipoprotein complex. J. Biol. Chem. 253: 354–359 [PubMed] [Google Scholar]
- 28.Morrison J., Fidge N. H., Tozuka M. 1991. Determination of the structural domain of apoA1 recognized by high density lipoprotein receptors. J. Biol. Chem. 266: 18780–18785 [PubMed] [Google Scholar]
- 29.Mori T., Kitani Y., Ogihara J., Sugiyama M., Yamamoto G., Kishida O., Nishimura K. 2012. Histological and MS spectrometric analyses of the modified tissue of bulgy form tadpoles induced by salamander predation. Biol. Open. 1: 308–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ICH Harmonised Tripartite Guideline. 2005 Validation of analytical procedures: text and methodology Q2(R1). Proceedings of the International Conference on Hormonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use in London, UK. [Google Scholar]
- 31.Nanjee M. N., Brinton E. A. 2000. Very small apolipoprotein A-I-containing particles from human plasma: isolation and quantification by high-performance size-exclusion chromatography. Clin. Chem. 46: 207–223 [PubMed] [Google Scholar]
- 32.Köhler G., Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 256: 495–497 [DOI] [PubMed] [Google Scholar]
- 33.Wolfrum C., Poy M. N., Stoffel M. 2005. Apolipoprotein M is required for prebeta-HDL formation and cholesterol efflux to HDL and protects against atherosclerosis. Nat. Med. 11: 418–422 [DOI] [PubMed] [Google Scholar]
- 34.Mulya A., Seo J., Brown A. L., Gebre A. K., Boudyguina E., Shelness G. S., Parks J. S. 2010. Apolipoprotein M expression increases the size of nascent pre beta HDL formed by ATP binding cassette transporter A1. J. Lipid Res. 51: 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christoffersen C., Jaujiainen M., Moser M., Porse B., Ehnholm C., Boesl M., Dahlback B., Nielsen L. B. 2008. Effect of apolipoprotein M on high density lipoprotein metabolism and atherosclerosis in low density lipoprotein receptor knock-out mice. J. Biol. Chem. 283: 1839–1847 [DOI] [PubMed] [Google Scholar]
- 36.Takayama M., Itoh S., Nagasaki T., Tanimizu I. 1977. A new enzymatic method for determination of serum choline-containing phospholipids. Clin. Chim. Acta. 79: 93–98 [DOI] [PubMed] [Google Scholar]
- 37.Chau P., Nakamura Y., Fielding C. J., Fielding P. E. 2006. Mechanism of prebeta-HDL formation and activation. Biochemistry. 45: 3981–3987 [DOI] [PubMed] [Google Scholar]
- 38.Ji A., Wroblewski J. M., Cai L., de Beer M. C., Webb N. R., van der Westhuyzen D. R. 2012. Nascent HDL formation in hepatocytes and role of ABCA1, ABCG1, and SR-BI. J. Lipid Res. 53: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Favari E., Calabresi L., Adorni M. P., Jessup W., Simonelli S., Franceschini G., Bernini F. 2009. Small discoidal pre-beta1 HDL particles are efficient acceptors of cell cholesterol via ABCA1 and ABCG1. Biochemistry. 48: 11067–11074 [DOI] [PubMed] [Google Scholar]
- 40.Asztalos B. F., Schaefer E. J., Horvath K. V., Yamashita S., Miller M., Franceschini G., Calabresi L. 2007. Role of LCAT in HDL remodeling: investigation of LCAT deficiency states. J. Lipid Res. 48: 592–599 [DOI] [PubMed] [Google Scholar]
- 41.Davidson W. S., Thompson T. B. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282: 22249–22253 [DOI] [PubMed] [Google Scholar]
- 42.Asztalos B. F., Sloop C. H., Wong L., Roheim P. S. 1993. Comparison of apo A-I-containing subpopulations of dog plasma and prenodal peripheral lymph: evidence for alteration in subpopulations in the interstitial space. Biochim. Biophys. Acta. 1169: 301–304 [DOI] [PubMed] [Google Scholar]
- 43.Sloop C. H., Dory L., Hamilton R., Krause B. R., Roheim P. S. 1983. Characterization of dog peripheral lymph lipoproteins: the presence of a disc-shaped “nascent” high density lipoprotein. J. Lipid Res. 24: 1429–1440 [PubMed] [Google Scholar]
- 44.Gwynne J., Brewer B., Jr, Edelhoch H. 1974. The molecular properties of apoA-1 from human high density lipoprotein. J. Biol. Chem. 249: 2411–2416 [PubMed] [Google Scholar]
- 45.Nakamura Y., Kotite L., Gan Y., Spencer T. A., Fielding C. J., Fielding P. E. 2004. Molecular mechanism of reverse cholesterol transport: Reaction of pre-beta-migrating high-density lipoprotein with plasma lecithin/cholesterol acyltransferase. Biochemistry. 43: 14811–14820 [DOI] [PubMed] [Google Scholar]
- 46.Duong P. T., Weibel G. L., Lund-Katz S., Rothblat G. H., Phillips M. C. 2008. Characterization and properties of pre beta-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 49: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miida T., Kawano M., Fielding C. J., Fielding P. E. 1992. Regulation of the concentration of pre beta high-density lipoprotein in normal plasma by cell membranes and lecithin-cholesterol acyltransferase activity. Biochemistry. 31: 11112–11117 [DOI] [PubMed] [Google Scholar]
- 48.Duong P. T., Collins H. L., Nichel M., Lund-Katz S., Rothblat G. H., Phillips M. C. 2006. Characterization of nascent HDL particles and microparticles formed by ABCA1-mediated efflux of cellular lipids to apoA-1. J. Lipid Res. 47: 832–843 [DOI] [PubMed] [Google Scholar]
- 49.Vedhachalam C., Duong P. T., Nickel M., Nguyen D., Dhanasekaran P., Saito H., Rothblat G. H., Lund-Katz S., Philips M. C. 2007. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-1 and formation of high density lipoprotein particles. J. Biol. Chem. 282: 25123–25130 [DOI] [PubMed] [Google Scholar]
- 50.Miyazaki O., Fukamachi I., Mori A., Hashimoto H., Kawashiri M., Nohara A., Noguchi T., Inazu A., Yamagishi M., Mabuchi H., et al. 2009. Formation of prebeta1-HDL during lipolysis of triglyceride-rich lipoprotein. Biochem. Biophys. Res. Commun. 379: 55–59 [DOI] [PubMed] [Google Scholar]
- 51.Oram J. F., Lawn R. M. 2001. ABCA1. The gatekeeper for eliminating excess tissue cholesterol. J. Lipid Res. 42: 1173–1179 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.