Abstract
Cyclic GMP-AMP synthase (cGAS) is a cytosolic DNA sensor mediating innate antimicrobial immunity. It catalyzes the synthesis of a noncanonical cyclic dinucleotide 2′,5′ cGAMP that binds to STING and mediates the activation of TBK1 and IRF-3. Activated IRF-3 translocates to the nucleus and initiates the transcription of the IFN-β gene. The structure of mouse cGAS bound to an 18 bp dsDNA revealed that cGAS interacts with dsDNA through two binding sites, forming a 2:2 complex. Enzyme assays and IFN-β reporter assays of cGAS mutants demonstrated that interactions at both DNA binding sites are essential for cGAS activation. Mutagenesis and DNA binding studies showed that the two sites bind dsDNA cooperatively and site B plays a critical role in DNA binding. The structure of mouse cGAS bound to dsDNA and 2′,5′ cGAMP provided insight into the catalytic mechanism of cGAS. These results demonstrated that cGAS is activated by dsDNA-induced oligomerization.
INTRODUCTION
Nucleic acids from bacteria and viruses induce potent immune responses in infected mammalian cells (Barber, 2011a; Hemmi et al., 2000; Kato et al., 2011; Keating et al., 2011). Microbial DNA in the cytosol has long been known to induce innate immune responses by stimulating the secretion of type-I interferons (Barber, 2011a; Hornung and Latz, 2010; Ishii et al., 2006; Stetson and Medzhitov, 2006). The endoplasmic reticulum (ER) membrane-localized adaptor protein STING is essential for the responses to cytosolic DNA (Barber, 2011b; Ishikawa and Barber, 2008; Ishikawa et al., 2009; Sun et al., 2009; Zhong et al., 2009). Recent studies have identified the cyclic GMP-AMP (cGAMP) synthase (cGAS) as a cytosolic DNA sensor upstream of STING (Sun et al., 2013). cGAS is activated upon DNA binding and catalyzes the synthesis of a cyclic dinucleotide c[G(2′,5′)pA(3′,5′)p] (referred to as 2′,5′ cGAMP hereafter) from ATP and GTP (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a). This dinucleotide in turn serves as a second messenger to stimulate the induction of IFN-β via STING (Ablasser et al., 2013; Diner et al., 2013; Wu et al., 2013). In addition, STING is also a direct sensor of bacterial cyclic dinucleotides such as c-di-GMP and c-di-AMP, which are structurally similar to 2′,5′ cGAMP (Burdette et al., 2011; Burdette and Vance, 2013; Danilchanka and Mekalanos, 2013). Ligand binding by STING induces the recruitment of the protein kinase TBK1 and transcription factor IRF-3 to the signaling complex (Tanaka and Chen, 2012). Phosphorylation of IRF-3 by TBK1 at the signaling complex on STING promotes the oligomerization of IRF-3 and its translocation into the nucleus, where it activates the transcription of the IFN-β gene together with the transcription factor NF-κB (Fitzgerald et al., 2003; Tanaka and Chen, 2012). The critical role of cGAS in antiviral immunity and cytosolic DNA sensing has been confirmed by recent studies of cGAS-deficient mice (Li et al., 2013).
Although the identity of the cGAS product has been intensively studied (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a) and the structures of mouse and porcine cGAS bound to dsDNA have been determined (Civril et al., 2013; Gao et al., 2013a), the mechanism of cGAS activation by dsDNA is still not fully understood. To address this question, we have determined the crystal structures of human cGAS (hcGAS) in isolation and mouse cGAS (mcGAS) bound to an 18 bp dsDNA. Our structures showed that cGAS forms a 2:2 complex with dsDNA instead of the 1:1 complex as described in previous studies (Civril et al., 2013; Gao et al., 2013a). We have confirmed the self-assembly of cGAS in the presence of dsDNA in solution by analytical ultracentrifugation (AUC) and small angle X-ray scattering (SAXS). Enzyme assays and DNA binding studies of cGAS mutants demonstrated that dsDNA binding-induced dimerization is required for cGAS activation. These results provide insight into the mechanism of cGAS activation by dsDNA, which is central to its function in innate immunity. In addition, our structure of mcGAS bound to dsDNA and 2′,5′ cGAMP provided insight into the catalytic mechanism of the enzyme. Furthermore, we have demonstrated that 2′,5′ cGAMP binds to STING with higher affinity compared to canonical cyclic dinucleotides and potently stimulates the expression of IFN-β in cells.
RESULTS
cGAS is activated by dsDNA and catalyzes the synthesis of a high-affinity ligand for STING
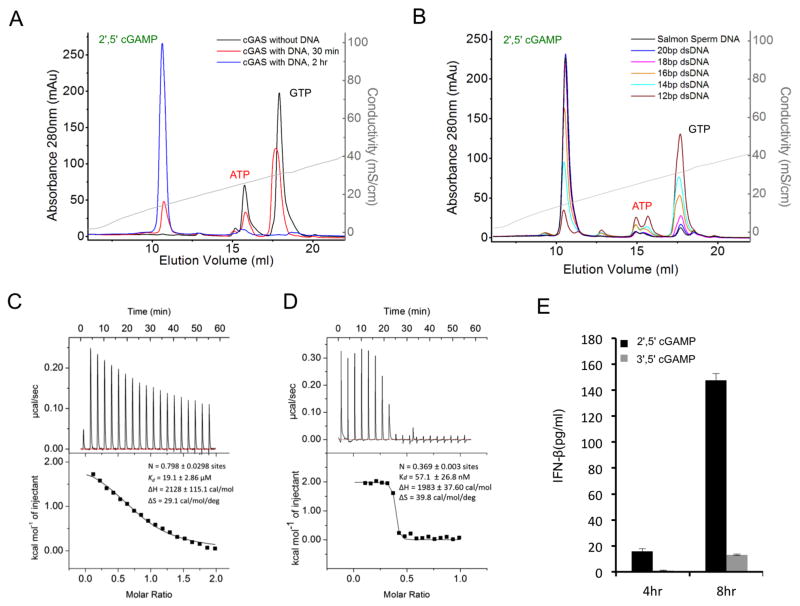
We have expressed and purified both human and mouse cGAS catalytic domains and conducted extensive biochemical characterization of the enzymes. First, we conducted enzymatic activity assays of mcGAS with and without dsDNA. mcGAS catalyzed the synthesis of a new dinucleotide in the presence of ATP, GTP, and dsDNA (Figure 1A). In contrast, no dinucleotide was produced in the absence of dsDNA. Consistent with previous studies, mass spectrometry analysis showed that the mass of the product is ~675 Dalton. We observed that the cGAS product did not elute at the position expected for 3′, 5′ cGAMP on a MonoQ ion exchange column (Figure S1A), indicating that the product of cGAS is not 3′, 5′ cGAMP. Consistent with this observation, others have shown that the product of cGAS is 2′,5′ cGAMP instead of 3′,5′ cGAMP (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a).
Figure 1.
cGAS is activated by dsDNA and catalyzes the synthesis of 2′,5′ cGAMP, a high affinity ligand for STING. (A). cGAS activity assay by ion exchange chromatography. The reaction product was first purified by ultrafiltration and then analyzed using a MonoQ 5/50 GL column. Salmon sperm DNA at 0.2 mg/ml was used to stimulate mcGAS catalytic domain at 10 μM concentration. (B). The catalytic activity of cGAS is dependent on the length of dsDNA. Reaction products from mcGAS stimulated with different dsDNA were analyzed by ion exchange chromatography. The enzyme and DNA concentrations used in these assays are 10 and 50 μM, respectively (C). Binding study of mcGAS catalytic domain with a 20 bp dsDNA by isothermal titration calorimetry (ITC). (D). Binding study of 2′,5′ cGAMP with human STING ligand binding domain by ITC. (E). 2′,5′ cGAMP stimulates the secretion of IFN-β in human THP-1 cells. The concentration of IFN-β in the culture supernatant was analyzed by ELISA at 4 and 8 hours post stimulation. 3′,5′ cGAMP was used as a control. The concentration of 2′,5′ or 3′,5′ cGAMP used in these assays is 25 μg/ml. The error bars represent the standard deviations of signals from four independent measurements. See also Figure S1.
Next, we examined whether cGAS activity is dependent on the length of dsDNA. We found that dsDNA of 12 bp in length did not activate mcGAS efficiently (Figure 1B), whereas dsDNA of 18 bp in length had about 90% activity compared to salmon sperm DNA (Figure 1B), which is used routinely to study dsDNA-mediated immune responses in transfected cells. Longer dsDNA oligonucleotides of 20 bp had activity comparable to salmon sperm DNA (Figure 1B). Isothermal titration calorimetry (ITC) showed that mcGAS catalytic domain binds a 20 bp dsDNA with an affinity of ~20 μM (Figure 1C). Due to the relatively low affinity, we did not observe DNA binding by cGAS catalytic domain by gel filtration chromatography. Furthermore, we used gel filtration chromatography to study the binding of 2′,5′ and 3′,5′ cGAMP to the ligand-binding domain of human STING. We observed almost stoichiometric binding of 2′,5′ cGAMP to STING (Figure S1B), whereas we consistently observed a significant population of free 3′, 5′ cGAMP in the binding studies (Figure S1C). Consistent with these observations, binding studies by ITC showed that STING binds 2′,5′ cGAMP at ~60 nM affinity (Figure 1D), whereas 3′,5′ cGAMP binds STING at ~3 μM affinity (Figure S1D). 2′,5′ cGAMP also induced higher amounts of IFN-β in human THP-1 cells compared to 3′,5′ cGAMP (Figure 1E). These studies demonstrated that cGAS is activated by dsDNA and catalyzes the synthesis of 2′,5′ cGAMP, a high affinity ligand for STING.
cGAS exhibits a similar fold as OAS1 and forms a dimer in the crystal
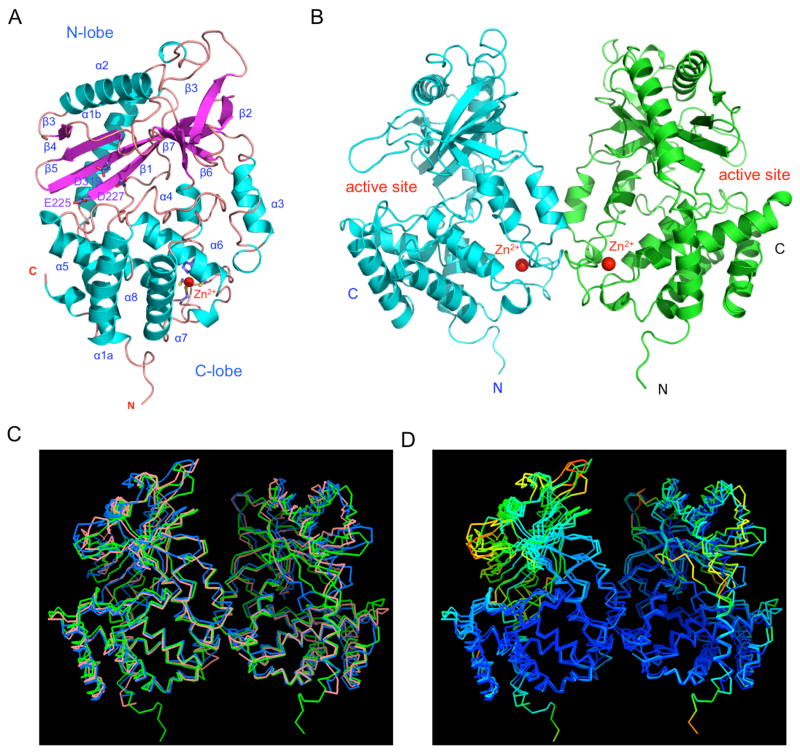
To elucidate the structural basis of cGAS activation by dsDNA, we have crystallized hcGAS catalytic domain in two crystal forms and determined their structures at ~2.0 Å resolution by single-wavelength anomalous dispersion (SAD) using Se-Met derivative crystals. Statistics of the crystallographic analysis are shown in Table S1. The structure of the cGAS catalytic domain (referred to as “cGAS” hereafter) has a bilobed structure (Figure 2A), similar to that of 2′–5′-oligoadenylate synthase (OAS1, r.m.s.d of 3.1 Å) (Hartmann et al., 2003). The overall structures of the four cGAS molecules in the two crystal forms are similar, with r.m.s.d of 0.6 to 1.1 Å in pairwise comparisons.
Figure 2.
Structure of human cGAS catalytic domain. (A). Ribbon representation of the structure of hcGAS catalytic domain. The Zn2+ ion is shown by the red sphere. (B). Structure of the hcGAS dimer in the crystallographic asymmetric unit. (C). Comparison of ligand-free human (green, PDB 4LEV), mouse (blue, PDB 4K8V), and porcine (salmon, PDB 4JLX) cGAS dimer structures. (D). Superposition of ligand-free human, mouse, and porcine cGAS dimers rainbow-colored according to B-factors of the Cα atoms. Atoms in red have higher B-factors and atoms in blue have lower B-factors.
The N-lobe of cGAS has a canonical nucleotidyltransferase (NTase) fold (Figure 2A). The C-lobe contains a tight five-helix bundle (Figure 2A). The long helices (α1a and α1b) at the N-terminus of the cGAS catalytic domain span the two lobes. The active site of cGAS is located on the edge of the deep groove between the N-lobe and C-lobe (Figure 2A and B). Mutations of any of the three conserved catalytic residues Glu225, Asp227, or Asp319 to alanine disrupted the catalytic activity of cGAS (data not shown). Unlike OAS1, cGAS has a unique zinc-binding site in a long loop between helices α4 and α5 (Figure 2A). This loop connects the two lobes and mediates the dimerization of two cGAS molecules in the crystal (Figure 2B). Residues His390, Cys396, Cys397, and Cys404 coordinate Zn2+ with tetrahedral geometry. The identity of this structural zinc was confirmed by inductively coupled plasma mass spectrometry (ICP-MS).
Although hcGAS catalytic domain is monomeric in solution, it forms noncrystallographic symmetry-related dimers in the two forms of crystals (Figure 2B). Similar dimers were also observed in a hexagonal crystal obtained from a high salt (~1.5 M sodium citrate) condition. In addition, the previously described ligand-free mouse and porcine cGAS form similar dimers in the crystals (Figure 2C). The overall structures of DNA-free human, mouse and porcine cGAS dimers are similar, with some structural differences in the NTase domains (Figure 2C). The structures of the NTase domains of hcGAS in both crystal forms are quite flexible and have average B-factors almost twice that of the C-lobes (Figure 2D). Similar to human cGAS, the NTase domains of mouse and porcine cGAS are also quite flexible and have significantly higher B-factors compared to C-lobes (Figure 2D).
The total buried surface area at the hcGAS dimer interface is ~1330 Å2, which is within the range (1600 ± 400 Å2) for typical protein-protein interfaces (Lo Conte et al., 1999). Interactions between the two cGAS molecules are mediated by 15 main-chain and side-chain hydrogen bonds. Residues Asn389 through Glu398 in the loop containing the Zn2+ binding site contribute to dimer formation. In addition, residues Ala346 and Lys347 form two hydrogen bonds with the sidechain of Glu398 via their main chain amine groups. Residues Lys347, Lys394, and Glu398 that mediate dimer formation are conserved in the sequences of hcGAS and mcGAS. These structural studies showed that cGAS catalytic domain exhibits a similar fold as OAS1 and forms a dimer in the crystals.
The structural basis of cGAS activation by dsDNA
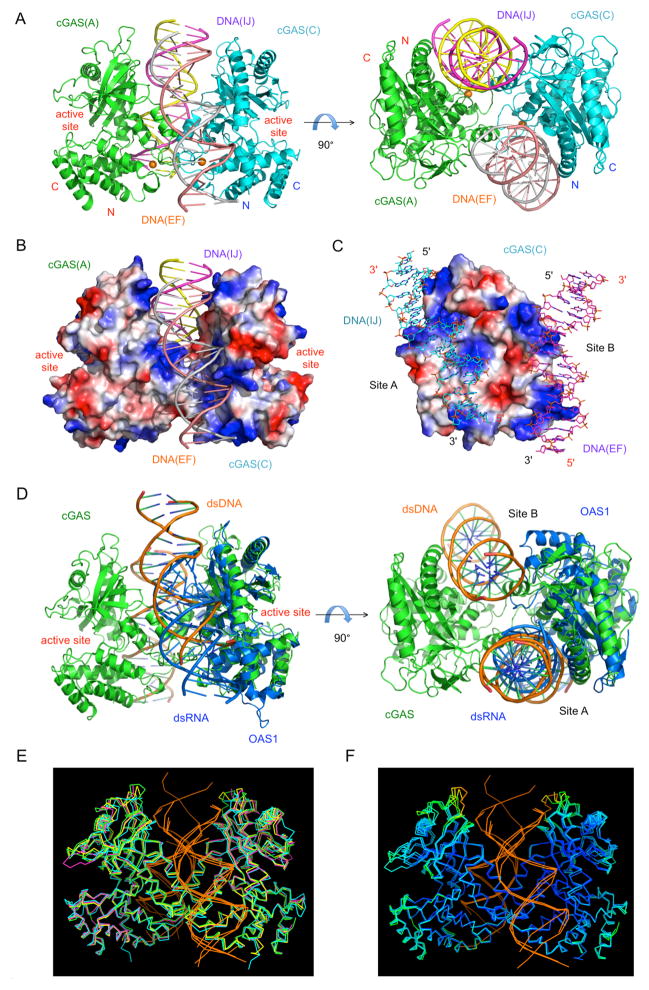
To elucidate the structural basis of cGAS activation by DNA, we have crystallized the catalytic domain of mcGAS bound to an 18 bp palindromic dsDNA and determined the structure of the complex at 2.5 Å resolution (Table S1). Two 2:2 mcGAS:dsDNA complexes with similar structures were observed in the crystallographic asymmetric unit (Figure 3A). DNA binding by cGAS is not sequence dependent. The four mcGAS molecules in the two 2:2 complexes bind to three different regions of the 18 bp dsDNA (Figures S2A to C). Superposition of the four cGAS:dsDNA complexes formed at the primary DNA binding site (site A) shows that each of the cGAS molecules interacts with the dsDNA in a similar manner but contacts different sequences of the 18 bp dsDNA. Two dsDNA molecules crosslink a cGAS dimer, forming a 2:2 complex (Figure 3A). Each cGAS molecule interacts with the two dsDNA molecules through two binding sites (Figures 2A and S2D). The buried surface areas between cGAS(A) and the two dsDNA duplexes are 1380 Å2 (dsDNA EF, site A) and 930 Å2 (dsDNA IJ, site B), respectively. Similarly, the buried surface areas between cGAS(C) and the two dsDNA duplexes are 1540 Å2 (dsDNA IJ, site A) and 790 (dsDNA EF, site B) Å2, respectively. Site B contributes 34~40% of the total buried surface area between cGAS and the dsDNA. The surface electrostatic potential of the cGAS dimer shows complementarity to the two negatively charged dsDNA (Figure 3B and C). Although the cGAS dimers are nearly symmetrical, they bind to the dsDNA in an asymmetrical manner (Figure 3A). Rotation of the complex structures along the two-fold axis of cGAS dimer shows that the two dsDNA fragments bound to cGAS are related to each other by translations of 4 bp (dsDNA EF and IJ) or 5 bp (dsDNA KL and GH) along the helical axis of the DNA. This arrangement of the dsDNA relative to the cGAS dimers avoids clashes between the two dsDNA duplexes within the complex (Figure S2E). This also suggests that the cGAS dimer cannot bind to the middle of two long dsDNA without clashes between the two DNA molecules. Instead, the cGAS dimers likely bind to the terminal regions of two dsDNA pointing in different directions (Figure S2E). The 2:2 mcGAS:dsDNA complex structure differs significantly from the OAS1:dsRNA complex. OAS1 binds to dsRNA in a similar way as cGAS binds to dsDNA at site A (Figure 3D), but only forms a 1:1 complex (Donovan et al., 2013). No higher order structure was observed in the OAS1:dsRNA complex structure (Donovan et al., 2013).
Figure 3.
The structural basis of cGAS activation by dsDNA. (A). Crystal structure of mcGAS catalytic domain bound to an 18 bp dsDNA. Ribbon representations of the 2:2 mcGAS:dsDNA complex structure in two different orientations. (B). Electrostatic surface potential of the mcGAS dimer in the mcGAS:dsDNA complex. Positively charged surface is colored blue, and negatively charged surface red. The two dsDNA bound to the cGAS dimer are shown by ribbon representations. (C). Electrostatic surface potential at the DNA binding sites of a mcGAS monomer. The two dsDNA bound to mcGAS are shown by the cyan and purple stick models. (D). Comparison of the 2:2 mcGAS:dsDNA complex structure with the 1:1 OAS1:dsRNA complex structure (PDB, 4IG8). (E). Superposition of four cGAS:dsDNA complex structures. Structure of mcGAS bound an 18 bp dsDNA (this work, PDB 4LEY) is in green; mcGAS bound to a 16 bp dsDNA (PDB, 4K96) in yellow; mcGAS bound to a 16 bp dsDNA and pppG(2′,5′)pG (PDB, 4K98) in magenta; porcine cGAS bound to a 14 bp dsDNA (PDB, 4KB6) in cyan. The dsDNA bound to cGAS are shown by the orange ribbons. (F). Superposition of three mcGAS:dsDNA complex structures (PDB, 4LEY, 4K96, and 4K98). The proteins are rainbow-colored according to B-factors of the Cα atoms. Atoms in red have higher B-factors and atoms in blue have lower B-factors. See also Figures S2.
More compelling evidence about DNA binding-induced dimerization of cGAS comes from the seven different structures of mouse and porcine cGAS bound to dsDNA published recently (Civril et al., 2013; Gao et al., 2013a). In all these structures, cGAS forms 2:2 complexes with dsDNA that are similar to those observed in our mcGAS:DNA complex structures through either crystallographic (PDB 4K97, 4K98, 4K99, 4K9A, 4K9B, and 4KB6) or non-crystallographic (PDB 4K96) packing contacts (Figure 3E). Together with two of our structures (PDB 4LEY and 4LEZ), in which mcGAS forms 2:2 dimers with DNA through non-crystallographic packing contacts, nine structures in five different space groups showed that cGAS and dsDNA complexes form similar 2:2 dimers in crystals. These structures suggest that it is unlikely that dsDNA-induced cGAS dimerization is an artifact of crystal packing interactions. Unfortunately, the functional significance of DNA-induced dimerization of cGAS as well as DNA binding at site B were not explored in any previous studies (Civril et al., 2013; Gao et al., 2013a), perhaps due to the fact that six out of the seven published 2:2 cGAS:DNA dimer structures are formed by crystallographic symmetry-related molecules.
cGAS interacts with the dsDNA through extensive electrostatic interactions and hydrogen bonds (Figure S2D). At the interface between cGAS(C) and dsDNA (IJ, site A), residues Lys151, Arg158, Arg180, Lys184, Lys372 and Lys395 form salt bridges with the phosphate backbone of the DNA (Figure S2D). In addition, the sidechain of Arg161 reaches into the minor groove of the DNA. Moreover, the sidechains of Ser165, Asn196, and Tyr200 form three hydrogen bonds with the phosphate backbone. Residues Lys160, Ala168 and His203 exhibit van der Waals contacts with the DNA. Most of these interactions are mediated by base pairs 2 to 12 of the dsDNA (Figure S2D). Interactions between cGAS(A) and DNA (IJ, site B) are less extensive as reflected by the smaller buried surface area at this interface (Figure S2D). Residues Arg222 and Lys335 form two salt bridges with the phosphate backbone of the DNA (Figure S2D). In addition, the sidechain of Arg342 reaches into the minor groove of the dsDNA, but does not interact with the phosphate backbone (Figure S2D). Moreover, Thr338 forms a hydrogen bond with the phosphate group of dA9. Interactions between cGAS dimer (AC) and the other dsDNA (EF) are similar to its interactions with dsDNA (IJ). The dimerization of mcGAS is mediated by hydrogen bonds at the dimer interface. Residues Lys382 and Glu386 at the Zinc-binding loop mediate the dimerization of mcGAS.
Comparison of the DNA-bound mcGAS structure and the ligand-free mcGAS structure revealed that DNA binding induced substantial conformational changes in the active site of cGAS (Figure S2F). Residues Glu211, Asp213, and Asp307 in the active site of mcGAS are realigned upon dsDNA binding (Figure S2G). In addition, the loop connecting helix α1b and β1 is restructured upon DNA binding. It has been shown that residue Ser199 in this restructured loop interacts with the γ phosphate group of the substrate ATP or GTP (Gao et al., 2013). These conformational changes in response to DNA binding likely contribute to the activation of cGAS. Furthermore, comparison of the ligand-free and DNA-bound mcGAS dimer structures shows that DNA binding opens instead of closes the cGAS dimer structure (Figure S2H). As indicated by the uniform distribution of B-factors in the NTase domains and C-lobes of three mcGAS:DNA complex structures (Figure 3F), DNA binding dramatically reduced the structural flexibility of the NTase domain. Crosslinking of the cGAS dimer by dsDNA appears necessary for the activation of cGAS by stabilizing the catalytically active conformation of the enzyme. In summary, the structure of mcGAS bound to the 18 bp dsDNA revealed that crosslinking of cGAS dimers by dsDNA molecules mediates the activation of cGAS.
DNA binding by cGAS induces higher-order complex formation in solution
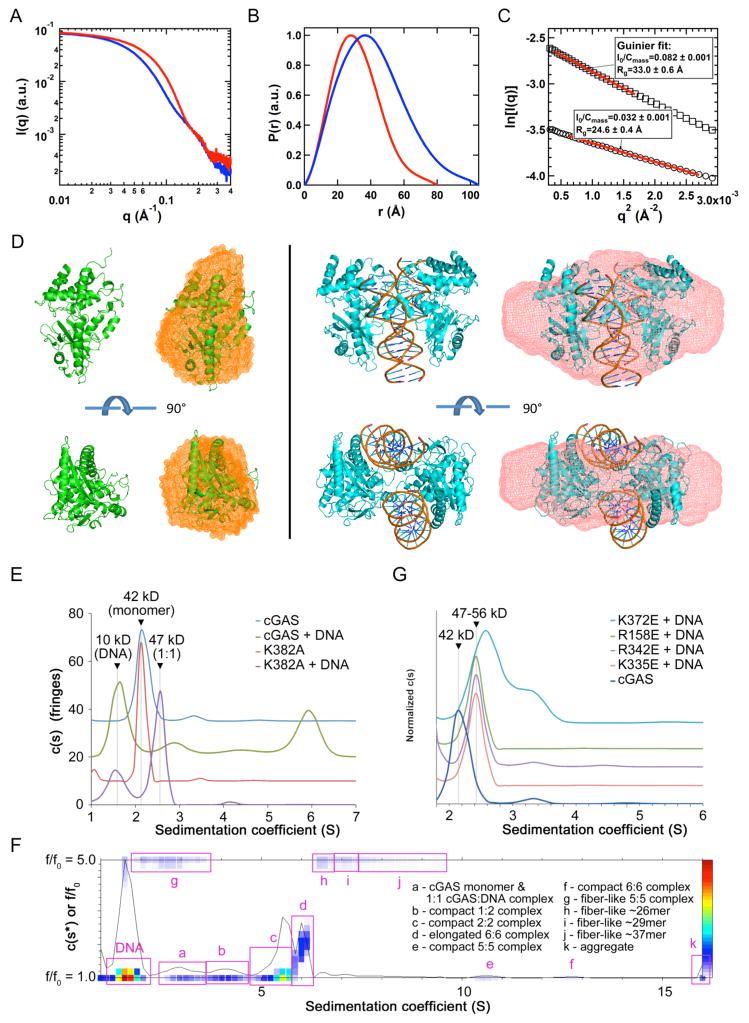
To determine whether cGAS catalytic domain forms the 2:2 complex in solution as was observed in the crystal structures, we conducted small angle X-ray scattering (SAXS) studies of mcGAS in isolation and in complex with a 20 bp dsDNA. The SAXS data showed a significant difference in the small-angle X-ray scattering of mcGAS with and without the 20 bp dsDNA (Figure 4A). In the presence of dsDNA, the pair distance distribution function shifts to longer distances (Figure 4B), indicating the formation of a larger size complex. The radii of gyration (Rg) for cGAS in isolation and the cGAS:DNA complex are 24.6 and 33.0 Å, respectively (Figure 4C). The calculated Rg values from the crystal structures are 22.2 Å for cGAS in isolation, and 30.4 Å for the cGAS:DNA complex (Figure 4C), both of which are close to experimental values but slightly smaller partially due to omitting the possible solvation layers in the calculations. The concentration-normalized X-ray forward scattering, i.e. Io/Cmass, is an indicator of molecular weight, where Io is the forward scattering and Cmass is the sample concentration in mg/ml. The ratio of concentration-normalized forward scattering between samples of cGAS with and without dsDNA is about 2.5 (Figure 4C), which is close to the theoretical value of 2.8 for the 2:2 cGAS:DNA complex vs cGAS in isolation. These results support that mcGAS forms a 2:2 complex with dsDNA in solution. The molecular envelope of cGAS in isolation matches the structure of monomeric cGAS (Figure 4D, left). The molecular envelope for the cGAS:DNA complex matches well with the crystal structure of the 2:2 mcGAS:DNA complex (Figure 4D, right).
Figure 4.
DNA binding by cGAS induces higher-order complex formation in solution (A). Small angle X-ray scattering (SAXS) data of mcGAS catalytic domain (red) and its complex with a 20 bp dsDNA (blue). (B). Normalized pair distance distribution function (PDDF) for mcGAS alone (red) and mcGAS:DNA complex (blue). (C). Guinier fits (red lines) of SAXS data for mcGAS alone (open cycles) and its complex with DNA (open squares). The SAXS data were normalized by the mass concentration of the samples. The fitted q ranges were limited to qmax*Rg < 1.3. Io is the forward scattering, i.e., scattering intensity at q=0. Cmass is the concentration of the samples in mg/ml. (D). Overlays of the crystal structures (cartoon) and SAXS molecular envelope (mesh) for mcGAS in isolation (left) and mcGAS:DNA complex (right). (E). Sedimentation velocity AUC analysis of wt and K382A mutant of mcGAS in the absence and presence of a 20 bp dsDNA, with experimentally determined mass estimates listed. The K382A dimer interface mutant formed a 1:1 complex with dsDNA but did not oligomerize. (F). Size-and-shape distribution analysis of the wt mcGAS:dsDNA sample reveals a complicated mixture of compact and elongated oligomeric species. Putative oligomeric species are identified based on experimentally determined molecular weights in Table S2. (G). Normalized sedimentation velocity AUC analysis of site A (K372E and R158E) and site B (R342E and K335E) mcGAS mutants in the presence of a 20 bp dsDNA. The wt mcGAS distribution is shown for comparison.
Analytical ultracentrifugation (AUC) analysis showed that mcGAS in isolation at ~200 μM is mostly monomeric with a very small fraction of dimers (Figure 4E). When cGAS (~150 μM) bound to dsDNA, it forms a range of species including 2:2 dimers and higher oligomers (Figures 4E and F, and Table S2). Oligomer formation is sensitive to both protein and DNA concentration, as a five-fold dilution of the sample (~30 μM of cGAS) results in dissociation of the oligomers, reflecting weak dimerization as well as the low affinity between cGAS and dsDNA. Clues into the formation of higher-order cGAS:dsDNA oligomers are suggested by the observed crystal packing. Each DNA duplex in the crystal stacks end-to-end with another duplex to mimic a continuous, 36 bp dsDNA (Figure S2E). In addition, we have tested how mutations at the dimer interface and the DNA binding sites affect the oligomerization of cGAS. The replacement of Lys382 by alanine at the dimer interface disrupted the dsDNA-induced oligomerization of cGAS (Figure 4E). Moreover, mutations K372E or R158E at site A and mutations R335E or R342E at site B also abolished the oligomerization of cGAS in presence of the 20 bp dsDNA (Figure 4G). These results confirmed that dsDNA binding induces the oligomerization of cGAS in solution. DNA binding at both site A and site B is essential for higher-order complex formation.
Mutations at the DNA binding sites and the dimer interface affect cGAS activity
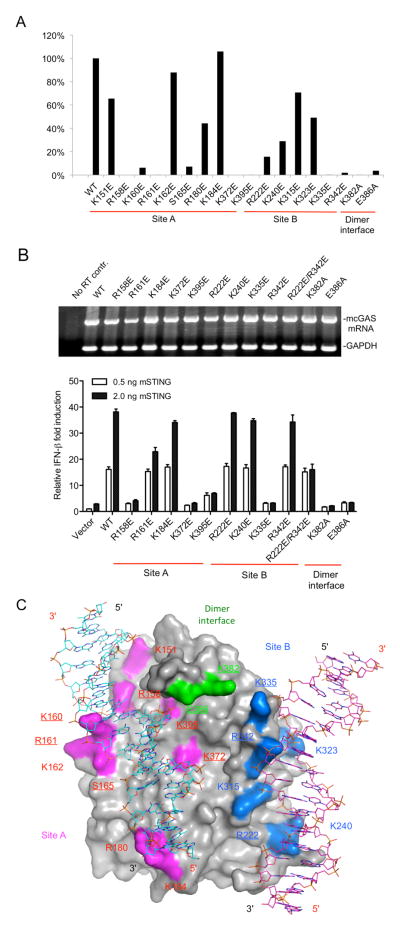
The crystal structure of the 2:2 mcGAS:dsDNA complex provides a new model of cGAS activation by dsDNA. To test this model, we generated 18 mutants of the mcGAS catalytic domain, purified each of the mutant proteins, and conducted enzyme activity assays. Replacement of Arg158, Lys 160, Arg161, Ser165, Lys372 and Lys395 at site A with glutamates significantly reduced cGAS activity (Figures 5A and S3A). In contrast, the replacement of Lys151, Lys162, and Arg180 at site A only moderately reduced the activity of cGAS (Figure 5A and S3A). A substitution of Lys184, which does not contact the DNA directly, had no affect the catalytic activity of cGAS (Figures 5A and S3A). Replacement of Arg222, Lys240, and Lys323 at site B also resulted in more than 50% reduction of cGAS activity (Figures 5A and S3A). In contrast, the replacement of Lys315, which does not interact with DNA directly, only resulted in ~30% reduction of activity (Figures 5A and S3A). Strikingly, substitutions of either Lys335 or Arg342 at site B with glutamate nearly abolished the activity of cGAS (Figures 5A and S3A), demonstrating that DNA binding at site B is also required for the activation of cGAS.
Figure 5.
Mutations at the DNA binding sites and the dimer interface affect cGAS activity. (A). Enzyme activities of mcGAS mutants. The activities of wild-type and mutant mcGAS were analyzed by ion exchange chromatography. The activities of the mutants relative to the wild-type enzyme were calculated based on the peak areas of 2′,5′ cGAMP in the chromatograms. (B). IFN-β luciferase reporter assays in HEK 293T cells transfected with mouse STING and cGAS. Error bars represent the standard deviations of signals from three independent transfections. The upper panel shows the mRNA levels of mcGAS and its mutants in the transfected cells. The mRNA levels were determined after 20 cycles of RT-PCR amplification. GAPDH mRNA was amplified as an internal control. (C). Locations of the mutated residues on the surface of mcGAS and effects of these mutations on cGAS activity. The two dsDNAs bound to cGAS are shown by the cyan and purple stick models. Residues mutated at site A are colored magenta and those in site B are colored blue. Residues at the dimer interface are colored green. Mutations that caused more than 80% loss of enzyme activity (in Panel A) are underlined. See also Figures S3.
To test whether dimer formation is also needed for cGAS activity, we replaced Lys382 or Glu386 at the dimer interface with alanine. These two mutations resulted in nearly complete loss of cGAS activity (Figures 5A and S3A). The replacement of Lys394 in hcGAS, which corresponds to Lys382 in mcGAS, also abolished the catalytic activity (Figure S3B). Consistent with cGAS dimerization being required for enzymatic activity, AUC analysis of the K382A mutant of mcGAS shows that this mutant cannot form dimers or higher-order oligomers with a 20 bp dsDNA (Figure 4E). Notably, mutant K382A still binds dsDNA and forms a 1:1 complex (Figure 4E), which is obviously not sufficient for cGAS activation (Figure 5A). Moreover, the replacement of Lys335, which forms a salt bridge with Glu386 at the mcGAS dimer interface and contacts the DNA at site B, also disrupted cGAS activity (Figure 5A).
To determine the physiological relevance of our structural analysis, we examined whether mutations at the two DNA binding sites and the dimer interface of mcGAS affect signal transduction in cells. Wild-type mcGAS activated the IFN-β promoter in the presence of increased concentration of mSTING (Figure 5B). Three independent substitutions at site A (R158E, K372E, and K395E) dramatically decreased the mcGAS activities while mutant K184E had activity similar to that of wild-type. Two substitutions at site B (K335E, and R222E plus R342E) also decreased signaling to background while mutants R222E, K240E or R342E with single amino acid substitutions in site B did not affect reporter activity (Figure 5B). Importantly, substitutions at the dimer interface (K382A and E386A) were defective for the mcGAS activity (Figure 5B). In addition, all of these mutations affected firefly luciferase reporter activity from an NF-κB promoter in a similar manner to that of IFN-β reporter (Figure S3C). The activities of most of the mutants correlate with enzymatic activities of mcGAS catalytic domain, except mutants R222E and R342E, perhaps because the defects are not significantly strong to be detectable in cells. However, mutation of both residues at the same time disrupted cGAS-mediated signaling in cells (Figure 5B). The locations of the mutated residues on the surface of mcGAS and their effects on the activity of cGAS were summarized schematically in Figure 5C. In conclusion, these studies demonstrated that mutations at the two DNA binding sites and the dimer interface affect cGAS activity in vitro and signaling in cells, suggesting that dsDNA-induced oligomerization of cGAS is essential for its activity.
cGAS binds dsDNA cooperatively through the two binding sites
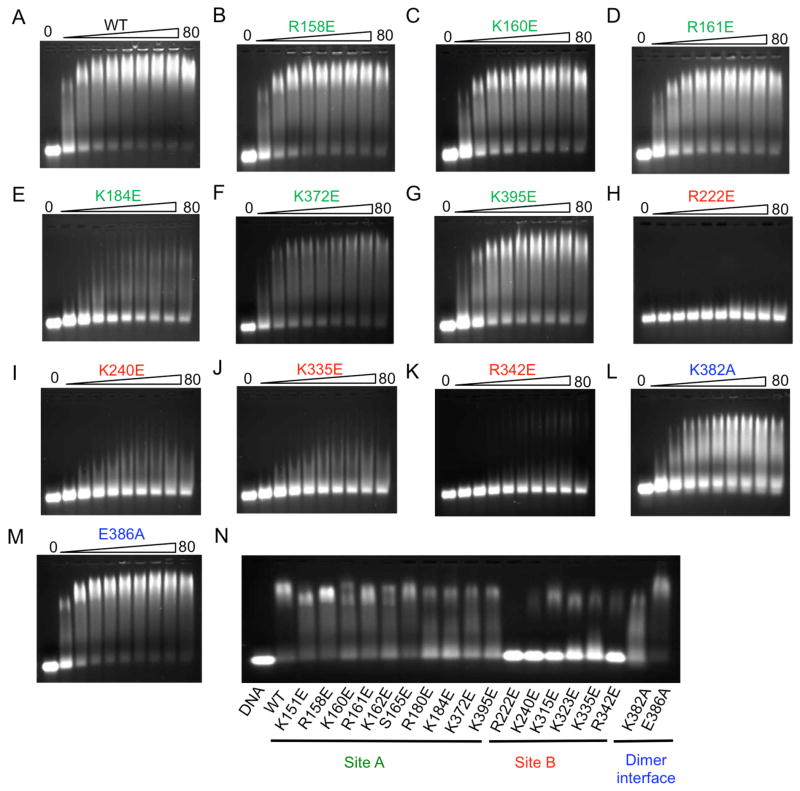
Although the results of biochemical and cell-based assays demonstrated that mutations at both site A and B affect cGAS function, it is not clear how these mutations affect DNA binding. To further explore the mechanism of cGAS activation by dsDNA, we have purified all the mutants of mcGAS catalytic domain described above and conducted DNA binding studies by electrophoretic mobility shift assays (EMSA) using a 45 bp interferon stimulatory DNA (ISD). First, we conducted DNA binding assays for wild-type mcGAS and each of the mutants individually by titrating 1 μM DNA with 5 to 80 μM of the protein (Figures 6A to M and S4A to H). The DNA binding studies showed that mutations K151E, R158E, and S165E at site A only slightly reduced DNA binding and did not affect dimer or higher order complex formation (Figures 6B, S4C and E). Mutations K160E, K162E, K372E, and K395E at site A reduced DNA binding, but did not disrupt dimer or higher order complex formation as well (Figures 6C, F, G, and S4D). In contrast, mutations R180E and K184E reduced DNA binding and higher order complex formation (Figures 6E and S4F). Since multiple residues are involved in DNA binding at site A, mutations of individual residues did not affect DNA binding or higher order complex formation dramatically. These results are consistent with the observation that mutations of hcGAS at site A do not affect DNA binding significantly (Civril et al., 2013). Unexpectedly, however, site B plays a much more important role in DNA binding compared to site A. Mutations of any residues at site B dramatically reduced DNA binding as well as higher-order complex formation (Figures 6H to K, S4G and H). Mutations R222E, K240E severely reduced DNA binding (Figures 6H and I). Mutations K315E, K323E, K335E, and R342E also dramatically reduced DNA binding and higher order complex formation (Figures 6J and K, S4G and H). In addition, mutation K382A at the dimer interface also reduced DNA binding and higher-order complex formation (Figure 6L). In contrast, mutation E386A at the dimer interface did not affect DNA binding significantly (Figure 6M). We have also conducted DNA binding studies for all the mutants together at DNA concentrations of 10, 20, and 50 μM and obtained similar results (Figures 6N, S4I and J). Furthermore, we observed that the inclusion of 5 mM EDTA in the binding studies abolished DNA binding by cGAS (Figure S4B), demonstrating that zinc binding is critical for the structural integrity of cGAS.
Figure 6.
cGAS binds dsDNA cooperatively through two binding sites. (A). DNA binding study of wt mcGAS catalytic domain by electrophoretic mobility shift assay (EMSA). The concentration of the 45 bp interferon stimulatory DNA (ISD) used in this study is 1 μM and the protein concentrations are from 5 to 80 μM. (B to G). DNA binding studies of site A mutants of mcGAS. (H to K). DNA binding studies of site B mutants. (L and M). DNA binding studies of mutants at the dimer interface. (N). DNA binding study of 18 mcGAS mutants together on one gel. The DNA concentration used in this study is 1 μM and mcGAS is at 20 μM. See also Figures S4.
Although wild-type full-length mcGAS is not stable and degrades readily during purification, we have successfully purified 8 mutants of full-length mcGAS and conducted DNA binding studies and enzyme assays (Figures S4K to T). Similar to the results from the DNA binding studies using mcGAS catalytic domain, mutations R158E or K372E at site A as well as mutations K382A or E386A at the dimer interface only have limited impact on DNA binding (Figures S4K, L, Q, R, and S). In contrast, mutations R222E, K240E, K335E, and R342E at site B dramatically reduced DNA binding and oligomerization of cGAS (Figures S4M to P, and 4S). Enzyme assays showed that mutations R158E, K372E at site A, K335E at site B, and K382A at the dimer interface dramatically reduced the activity of full-length mcGAS (Figure S4T). In agreement with results from the IFN-β reporter assays (Figure 5B), mutations R222E, K240E, and R342E at site B only had limited impact on the enzymatic activity (Figure S4T).
These mutagenesis and DNA binding studies showed that site B plays an important role in DNA binding and higher-order complex formation by cGAS. It is most likely that DNA binding to cGAS at sites A and B is cooperative and the dimerization of cGAS facilitates the cooperative DNA binding. Although mutations at site A did not influence the cooperative DNA binding significantly, mutations at site B dramatically reduced the cooperative binding of DNA. AUC studies of mcGAS mutants confirmed the importance of DNA binding at site B for cGAS assembly. The site B mutants K335E and R342E were incapable of assembly and only formed 1:1 complexes with DNA (Figure 4G). In contrast, site A mutant K372E could assemble into larger complexes, although higher assembly was diminished compared to wt cGAS (Figure 4G). Altogether, these results demonstrated that cGAS binds dsDNA cooperatively through two binding sites and site B plays a critical role in the cooperative DNA binding.
Structure of mcGAS bound to dsDNA and 2′,5′ cGAMP
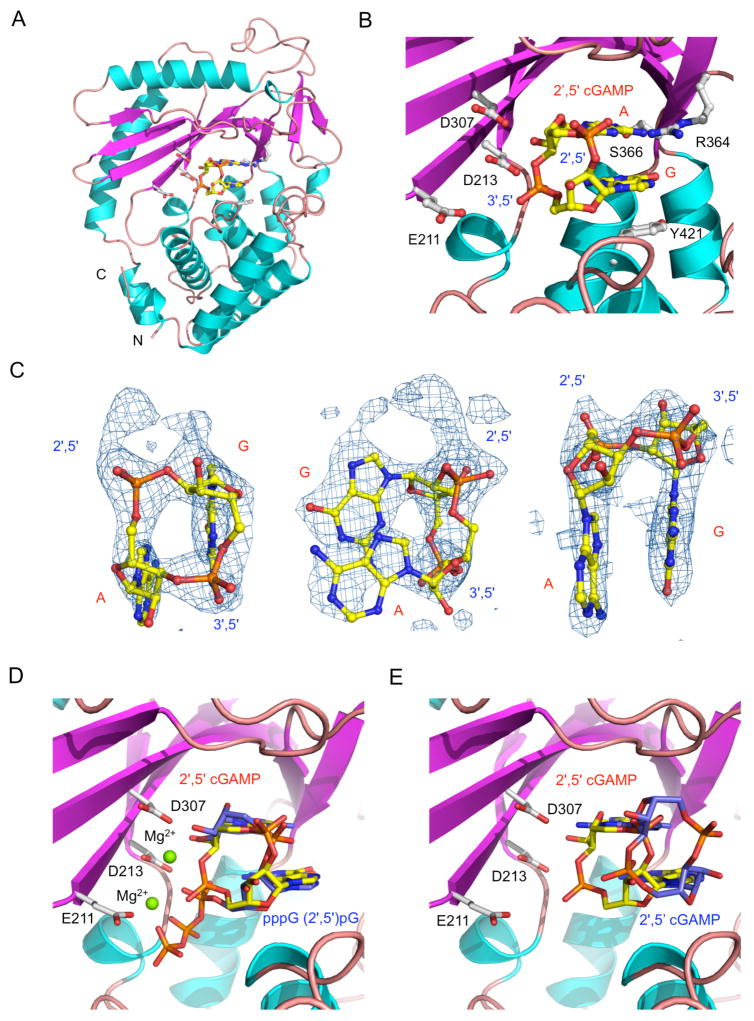
To elucidate the catalytic mechanism of cGAS, we have determined the 2.36 Å resolution structure of mcGAS catalytic domain bound to dsDNA and 2′,5′ cGAMP (Figures 7A, B, and S5A). The ternary complex crystallized in orthorhombic space group P212121 with one 2:2:2 cGAS:dsDNA:2′,5′ cGAMP complex in the asymmetric unit (Table S1). The difference (Fo - Fc) map shows well-defined electron density for 2′,5′ cGAMP at one binding site (Figure 7C), while the density at the second binding site is fragmented. The difference map shows clear features of the 2′,5′ phosphodiester bond between G and A and the 3′,5′ phosphodiester bond between A and G (Figure 7C). The adenine base forms three hydrogen bonds with the sidechains of Arg364 and Ser366. The 2′ hydroxyl of the adenylate forms two hydrogen bonds with catalytic residues Asp213 and Asp307. The 3′,5′ phosphodiester bond between A and G is positioned near the catalytic residues Asp213 and Asp307. Since the phosphate of the A:G linkage is close to the catalytic residue Asp307 (~3.15 Å, Figure 7B), this structure is likely to be unstable in solution. Consistent with this prediction, the second product-binding site is poorly occupied due to exposure to solvent. The structure and orientation of 2′,5′ cGAMP observed in our structure is similar to that of the 5′-pppG(2′,5′)pG, 5′-pppdG(2′,5′)pdG, and 5′-pG(2′,5′)pA (Gao et al., 2013a) bound to mcGAS reported previously (Figures 7D, S5B and C). However, our ternary complex structure with 2′, 5′ cGAMP is strikingly different from the structure reported by Gao et al. (Figure 7E). The product observed in the structure reported by Gao et al. has the guanine and adenine bases flipped relative to our structure and the phosphate ribose ring moved away from the active site of cGAS (Figure 7E). Superposition of the two structures clearly showed that the difference map we observed could not be interpreted by the 2′, 5′ cGAMP structure reported by Gao et al. In our structure, the guanine ring of 2′,5′ cGAMP stacks against the phenol ring of Tyr421 (Figure 7B), while in the structure reported by Gao et al, the adenine ring of 2′, 5′ cGAMP stacks against the sidechain of Tyr421. According to the proposed catalytic mechanism of cGAS (Gao et al., 2013a), it is likely that our structure shows how 2′, 5′ cGAMP binds to cGAS immediately after the formation of the 3′,5′ phosphodiester bond, while the structure observed by Gao et al likely shows how 2′, 5′ cGAMP binds cGAS before its release from the active site. In summary, the structure of cGAS bound to DNA and 2′,5′ cGAMP confirmed the identity of cGAS product and provided insight into the catalytic mechanism of the enzyme.
Figure 7.
Structure of mcGAS bound to dsDNA and 2′,5′ cGAMP. (A). Ribbon representation of mcGAS bound to an 18 bp dsDNA and 2′,5′ cGAMP. The dinucleotide is shown by the yellow ball-and-stick model. Catalytic residues and residues that interact with 2′,5′ cGAMP are shown by the gray ball-and-stick models. For clarity, the two dsDNA bound to mcGAS are not shown. (B). Close-up view of 2′,5′ cGAMP bound to mcGAS. (C). Difference maps of 2′,5′ cGAMP bound to mcGAS in three different orientations. The σA weighted Fo-Fc map calculated without the ligand is contoured at 3.5σ. 2′,5′ cGAMP is shown by the yellow ball-and-stick model. (D). Comparison of structures of pppG(2′,5′)pG (blue sticks) and 2′,5′ cGAMP (yellow sticks) bound to mcGAS. The figure was generated by superposition of mcGAS bound to pppG(2′,5′)pG (PDB, 4K98) onto the complex structure of mcGAS bound to 2′,5′ cGAMP (this work, PDB 4LEZ). The structure of mcGAS determined in this work is shown by the cyan and purple ribbons. Residues at the active site of mcGAS are shown by the gray stick models. (E). Comparison of structures of 2′,5′ cGAMP bound to mcGAS in two different orientations. This figure was generated by superposition of a previously determined cGAS:DNA:2′,5′ cGAMP complex structure (PDB, 4K9B) onto the complex structure from this work (PDB, 4LEZ). See also Figure S5.
DISCUSSION
Based on the crystal structures of cGAS catalytic domain in isolation, in complex with dsDNA, in complex with dsDNA and 2′,5′ cGAMP, and results from our biophysical, biochemical, and cell-based studies, we propose the following model for cGAS activation by dsDNA. cGAS is monomeric in isolation and does not have catalytic activity due to local destabilization of the structure of the NTase domain and the active site. DNA binding induces conformational changes in cGAS and crosslinks two cGAS molecules to form a 2:2 dimer or higher order complexes, resulting in the activation of cGAS. Site A plays a major role in inducing the conformational changes of cGAS in response to contacts with DNA, while site B contributes to the cooperative DNA binding by the cGAS dimer. Crosslinking of the cGAS dimer by dsDNA restricts the structural flexibility of cGAS and stabilizes the active conformation of the enzyme. Interactions at the cGAS dimer interface make additional contributions to the DNA binding-induced dimerization or oligomerization of cGAS.
Our cell-based reporter assays and enzyme assays using mcGAS mutants suggest that DNA binding at both site A and B as well as the dimerization of cGAS are needed to activate the enzyme. The two DNA binding sites likely have distinct contributions to cGAS function. Our mutational analysis showed that mutations at site A do not consistently affect DNA binding and the activities of the mutants do not correlate well with DNA binding. For example, mutations R158E, K160E, R161E, and S165E dramatically reduced enzyme activity, but only slightly reduced DNA binding. This suggests that residues involved in DNA binding at site A likely have two levels of function. Some residues mainly mediate DNA binding, while others mediate DNA binding-induced the conformational changes of cGAS. Since DNA binding at site A is more extensive and cGAS binds DNA cooperatively through two binding sites, mutations of individual residues at site A only have limited impact on DNA binding. However, mutations of key residues such as R158, K372, K395, and K335 that might mediate cGAS conformational changes have large impacts on the activity of the enzyme. In contrast, mutations of key residues at site B have much larger impacts on DNA binding, demonstrating the critical role of site B in cooperative DNA binding. Moreover, the impact of mutations at site B on DNA binding correlates with the impact of these mutations on enzyme activity, suggesting that site B has a primary role in DNA binding and crosslinking of the cGAS dimer. Since the enzyme activity assays were carried out under different enzyme and DNA concentrations using a different buffer, this may also contribute to the imperfect correlation between the results of enzyme assays and DNA binding studies for some residues. For example, mutations R222E and K240E dramatically reduced DNA binding, but these mutants still exhibit some catalytic activity in the context of cGAS catalytic domain and show almost full activity in cell-based assays with full-length cGAS. Since the primary roles of these two residues are DNA binding at site B, the higher DNA and enzyme concentrations used for the enzyme assays may overcome the effects of these two mutations. It is also likely that full-length cGAS exhibits higher affinity for dsDNA that helps to overcome the effect of these two mutations on cGAS activity in the cells.
It was proposed that cGAS is activated by dsDNA in a similar manner as OAS1 by dsRNA through a 1:1 cGAS:DNA complex via site A (Civril et al., 2013; Gao et al., 2013a). The critical roles of site B in DNA binding and cGAS activation were not investigated in previous studies. In addition to a requirement for site B, we found that dimerization is required for the DNA-dependent activity of cGAS. AUC analysis of mcGAS mutant K382A shows that mutation of Lys382 at the dimer interface does not affect the formation of a 1:1 cGAS:dsDNA complex, yet the enzyme is completely inactive. Notably, cGAS is activated by a mechanism distinct to that of OAS1 activation by dsRNA (Donovan et al., 2013). OAS1 does not contain the zinc-binding loop that mediates the dimerization of cGAS and dimer formation by OAS1 was not observed in the crystal structure of OAS1 bound to dsRNA (Donovan et al., 2013; Hartmann et al., 2003). Furthermore, if cGAS were to form a 1:1 complex with dsDNA only via site A, short dsDNA of 12 bp should also activate the enzyme efficiently. As seen in our cGAS:dsDNA complex structures, only when the length of the dsDNA is above 16 bp is it possible for the DNA to span the two binding sites in a cGAS dimer effectively, crosslink the cGAS dimer, and activate the enzyme. Taken together, we conclude that the 2:2 cGAS:dsDNA complex structure reported here reveals the mechanism of cGAS activation by dsDNA that has been overlooked by previous studies (Civril et al., 2013; Gao et al., 2013a).
The structures of cGAS bound to various nucleotides provided insight into how the enzyme may catalyze the synthesis of 2′,5′ cGAMP. Although a catalytic mechanism of cGAS has been proposed based on these structures (Gao et al., 2013a), this mechanism needs to be tested by further structural studies. For example, cGAS has high specificity for ATP and GTP as substrates, however, the specificity of the enzyme cannot be fully explained by the available structures. According to the proposed mechanism, the reaction intermediate needs to be flipped after the formation of the 2′,5′ phosphodiester linkage (Gao et al., 2013a). It is not clear how this could occur during the reaction. Moreover, how cGAS catalyzes the synthesis of two different kinds of phosphodiester bonds remains unknown. It is also not clear why the product is not released immediately after the formation of the 3′,5′ phosphodiester bond, and needs to undergo another flip over before its release from the active site.
We have demonstrated that 2′,5′ cGAMP exhibits higher affinity for STING compared to canonical bacterial cyclic dinucleotides and is more potent in stimulating IFN-β expression in cells. However, the mechanism of how 2′,5′ cGAMP contributes to enhanced STING binding remains to be established. In addition, the mechanism of how STING and its natural variants distinguish between the endogenous second messenger and bacterial analogs is also not clear. Further biochemical and biophysical characterization of 2′,5′ cGAMP binding to STING should provide insight into these questions. Moreover, although crystal structures of STING bound to c-di-GMP have been reported (Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012), crystallographic analysis of STING bound to 2′, 5′ cGAMP is technically challenging since STING forms a homodimer but the ligand is asymmetrical. Even though two structures of 2′,5′ cGAMP bound to STING have been reported (Gao et al., 2013b; Zhang et al., 2013), the electron density observed in these studies corresponds to the average electron density of 2′,5′ cGAMP in two distinct orientations. Therefore, it is still not completely clear how STING recognizes 2′,5′ cGAMP. Further binding studies by NMR or molecular simulations should provide additional insight into this question.
It has been reported that cGAS, also known as C6orf150, inhibits the replication of several viruses (Schoggins et al., 2011). Recent studies revealed that cGAS synthesizes 2′, 5′ cGAMP to carry out these functions (Diner et al., 2013). Since 2′, 5′ cGAMP is an endogenous second messenger with a molecular weight of only 675 dalton, it has the potential to be used as an antiviral reagent. Interestingly, our cell-based studies showed that 2′,5′ cGAMP added to the culture medium induced IFN-β production in THP-1 cells, suggesting that the cells can pick up 2′,5′ cGAMP added to the culture medium without the use of liposomes to deliver it into the cytosol. Since the enzymatic synthesis of 2′,5′ cGAMP is rather straightforward, this cyclic dinucleotide has the potential to be used as an immune modulator, such as adjuvant for vaccines, as well as antiviral or anticancer reagent.
EXPERIMENTAL PROCEDURES
Protein expression and purification
hcGAS and mcGAS were cloned into a modified pET-28(a) vector with an N-terminal SUMO tag. The proteins were expressed in E. coli BL21(DE3) at 15 °C and purified as described (Shu et al., 2012). Mutants of hcGAS and mcGAS were generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies). The cGAS mutants were expressed and purified the same way as the wild-type protein. Human STING ligand binding domain was expressed and purified as described (Shu et al., 2012).
cGAS activity assay
cGAS or its mutants were incubated with DNA in a reaction buffer containing 20 mM HEPES (pH 7.5), 5 mM MgCl2, 2 mM ATP, and 2 mM GTP at 37°C for 2 hours. The samples were centrifuged, passed through a 10 kD ultrafiltration filter (Millipore), and analyzed by ion exchange chromatography using a MonoQ column (GE Healthcare).
Crystallization, data collection, and structural determination
hcGAS in isolation and mcGAS in complex with DNA (with or without 2′, 5′ cGAMP) were crystallized by the hanging-drop vapor-diffusion method at 4 °C. Diffraction data for the Se-Met derivative crystals of hcGAS were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) and processed with the HKL2000 package (Otwinowski and Minor, 1997). The structures were determined by SAD using the Phenix package (Adams et al., 2010). Diffraction data for mcGAS bound to the 18 bp dsDNA were collected using an RAXIS IV++ detector and processed with the HKL2000 or the Mosflm package (Winn et al., 2011). The structure was determined by molecular replacement and refined against the twin_lsq_F target using Phenix. Diffraction data for the mcGAS bound to the 18 bp dsDNA and 2′,5′ cGAMP was collected using an RAXIS IV++ detector and processed with HKL2000. The structure was determined by molecular replacement using using Phenix.
Small angle X-ray scattering
Small-angle X-ray scattering (SAXS) experiments were performed at beamline 12ID-B of Advanced Photon Sources (APS). The 2-D scattering images were converted to 1-D SAXS curves through azimuthally averaging after solid angle correction and then normalizing with the intensity of the transmitted X-ray beam. The radius of gyration (Rg) was calculated using the Guinier equation. The pair distance distribution function (PDDF) was calculated using GNOM (Svergun, 1992). Three dimensional molecular envelopes were calculated using DAMMIN (Svergun, 1999). Molecular models of hcGAS in isolation and mcGAS in complex with DNA were docked into the molecular envelope manually using PyMol.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed in a Beckman ProteomeLab XL-I at 4°C using interference optics. Raw fringe data were deconvoluted into sedimentation coefficient or sedimentation coefficient-frictional ratio distributions using c(s) and c(s,f/f0) models, respectively, in the program Sedfit (Brown and Schuck, 2006). The c(s,f/f0) analysis deconvolutes complicated mixtures into a size-and-shape distribution that separately fits the sedimentation coefficient and frictional ratio for each species, allowing more accurate determination of putative species based on molecular weights. Values for buffer density and viscosity and partial specific volume of cGAS were calculated using Sednterp. The partial specific volume of the cGAS:DNA complex was calculated as a weight-averaged value of the partial specific volumes for cGAS and the dsDNA and corrected to 4°C using Sednterp.
Supplementary Material
Highlights.
cGAS is activated by dsDNA and catalyzes the synthesis of 2′,5′ cGAMP
cGAS is activated by dsDNA induced oligomerization
Mutations at the DNA binding sites and the dimer interface affect cGAS activity
cGAS binds dsDNA cooperatively through two binding sites
Acknowledgments
The diffraction data of the hcGAS crystals were collected the Stanford Synchrotron Radiation Lightsource (SSRL). The SAXS studies of cGAS were conducted at the Advanced Photon Source (APS). We thank Dr. Liang Tong from Columbia University for suggestions on structural determination and Dr. Jun-yuan Ji from Texas A&M University for critical reading of the manuscript and valuable discussions. This research was supported by the National Institute of Health (Grant AI 087741 to P.L.) and the Welch Foundation (Grant A-1816 to P.L.). This work is dedicated to the memory of professor Yongxing Wang (1914–2008) of Peking University.
Footnotes
Accession codes: Coordinates and structure factors for hcGAS, mcGAS in complex with the 18 bp dsDNA, mcGAS in complex with the 18 bp dsDNA and 2′, 5′ cGAMP have been deposited in the Protein Data Bank with accession codes: 4LEV, 4LEW, 4LEY, and 4LEZ.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011a;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. STING-dependent signaling. Nat Immunol. 2011b;12:929–930. doi: 10.1038/ni.2118. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:6. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154:962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nature Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013a;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, et al. Structure-Function Analysis of STING Activation by c[G(2′,5′)pA(3′,5′)p] and Targeting by Antiviral DMXAA. Cell. 2013b;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature structural & molecular biology. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nature Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science. 2013 doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. Journal of molecular biology. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nature structural & molecular biology. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature structural & molecular biology. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophysical journal. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Molecular cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.