Abstract
Purpose
Signal Transducers and Activators of Transcription (STATs) activate transcription in response to numerous cytokines, controlling proliferation, gene expression and apoptosis. Aberrant activation of STAT proteins, particularly STAT-3, is implicated in the pathogenesis of many cancers, including Globlastoma Multiforme (GBM), by promoting cell cycle progression, stimulating angiogenesis, and impairing tumor immune surveillance. Little is known about the endogenous STAT inhibitors, the Protein Inhibitors of Activated STATs (PIAS) proteins, in human malignancies. The objective of this study was to examine the expression of STAT-3 and its negative regulator, PIAS3, in human tissue samples from control and GBM brains.
Experimental Design
Control and GBM human tissues were analyzed by immunoblotting and immunohistochemistry to determine the activation status of STAT-3 and expression of the PIAS3 protein. The functional consequence of PIAS3 inhibition by siRNA or PIAS3 over-expression in GBM cells was determined by examining cell proliferation, STAT-3 transcriptional activity and STAT-3 target gene expression. This was accomplished using 3H-TdR incorporation, STAT-3 dominant-negative constructs, RT-PCR and immunoblotting.
Results and Conclusions
STAT-3 activation, as assessed by tyrosine and serine phosphorylation, was elevated in GBM tissue compared to control tissue. Interestingly, we observed expression of PIAS3 in control tissue, while PIAS3 protein expression in GBM tissue was greatly reduced. Inhibition of PIAS3 resulted in enhanced glioblastoma cellular proliferation. Conversely, PIAS3 over-expression inhibited STAT-3 transcriptional activity, expression of STAT-3 regulated genes, and cell proliferation. We propose that the loss of PIAS3 in GBM contributes to enhanced STAT-3 transcriptional activity and subsequent cell proliferation.
Keywords: STATs, PIAS3, Glioma, Signal Transduction, Cancer
INTRODUCTION
World Health Organization Grade IV glioblastoma multiforme [GBM] is the most aggressive malignant astrocytic glioma because of the high degree of cellularity, vascular proliferation, and necrosis. Patients diagnosed with GBMs have a median life expectancy of less than one year (1). GBMs are characterized by their propensity to infiltrate throughout the brain, which results in the inability of surgery to cure patients even when surgical resection is possible. In addition, the majority of GBMs are resistant to standard radiotherapy and chemotherapy (1).
Aberrant signaling through receptor tyrosine kinases, including the epidermal growth factor receptor [EGFR] and platelet derived growth factor receptor [PDGFR], is a hallmark of GBM (2). Constitutive activation of EGFR and PDGFR promotes cell growth and evasion of apoptosis, events that lead to maintenance of a tumor-promoting environment. Deregulated signaling through the Mitogen Activated Protein Kinase, PI3 Kinase/AKT, Protein Kinase C, NF-κB and JAK-STAT pathways has also been implicated in glioma development and progression (2, 3).
The Janus Kinase [JAK] family of receptor-associated tyrosine kinases are activated by phosphorylation after ligand binding, and activate Signal Transducers and Activators of Transcription [STAT] proteins to induce gene expression (4). The STAT family consists of seven members (STAT-1, 2, 3, 4, 5A, 5B, 6) and is activated by many stimuli, including the interleukin-6 [IL-6] cytokine family. Members include IL-6, Oncostatin M [OSM], Leukemia Inhibitory Factor, Ciliary Neurotrophic Factor, and interleukin-11 (5). IL-6 cytokines preferentially activate STAT-3, leading to dimerization, nuclear translocation, and binding to Interferon-Gamma [IFN-γ] Activated Site-like DNA elements (4). STAT-3 induces expression of genes that regulate anti-apoptotic behavior and proliferation such as Survivin, VEGF, c-Myc, and Cyclin D1 (6–8).
Aberrant activation of STAT-3 is observed in primary cancers (9). In GBM and medulloblastoma tumors, STAT-3 was shown to be constitutively active, as assessed by tyrosine phosphorylation status (10, 11). Activated STAT-3 localized to the vascular endothelial growth factor receptor-2 in tumor endothelial cells (11), and immunohistochemical studies of GBM demonstrated that tumor cells also contained tyrosine phosphorylated STAT-3 (10, 12, 13). In addition, experimental mouse gliomas express constitutively activated STAT-3 (13). Due to their ability to activate STAT-3, IL-6 cytokines have been implicated in progression of brain tumors (10, 11, 13). Human GBMs express elevated levels of OSM and IL-6 (8, 14). Up-regulation of IL-6 and OSM in glioblastoma cells regulates VEGF promoter activity, potentially promoting angiogenesis (8, 15).
Because JAK-STAT signaling is dysregulated in GBM, it is important to characterize negative regulators of this pathway in order to identify potential therapeutic targets. The Protein Inhibitors of Activated STATs (PIAS) family of proteins (PIAS1, PIAS3, PIASx, PIASy) regulate STAT activity (16). Upon cytokine stimulation, PIAS1 and PIAS3 bind activated STAT-1 and STAT-3, respectively, and prevent their ability to bind DNA (16). There are limited studies on the expression or function of PIAS proteins in disease states. Increased PIAS3 expression was detected in 100 out of 103 human cancers, including lung, breast, and brain tumors (17). In contrast, PIAS3 was not detected in most samples of anaplastic lymphoma kinase-positive T/Null-Cell lymphoma cells, which correlated with heightened STAT-3 activation (18). Over-expression of PIAS3 induced apoptosis in prostate cancer cells (19) and suppressed growth of human lung cancer cells (20). PIAS1 and PIAS3 enhanced transcriptional activity of the androgen receptor (AR) in prostate cancer cells, while PIASy inhibited AR-mediated gene expression (21). These results indicate differences in PIAS expression/function depending on the system under investigation.
This study sought to determine the status and functional relevance of PIAS3 expression in GBMs. We established a relation between elevated levels of activated STAT-3 in GBM and the presence/absence of PIAS3. Additionally, we demonstrated that loss of PIAS3 promoted proliferation of GBM cells. We propose that PIAS3 loss contributes, in part, to the increased STAT-3 activation observed in GBM, and proliferation and survival of glioma cells.
MATERIALS AND METHODS
Cells
U251-MG, U87-MG, SNB-19, M059K-MG, U138-MG, and U118-MG human astroglioma cells were cultured as previously described (8). Primary murine astrocytes were >97% positive for GFAP, and microglia >90% positive for Mac1, as previously described (22). Neuronal cultures were prepared by isolating cerebral hemispheres from P0 mice and removing the meninges, as described (23). GBM cell cultures from four patients were received from the UAB Brain Tumor Bank of the Cooperative Tissue Network, in accordance with the UAB Human Tissue Committee policies, IRB Exemption #X050415007. GBM primary cells were obtained after 30 days in culture and were grown in DMEM/F-12 medium supplemented with 2 mM L-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% HI-FBS, as previously described (24).
Reagents
Recombinant human OSM, IL-6, and sIL-6R were obtained from R&D Systems (Minneapolis, MN). PMA was purchased from Calbiochem (San Diego, CA). Antibodies to STAT-3, PIAS3 (C-terminus, sc-14017), and p21 were obtained from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Anti-PIAS3 (N-terminus, AP-1245a) antibody was purchased from Abgent (San Diego, CA). Antibodies to phospho-serine STAT-3, phospho-tyrosine STAT-3, and STAT-3 were from Cell Signaling Technology (Beverly, MA). Anti-SOCS-3 antibody was from Zymed (Carlsbad, CA) and anti-actin antibody was from Sigma (St. Louis, MO). The 3xLy6epZLuc-TK (STAT-3) luciferase construct was obtained from Addgene (Cambridge, MA). The 1556-bp SOCS-3 promoter has been previously characterized (25). The MMP-9-Luc luciferase reporter plasmid containing 670 bp of the human MMP-9 promoter was a generous gift from D. Boyd (MD Anderson Cancer Center, Houston, TX, USA). The STAT-3F dominant-negative expression construct has been previously described (26). Human GAPDH primers were purchased from Clontech (Mountain View, CA). Human PIAS3 cDNA (Clone ID: 3528679; Accession: BC001154) was purchased from Open Biosystems (Huntsville, AL) and cloned into the pcDNA3 vector. MG-132 was purchased from Cal Biochem (Gibbstown, NJ).
Human Tissues
Tissues were received from the UAB Brain Tumor Bank of the Cooperative Tissue Network, in accordance with the UAB Human Tissue Committee policies, IRB Exemption #X050415007. The non-neoplastic brain tumor biopsy samples were obtained from patients diagnosed with epilepsy. The tumor biopsies were diagnosed as GBM (WHO Grade IV tumors).
siRNA Transfection
U87-MG cells were untreated or transiently transfected using Dharmafect 1 reagent (Dharmacon) with either 100 nmol PIAS3 siPOOL Reagent or 100 nmol of negative control siRNA (Ambion), according to the manufacturer’s instructions to achieve approximately 85% transfection efficiency. Twenty-four hours after transfection, cells were collected and analyzed for target knockdown by RT-PCR with primers for human PIAS3 and GAPDH. Cells were collected 24, 48, and 72 h post-transfection and analyzed by immunoblotting for PIAS3 and actin.
Immunoblotting
Non-neoplastic brain biopsy (controls) and GBM biopsy samples were lysed, as described previously (8). Total protein (40 μg) was resolved on an 8% SDS-PAGE gel, transferred to nitrocellulose membrane, and blocked for 1 h in 5% milk. The membranes were then probed with antibodies against phospho-tyrosine STAT-3, phospho-serine STAT-3, total STAT-3 (1:1000), PIAS3 (Abgent) (1:100 dilution), SOCS-3 (1:500) as well as actin (1:1000 dilution) to control for loading. The ECL method was used for protein detection, as previously described (25). For SOCS-3, 100 μg of protein was used.
U251-MG cells were treated with either 50 ng/ml OSM or 10 ng/ml IL-6 plus 50 ng/ml sIL-6R for various time periods. Cells were lysed as previously described (25). Total cell lysate (40 μg) was resolved on an 8% SDS-PAGE gel, transferred to nitrocellulose membrane, and then blocked for 1 h in 5% milk. Membranes were probed as described above.
For proteosome inhibition, U251-MG, SNB-19 and GBM primary cells were incubated with MG-132 (10 μM) for various times and then collected as described above. Forty μg of total cell lysate was resolved on an 8% SDS-PAGE gel, and analyzed as described above. Densitometry was used to quantify the immunoblotting results, and actin served as an internal control for sample loading. All results were normalized by the respective actin values.
RNA Isolation, Quantitative Real-Time PCR, and Reverse-Transcriptase PCR
Total RNA from human tissues was extracted using a PowerGen 125 with Trizol. The Gene Amp 7700 Sequence Detection System (Applied Biosystems, Foster City, CA) was used for detection of real-time PCR products amplified from reverse-transcribed total RNA (25 ng). Total RNA from U251-MG cells was extracted using Trizol. PCR reactions were done in triplicate for each sample on a 96-well plate that included separate wells to quantify an internal RNA control (S9) and a standard curve. The cycling conditions were 48°C for 30 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. FAM-conjugated Taqman probes (Applied Biosystems, Foster City, CA) were synthesized as follows: PIAS3: 5′-CACTCGCGGGCACTGATCAAGGAGA-3′, SOCS-3: 5′-ACCAGCTGGTGGTGAACGCAGTGCG -3′, GAPDH: 5′ TGGAGGGCAAGTCTGGTGCCAGCAG-3′, and S9: 5′-AGCAGGTGGTGGTGAACATCCCGTCCTT-3′. For Reverse-Transcriptase PCR (RT-PCR), 2 μg of RNA was reverse-transcribed to cDNA, which was amplified using primers specific for human PIAS3 (Forward: 5′-ATTGACTGCTGACCCTGACA-3′, Reverse: 5′-GGGACAGCGAAGTTTCCATA-3′), human Bcl-xL (Forward: 5′-GGAGCTGGTGGTTGACTTTC -3′, Reverse: 5′-ACAATGCGACCCCAGTTTAC-3′), human Survivin (Forward: 5′-TTTCTGCCACATCTGAGTCG-3′, Reverse: 5′-TGTCGAGGAAGCTTTCAGGT-3′) and GAPDH (Clontech). Densitometry was used to quantify the PCR results. GAPDH mRNA served as an internal control for sample loading and mRNA integrity, and all results were normalized by the respective GAPDH values.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissues were obtained from the UAB Brain Tumor Bank, in accordance with the UAB Human Tissue Committee policies, IRB Exemption #X050415007. Briefly, tissue sections were deparaffinized, and endogenous peroxidase activity blocked with 3% hydrogen peroxide in PBS. Sections were then incubated in PBS-blocking buffer for 30 min at room temperature. PIAS3 antibody (Abgent, AP-1245a) (1:300) was applied to sections overnight at 4°C. After washes with PBS, sections were incubated with a donkey anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:1500) (Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. Tyramide Signal Amplification [TSA] detection followed and biotin-tyramide was applied (1:500) to the slides for 10 min. Tissues were then subjected to incubation for 30 min with the Vectastain ABC Kit (Vector Labs, Burlingame, CA). Immunostaining was detected using a metal-enhanced DAB substrate kit (Pierce, Rockford, IL) according to the manufacturer’s instructions.
Transfection and Luciferase Assay
U251-MG cells were transiently transfected using the Fugene-6 reagent (Roche Diagnostics) with either 500 ng of 3xLy6epZLuc-TK, which contains 3 STAT-3 sites (27), 500 ng of the 1556-bp SOCS-3 promoter (25), or 200 ng of the MMP-9 promoter. Cells were also transfected with either the STAT-3F plasmid or the PIAS3-pcDNA (100–500 ng). Cells were incubated for 16 h in the absence or presence of either OSM or PMA, then cell lysates were assayed in triplicate for luciferase activity (28), and normalized to total protein. The luciferase activity from the untreated condition was arbitrarily set at 1 for calculation of fold induction.
[3H]-TdR Incorporation
[3H]-TdR incorporation was performed as previously described (24). Briefly, U87-MG and U251-MG cells were labeled with 1 μCi of [3H]-TdR for 16 h before terminating the culture. Radioactivity was counted in duplicate samples using a scintillation counter (Beckman, Fullerton, CA).
Statistical Analysis
Levels of significance for comparison between samples were determined by the Student’s t-test distribution. p values of ≤ 0.05 were considered to be statistically significant.
RESULTS
The STAT-3 Protein is Constitutively Activated and PIAS3 Protein Expression is Diminished in Human GBM Tissue
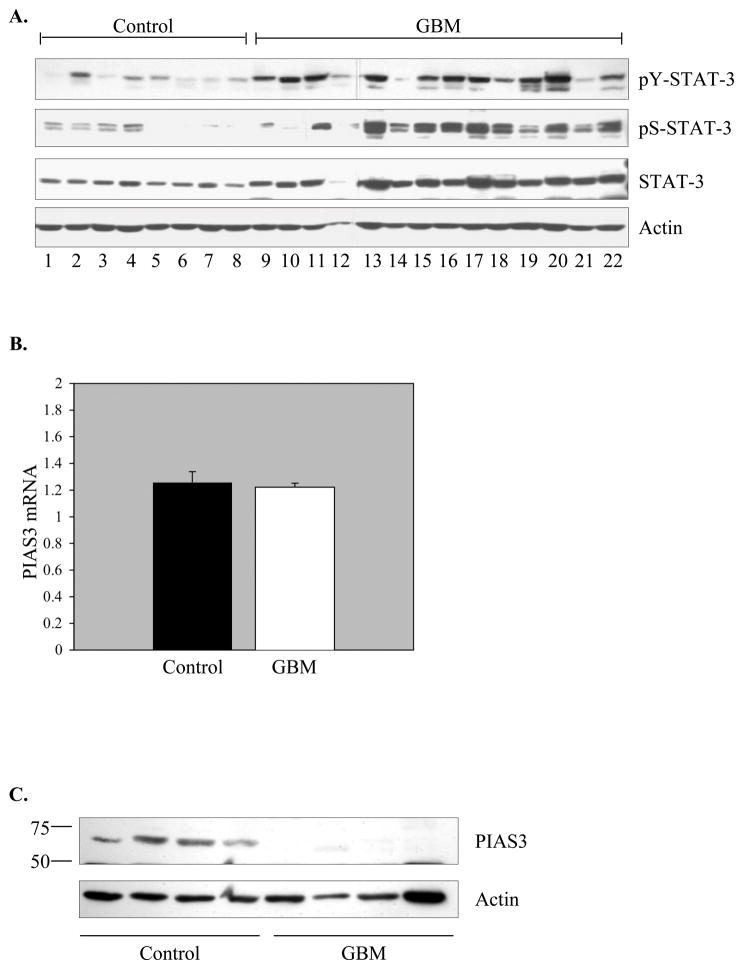
There are conflicting reports about the activation status of STAT-3 in GBM. While tyrosine phosphorylated STAT-3 was detected in GBM samples by several investigators (10–13), a tissue microarray study failed to confirm these findings (29). STAT-3 activation is reflected by phosphorylation of both tyrosine and serine residues; thus, we examined both parameters in this study. Tissue samples were analyzed by immunoblotting, and higher levels of tyrosine and serine phosphorylated STAT-3 protein were detected in GBM tissue compared to controls (Figure 1A). This is the first demonstration of serine phosphorylated STAT-3 in GBM tissues. These results indicate that GBM samples contain elevated levels of activated STAT-3.
Figure 1. STAT-3 Protein Activation is Elevated and PIAS3 Protein Expression is Reduced in GBM Tissues Compared to Control Brain Tissue.
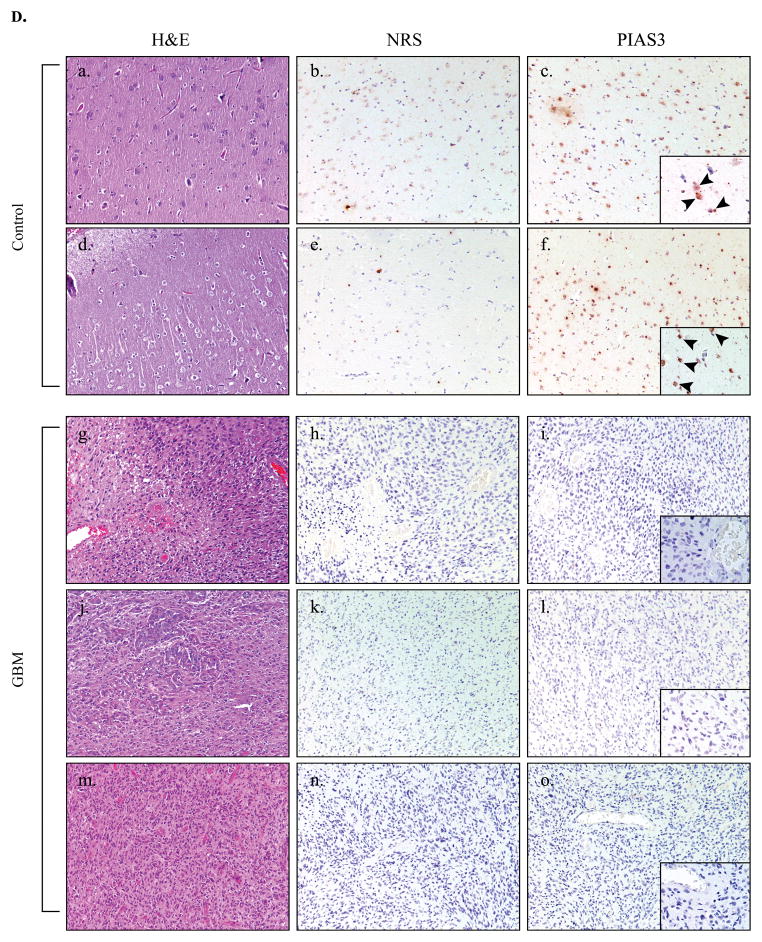
A. Immunoblot of phospho-Y (705) STAT-3, phospho-S (727) STAT-3, total STAT-3 and actin levels in control tissues (n=8) and GBM tissues (n=14). Representative of three experiments. B. Quantitative real-time PCR was performed for PIAS3 mRNA in control samples and GBM samples. PIAS3 mRNA expression is represented as a ratio to the housekeeping gene, S9. Results are the mean ± S.D. of three experiments on control (n=11) and GBM (n=11) tissue samples. C. Immunoblot of PIAS3 protein (Abgent, AP-1245a) in control and GBM tissues. Blots were re-probed for actin to verify protein loading between samples. Representative of four experiments. D. Human control (a–f) (n=2) and GBM (g–o) (n=3) tissues were analyzed by histochemistry. a, d, g, j, m, Tissue integrity was verified by H&E staining. b, e, h, k, n, Immunohistochemistry using normal rabbit serum served as a negative control for staining in control and GBM tissues. c, f, i, l, o, Control and GBM tissues were stained with anti-PIAS3 antibody. Nuclei were counter-stained with hematoxylin. Low power photomicrographs X 100 magnification. Insets: X 400.
PIAS3 functions as a negative regulator of STAT-3 signaling by interfering with its ability to bind DNA (16). Using tissue samples from both GBM and control brains, PIAS3 mRNA and protein levels were examined by real time RT-PCR and immunoblotting, respectively. Similar levels of PIAS3 mRNA expression were observed in control and GBM tissues (Figure 1B). Due to the instability in housekeeping gene expression in human tumor tissues, the levels of PIAS3 were compared to both S9 (Figure 1B) and GAPDH (data not shown), and comparable results were obtained. The 68 kDa PIAS3 protein was expressed in all control tissues tested, however, little to no expression of the PIAS3 protein was detected in GBM tissues (Figure 1C). In 35 GBM samples tested, PIAS3 protein expression was detected in only 4 samples, translating to 89% of GBM samples with loss of PIAS3 protein expression, while 100% of control samples (n=33) expressed PIAS3 (data not shown). Because of the striking difference in PIAS3 protein levels between control and GBM groups, a second antibody recognizing a different epitope on the PIAS3 protein was used to confirm the results (data not shown). To determine if any correlates could be established between the absence/presence of PIAS3 in GBM samples and STAT-3 activation, we compared these two parameters. Of the 14 GBM samples analyzed in Figure 1A, we were able to obtain PIAS3 status on 11 samples. Two of the 11 GBM samples expressed PIAS3 (samples 14 and 18), and those samples also had low levels of activated STAT-3 (see Figure 1A). Samples 9, 10, 11, 17, 19, 20, 21 and 22 lacked PIAS3 expression, and had high levels of activated STAT-3, except for sample 21 (see Figure 1A). These results suggest that a lack of PIAS3 in GBMs may contribute to constitutive STAT-3 activation.
Tissues were examined using immunohistochemistry, and sections analyzed by a neuropathologist blinded to the antibodies used. Strong nuclear staining for PIAS3 was observed in 2 control brain specimens (Figure 1D c, f, see arrowheads). Positive staining appeared mainly in neurons, with weaker staining in astrocytes and endothelial cells. Human GBM (n=3) tissues had little to no positive reactivity for PIAS3 (Figure 1D i, l, o). These data indicate that loss of PIAS3 protein expression in GBM samples is frequent.
The PIAS3 Protein is Constitutively Expressed In Vitro
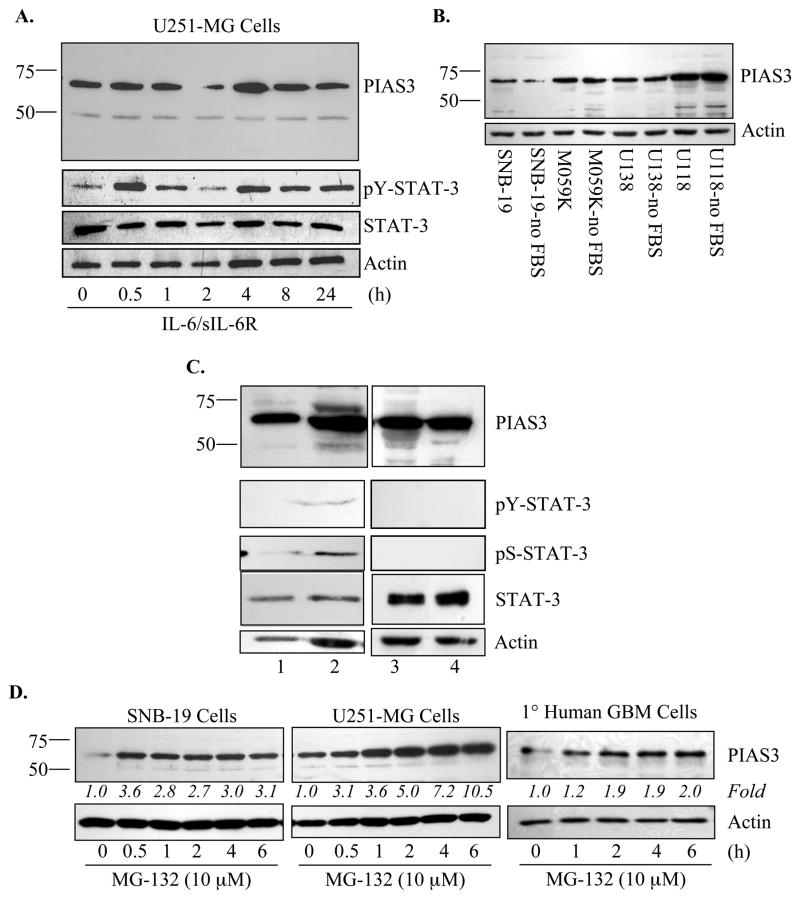
Because the expression levels of PIAS3 were substantially different between control and GBM samples, we examined the expression of PIAS3 in human glioblastoma cells. Constitutive expression of PIAS3 was observed in U251-MG cells (Figure 2A). IL-6/sIL-6R and OSM treatment induced STAT-3 tyrosine phosphorylation, but had little effect on PIAS3 expression (Figure 2A and data not shown). Comparable results were obtained with U87-MG cells (data not shown). Whole cell extracts from four additional glioblastoma lines, SNB-19, M059K, U138-MG, and U118-MG, demonstrated constitutive, although varying, levels of PIAS3 expression, which were unaffected by the presence of serum (Figure 2B). Primary human GBM cultures expressed PIAS3, and had non-detectable to low constitutive STAT-3 activation (Figure 2C), which is in contrast to the GBM tissues. These data suggest that regulation of the PIAS3 protein differs between GBM tissues and the in vitro systems tested, including cultures established from resected GBM tissues.
Figure 2. PIAS3 is Constitutively Expressed in Glioblastoma Cell Lines and Primary CNS Cells.<.
br>A. U251-MG cells were incubated in the absence or presence of IL-6 (10 ng/ml) plus sIL-6R (50 ng/ml) for up to 24 h, then cell lysates were assessed by immunoblotting for phospho-Y (705) STAT-3, STAT-3 and PIAS3 (Abgent, AP-1245a) protein expression. Immunoblots were re-probed for actin. Representative of three experiments. B. Constitutive PIAS3 protein expression in four glioblastoma cell lines, SNB-19, M059K, U113-MG and U118-MG, is demonstrated by immunoblotting (Abgent, AP-1245a). Cells were maintained in 10% FBS, or serum starved (no FBS) prior to collection. Blots were stripped and re-probed for actin to control for protein loading. C. Four primary human GBM cultures were analyzed for phospho-Y (705) STAT-3, phospho-S (727) STAT-3, STAT-3 and PIAS3 (Abgent, AP-1245a) protein expression by immunoblotting. Resected human GBM tissues were cultured, as described in Materials and Methods, and lysates collected for analysis. Anti-actin antibody was used to control for protein loading. D. SNB-19, U251-MG, and primary human GBM cells were incubated in the absence or presence of MG-132 (10 μM) for up to 6 h, then cell lysates were assessed by immunoblotting for PIAS3 (Abgent, AP-1245a) expression. Immunoblots were re-probed for actin. Data are representative of three independent experiments. Fold induction is calculated based on densitometry measurements, normalized to actin and compared to the untreated sample.
To further examine the in vitro expression pattern of PIAS3, primary murine astrocytes, murine microglia, and rat cortical neurons were examined (Supplemental Figure 1). In all CNS cell types, expression of PIAS3 was detected. Collectively, these data indicate that in vitro, human glioblastoma cell lines, as well as primary glial and neuronal cultures, show constitutive PIAS3 expression.
The discrepancy of PIAS3 protein expression patterns between primary GBM tissues and cell lines suggested that the absence of PIAS3 in vivo might be due to degradation. We tested this hypothesis utilizing the SNB-19 and U251-MG cell lines, as well as primary human GBM cultures. Treatment with MG-132, a classical inhibitor of the proteosome, resulted in an accumulation of the PIAS3 protein (Figure 2D), that increased over time in U251-MG cells. These results demonstrated that expression of PIAS3 in vitro is enhanced by inhibition of the proteosome, suggesting that ubiquitin-mediated degradation may promote the loss of PIAS3 in vivo.
STAT-3 is Activated by OSM in U251-MG Cells
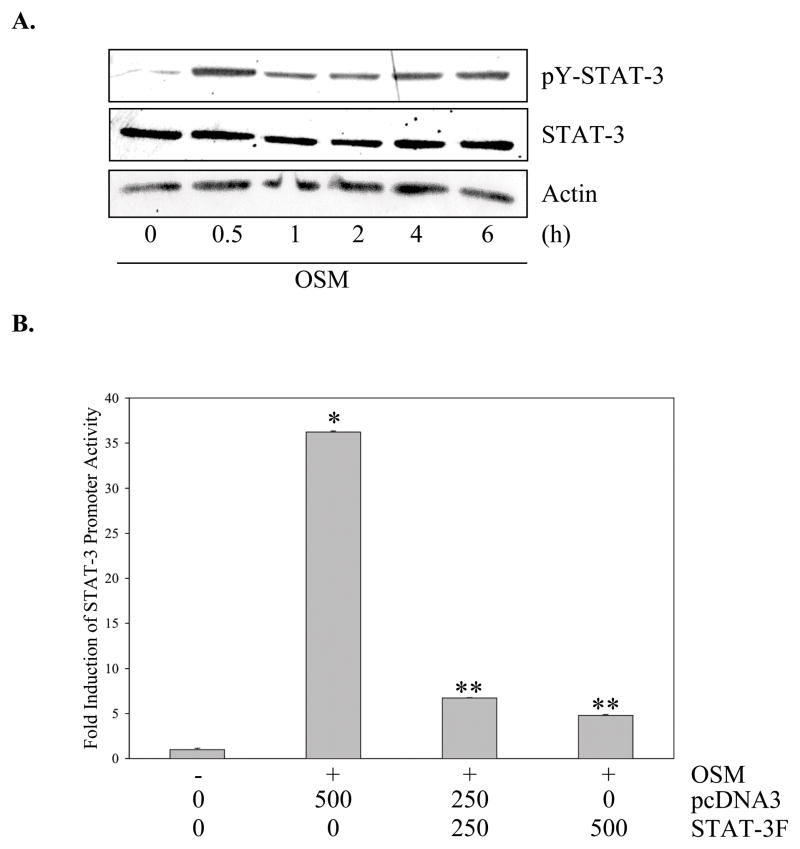
Having illustrated the activation of STAT-3 and loss of PIAS3 in vivo, the activation status of this signaling pathway was analyzed in vitro. In untreated U251-MG cells, STAT-3 had little to no basal tyrosine phosphorylation. Treatment with OSM induced STAT-3 tyrosine phosphorylation at 30 min, which persisted to 6 h (Figure 3A). Comparable results were observed using IL-6/sIL-6R (Figure 2A). Other GBM cell lines, including U87-MG, CH235-MG, and D54-MG, were tested for STAT-3 activation, and similar results were observed (data not shown). A measure of the transcriptional activity of STAT-3 is its ability to activate a STAT-3 driven promoter. Upon treatment with OSM, U251-MG cells demonstrated a 35-fold increase in STAT-3 promoter activity (Figure 3B), which was inhibited by a dominant-negative STAT-3 construct, STAT-3F (Figure 3B). Collectively, these data demonstrated that basal levels of STAT-3 activation in glioblastoma cells were minimal. Importantly, activation was observed upon OSM or IL-6/sIL-6R treatment, which resembled the pattern observed in human GBM tissues.
Figure 3. OSM Activates STAT-3 in Human Glioma Cells.
A. U251-MG cells were incubated in the absence or presence of OSM (50 ng/ml) for up to 6 h and cell lysates assessed by immunoblotting with phospho-Tyr (705) STAT-3 antibody. Blots were reprobed for total STAT-3 and actin. Representative of four experiments. B. U251-MG cells were transiently transfected with the 3xLy6epZLuc-TK (STAT-3) luciferase construct (500 ng) either alone or with pcDNA3 and/or STAT-3F (0–500 ng), and then recovered overnight before treatment with medium or OSM for 16 h. Lysates were analyzed in triplicate for luciferase activity and normalized to protein levels. Data shown are the mean ± S.D. from one of three independent experiments. *p≤0.002, compared to medium only condition. **p≤0.05, compared to OSM plus pcDNA3 (500 ng) condition.
siRNA Mediated Inhibition of PIAS3 Increases Proliferation
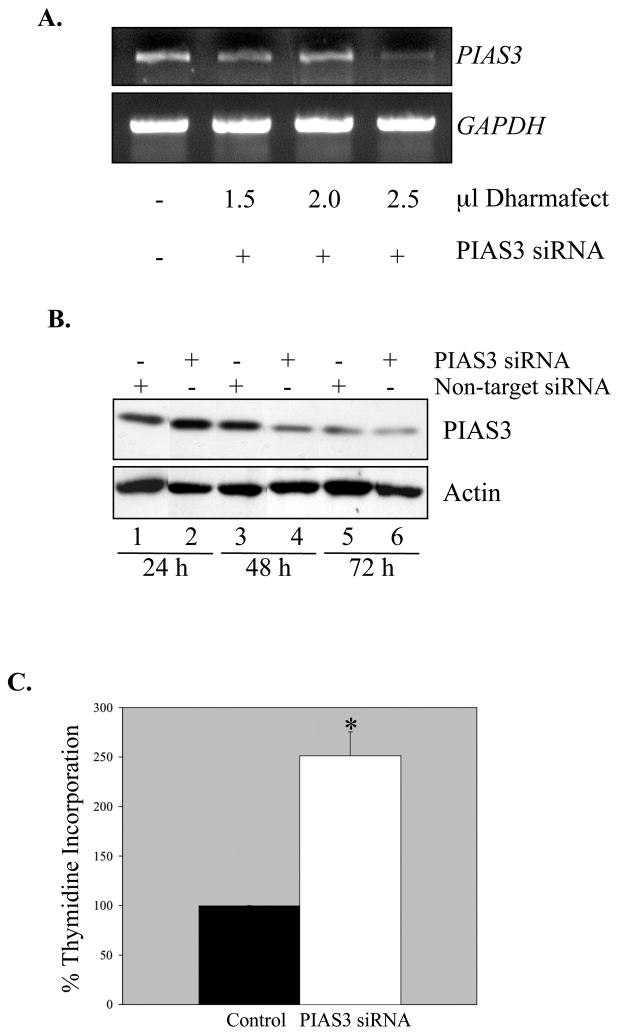
To mimic the observed in vivo loss of PIAS3, siRNA was used to inhibit PIAS3 expression in U87-MG cells. These cells were selected as they were the most amenable to siRNA knockdown. Transient transfection using PIAS3 siRNA demonstrated specific inhibition of PIAS3 mRNA at 24 h (Figure 4A). PIAS3 protein levels decreased over time in culture (Figure 4B, lanes 1, 3, and 5), however, PIAS3 knockdown was detected at 48 and 72 h (lanes 4 and 6). STAT-3 activation in the PIAS3 knockdown cells was examined, and minimal to no enhancement of basal tyrosine-phosphorylated STAT-3 was observed (data not shown), indicating that in cells with reduced PIAS3 expression, levels of phosphorylated STAT-3 remain relatively unchanged. Although activation of STAT-3 is measured by tyrosine and serine phosphorylation, one functional indicator of STAT-3 transcriptional activation is cell proliferation (30). We examined this parameter using the [3H]-TdR incorporation assay. Knockdown of PIAS3 resulted in a significant increase in proliferation of U87-MG cells (Figure 4C), indicating that in the absence of PIAS3, GBM cell proliferation is enhanced.
Figure 4. PIAS3 Inhibition Increases Proliferation of U87-MG Cells.
A. U87-MG cells were transfected using increasing amounts of transfection reagent and 100 nmol PIAS3 siPOOL reagent. 24 h post-transfection, RNA was collected and reverse transcribed to cDNA. Semi-quantitative PCR was performed to analyze levels of PIAS3and GAPDH mRNA. B. U87-MG cells were transfected using 2.5 μl of transfection reagent plus either 100 nmol non-target siRNA or 100 nmol PIAS3 siPOOL reagent. 24, 48, and 72 h post-transfection, lysates were assessed for PIAS3 protein by immunoblotting (Abgent, AP-1245a). Blots were reprobed for actin as loading and specificity controls. C. U87-MG cells were transfected as described above with either non-target siRNA (Control) or PIAS3 siRNA. 24 h post-transfection, cells were incubated in serum free media containing 1 μCi [3H]-TdR for 16 h, and then cell proliferation determined using the [3H]-TdR incorporation assay. Data are the mean ± S.D. of two experiments. Values are expressed as a percentage of the control. *p≤0.02, compared to control condition.
PIAS3 Over-expression Inhibits STAT-3 Mediated Gene Expression and Cell Proliferation
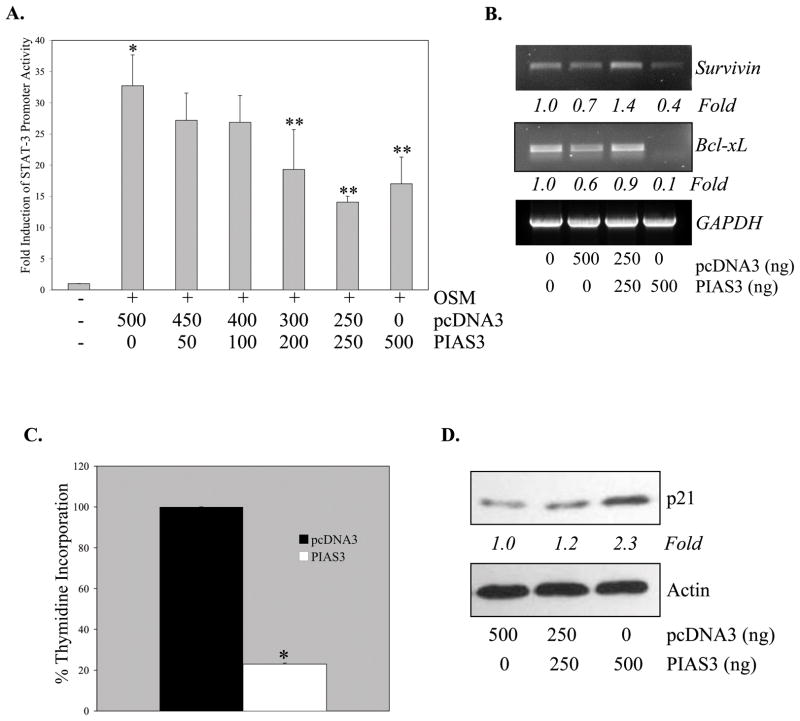
As a complement to PIAS3 knockdown, the effects of PIAS3 over-expression were examined. Ectopic expression of PIAS3 in U251-MG cells caused a significant dose-dependent inhibition of OSM-induced STAT-3 transcriptional activity (Figure 5A). STAT-3 in GBM cells is important for transcriptional induction of expression of anti-apoptotic genes, including Bcl-xL and Survivin (10, 31). Basal levels of both Survivin and Bcl-xL were inhibited by PIAS3 over-expression (Figure 5B). These results suggest that PIAS3 regulates the expression of genes important in cell survival.
Figure 5. Over-Expression of PIAS3 Inhibits STAT-3 Transcriptional Activity, STAT-3 Regulated Gene Expression and Cell Proliferation.
A. U251-MG cells were transiently transfected with the STAT-3 luciferase construct (500 ng) either alone or with pcDNA3 and/or PIAS3-pcDNA3 (0–500 ng), and recovered overnight before treatment with medium or OSM for 16 h. Lysates were analyzed in triplicate for luciferase activity and normalized to protein levels. Data shown are the mean ± S.D. from one of three independent experiments. *p≤0.002, compared to medium only. **p≤0.05, compared to OSM treatment. B. U251-MG cells were transiently transfected with pcDNA3 (0–500 ng) or PIAS3-pcDNA3 (0–500 ng). 24 h post-transfection, total RNA was collected and reverse transcribed to cDNA. Semi-quantitative PCR was performed to analyze levels of Survivin, Bcl-xL and GAPDH mRNA. Data are representative of two independent experiments. Fold induction is calculated based on densitometry measurements, normalized to GAPDH and compared to the untreated sample. C. U251-MG cells were transiently transfected with pcDNA3 or PIAS3-pcDNA3 (200 ng). 24 h post-transfection, cells were incubated in serum free media containing 1 μCi [3H]-TdR for 16 h, and then cell proliferation determined using the [3H]-TdR incorporation assay. Data are the mean ± S.D. of two experiments. Values are expressed as a percentage of the pcDNA3 control. *p≤0.0002, compared to pcDNA3 condition. D. U251-MG cells were transiently transfected with pcDNA3 (0–500 ng) or PIAS3-pcDNA3 (0–500 ng). 24 h post-transfection cell lysates were harvested and analyzed for p21 expression by immunoblotting. Immunblots were re-probed with actin. Data are representative of three independent experiments. Fold induction is calculated based on densitometry measurements, normalized to actin and compared to the pcDNA sample.
To test the functional significance of PIAS3 over-expression, cell proliferation was examined. Ectopic expression of PIAS3 resulted in ~80% reduction of cell proliferation (Figure 5C). To further explore the effects of PIAS3 on the cell cycle, the expression of p21, the cyclin dependent kinase inhibitor, was evaluated. Expression of the p21 protein was enhanced by approximately 2-fold upon PIAS3 ectopic expression (Figure 5D). Collectively, these data indicated that PIAS3 over-expression attenuated STAT-3 transcriptional activity, inhibited the expression of Survivin and Bcl-xL, and reduced the proliferation of glioma cells. The inhibition of cell proliferation may be due to enhanced expression of p21, which regulates the cell cycle at the transition from G1 to S phase.
PIAS3 Over-expression Inhibits SOCS-3, and SOCS-3 Expression is Elevated in Human GBM Tissues
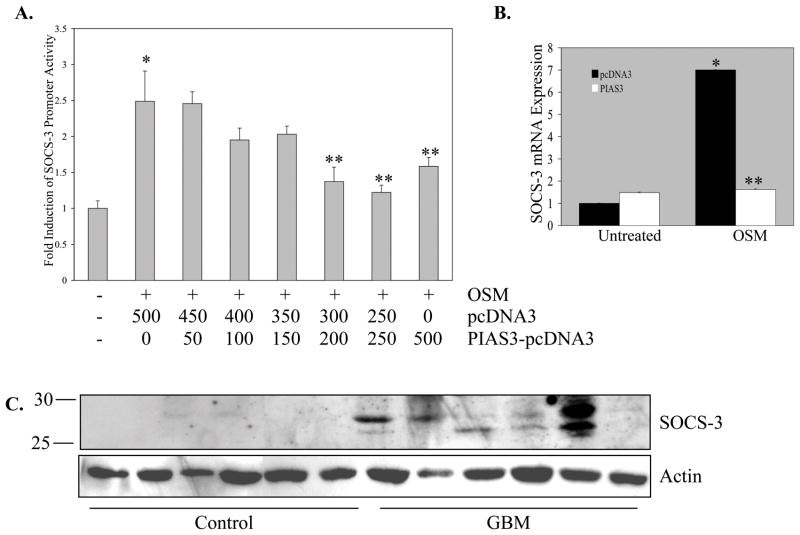
Unlike the constitutively expressed PIAS proteins, SOCS proteins are cytoplasmic, inducibly expressed proteins that act in a negative feedback loop to inhibit JAK activation. SOCS-3 is not only an endogenous inhibitor of STAT-3 (32), but also a STAT-3 transcriptional target (28). Recent investigations have revealed a role for SOCS-3 in cancer cells (33, 34). Ectopic PIAS3 expression repressed OSM activation of the SOCS-3 promoter, in a dose-dependent manner (Figure 6A). In addition, levels of OSM-induced SOCS-3 mRNA were significantly inhibited upon PIAS3 over-expression (Figure 6B). These results indicated that over-expression of PIAS3 can reduce STAT-3 transcriptional activity and inhibit the expression of the STAT-3 target gene, SOCS-3.
Figure 6. PIAS3 Over-expression Inhibits SOCS-3 Promoter Activity and mRNA Expression, and SOCS-3 Protein Expression is Elevated in Human GBM Tissues.
A. U251-MG cells were transiently transfected with the SOCS-3 promoter construct (500 ng) either alone or with pcDNA3 and/or PIAS3-pcDNA3 (0–500 ng), and recovered overnight before treatment with medium or OSM for 16 h. Lysates were analyzed in triplicate for luciferase activity and normalized to protein levels. Data shown are the mean ± S.D. from one of three independent experiments ± S.D. *p≤0.002, compared to medium only. **p≤0.05, compared to OSM treatment. B. U251-MG cells were transiently transfected with pcDNA3 or PIAS3-pcDNA3 (200 ng) and recovered overnight before treatment with either medium or OSM for 1 h. Total RNA was isolated. RNA was reverse transcribed to cDNA, which was analyzed by quantitative real-time PCR. SOCS-3 mRNA expression is represented as a ratio to the housekeeping gene, S9. Fold induction is represented as fold over untreated, pcDNA3-transfected condition. Data shown are the mean ± S.D. from one of two independent experiments. *p≤0.005, compared to pcDNA3 condition. **p≤0.05, compared to OSM plus pcDNA3 (500 ng) condition. C. Immunoblot of SOCS-3 protein in control brain (n=6) and GBM (n=6) lysates. Blots were probed for actin as a loading control.
Given the observation that activated STAT-3 levels were elevated, and PIAS3 levels were diminished in GBMs, we examined the expression of SOCS-3 in vivo. Enhanced levels of the SOCS-3 protein were observed in GBM tissues compared to controls (Figure 6C), with variability in expression levels. SOCS-3 protein expression was observed in 16/18 GBM tissues, while only 1/8 control samples expressed SOCS-3 (data not shown). These data demonstrated that SOCS-3, a STAT-3 transcriptional target gene, was elevated at the protein level in GBM tissues.
PIAS3 Inhibits Transcriptional Activation of the NF-κB Regulated Gene, MMP-9
PIAS3 is classically defined as an inhibitor of the JAK-STAT pathway, but its expression has recently been shown to modulate a number of signal transduction pathways, many of which are implicated in glioma cell biology, such as the NF-κB and TGF-β pathways (16). To test the effect of PIAS3 on the NF-κB pathway, we examined transcriptional activation of the matrix metalloproteinase-9 (MMP-9) promoter, as we have previously shown that MMP-9 gene expression is dependent on the NF-κB signaling pathway (35). We observed that ectopic expression of PIAS3 in U251-MG glioma cells inhibited PMA-induced activation of the MMP-9 promoter, in a dose-dependent manner (Supplemental Figure 2). These results confirm that PIAS3 expression modulates the transcriptional activation of NF-κB target genes, and describes this effect on the MMP-9 gene for the first time in human glioma cells.
DISCUSSION
The role of PIAS3 in human disease states is largely unclear. Although PIAS3 expression has been shown to correlate with malignancy (17, 36), other reports describe deficient PIAS3 expression in various human cancers (18). PIAS3 function has been examined in vitro using transformed cell lines. Infection of prostate cancer cells with adenovirus encoding PIAS3 increased apoptosis (19). Furthermore, PIAS3 over-expression prevented growth of prostate tumor xenografts in nude mice (19). Over-expression of PIAS3 in lung cancer cells led to inhibition of cell growth, increased susceptibility to chemotherapeutic agents, and suppression of Akt activation (20). Additionally, PIAS3 over-expression in COS-7 cells suppressed gene induction by v-src activated STAT-3 (37).
This study examined PIAS3 expression in human GBM tissues. The striking absence of PIAS3 protein in GBM tissues compared to control tissues was observed by both immunoblotting and immunohistochemistry (Figures 1C and 1D) and was confirmed by two PIAS3 antibodies, targeting different PIAS3 epitopes. Our results contrast those of Wang and Banerjee (2004), who detected PIAS3 in GBM specimens (17). However, they examined a very small number of GBM tissues and utilized a different PIAS3 antibody, which may account for the differences observed. There were no differences between control and GBM tissue expression of PIAS3 mRNA (Figure 1B), thus we postulated that the differences seen at the protein level were most likely due to post-translational changes to PIAS3. In the GBM tissue microenvironment, it is possible that PIAS3 was quickly degraded by the ubiquitin-proteosome system, releasing constraints on STAT-3 signal transduction. MG-132 treatment of primary GBM cells and glioma cell lines induced an appreciable accumulation of the PIAS3 protein in vitro (Figure 2D), supporting the notion that PIAS3 is rapidly degraded in GBM tissues. PIAS3 protein stability was shown to be regulated by nitric oxide (NO), which promoted its association with an E3 ubiquitin ligase and subsequent degradation (38). NO metabolism is closely associated with glioma progression, and the enzyme responsible for NO production has been detected in human glioma tissues (39). The loss of PIAS3 protein, either by rapid degradation or another mechanism, may allow activated STAT-3 to sustain transcription of genes which control proliferation and the cell cycle. Proteins including p53 and p27Kip-1 are destabilized and degraded by ubiquitination in a variety of cancers (40). The loss of PIAS3 protein provides a mechanism to promote aberrant STAT-3 signal transduction in GBM tissues, which is a downstream target of mutated EGFR, VEGFR, and the IL-6 cytokine family (11, 12, 41). The constitutive expression of PIAS3 provides a unique role for this protein to act as a “buffer” to inhibit sustained STAT-3 transcriptional activity, and in the absence of PIAS3, in conjunction with aberrant growth factor signal transduction, activation of STAT-3 and enhanced STAT-3 transcriptional activity is observed in GBMs.
PIAS3 expression was inhibited in astroglioma cells using siRNA to mimic the in vivo loss observed in GBM tissue. PIAS3 knockdown resulted in a significant increase in proliferation of astroglioma cells (Figure 4C), but did not affect the overall levels of phosphorylated STAT-3 (data not shown). These data support the previous findings of Herrmann et. al. (42) which demonstrate that PIAS3 expression does not modulate cellular levels of phosphorylated STAT-3, but does influence its ability to activate transcription. Accordingly, over-expression of PIAS3 resulted in significant inhibition of STAT-3 transcriptional activity and down-regulation of Survivin and Bcl-xL (Figures 5A and 5B). Attenuation of cell proliferation (Figure 5C) and enhanced p21 protein expression (Figure 5D) was observed in cells that ectopically expressed PIAS3. These data support the hypothesis that PIAS3 influences the expression of genes which control cell proliferation and survival. Thus, the PIAS3 protein exhibits a number of anti-tumorigenic functional properties important for restraining GBM cell growth and gene expression.
Although SOCS-3 is a negative regulator of the STAT-3 signaling pathway, it is also a STAT-3 transcriptional target. Over-expression of the PIAS3 protein caused a significant reduction in SOCS-3 promoter activity and SOCS-3 mRNA expression (Figures 6A and 6B), demonstrating SOCS-3 transcriptional repression when PIAS3 is over-expressed. Because SOCS-3 promoter activity and mRNA expression were negatively affected by PIAS3 over-expression, we examined its expression in human tissues, and discovered SOCS-3 aberrant expression in GBM tissues (Figure 6C). SOCS-3 is a member of the SOCS protein family, which is comprised of CIS and SOCS-1-7 (32). SOCS-3 is a cytokine-inducible endogenous inhibitor of STAT-3 that works in a classical negative feedback loop (32), but it has recently been described to have a complicated role in cancer cell signaling. A tumor-suppressing function of SOCS-3 was suggested by reports describing hypermethylation of SOCS-3, and subsequent loss of expression, in a variety of cancers (43). This defective expression of SOCS-3, along with loss of feedback inhibition of STAT-3 activation, may contribute to tumor progression. However, other reports revealed that aberrant SOCS-3 expression may actually promote tumorigenesis. Elevated SOCS-3 expression was reported in human breast cancer and melanoma tissues, as well as primary lymphoma cells (44, 45). Further, growth and proliferation was markedly induced in cancer cells that over-expressed SOCS-3 (33). SOCS-3 overexpression was attributed to constitutive STAT-3 activation in various cancers (46). Interestingly, human GBM tissues were recently reported to constitutively express SOCS-3, which correlated with enhanced cell growth/survival and radioresistance (34). Because SOCS-3 is a transcriptional target of STAT-3, the parallel, elevated expression of both activated STAT-3 and SOCS-3 proteins in GBM is plausible, and may contribute to the anti-apoptotic and radioresistant phenotype of GBMs.
In many cancers, including GBMs, mechanisms to regulate STAT-3 activity have failed, thus enabling activated STAT-3 to function as a tumor-promoter (30). Interestingly, recent data showed that STAT-3 served conflicting roles either acting as a tumor suppressor or promoter, depending on the genetic profile of the tumor (47), furthering the customized, clinical use of STAT-3 inhibitors as potential therapeutics. Abrogation of STAT-3 activity by a variety of inhibitors such as AG490 and WP1066 inhibited GBM cell growth, promoted apoptosis, and suppressed expression of anti-apoptotic genes (3, 10, 48). The loss of PIAS3 expression observed in GBMs may be a contributing factor to the STAT-3 transcriptional activation observed in this cancer. More recently, PIAS3 was shown to regulate other signaling pathways, including the NF-κB, PI3-K/AKT and TGF-β pathways (16, 20). PIAS3 interacted with NF-κB p65 and repressed its transcriptional activity, thereby functioning as a negative regulator of NF-κB (49). Supporting this finding, we showed that transcriptional activation of the NF-κB regulated gene, MMP-9, is inhibited by PIAS3 over-expression in glioma cells (Supplemental Figure 2), linking PIAS3 expression to inhibition of NF-κB signaling for the first time in glioma cell biology. Furthermore, others have shown that PIAS3 interacted with AKT in vitro, suppressing its phosphorylation and activation (20). PIAS proteins were also implicated in either positively or negatively regulating SMAD transcriptional activity, which mediate TGF-β biological activities (16). The aberrant activation of the NF-κB, PI3-K, and TGF-β pathways have been correlated with poor prognosis in patients with GBMs (12, 29, 50). Thus, the loss of PIAS3 expression in GBMs has implications for multiple signaling pathways contributing to gliomagenesis.
Supplementary Material
CLINICAL RELEVANCE.
STAT-3 is a cytoplasmic transcription factor that becomes activated in response to a variety of cytokines, chemokines, and growth factors. STAT-3 is involved in mediating cell proliferation, immune evasion, and anti-apoptotic behavior. STAT-3 is aberrantly activated in a number of human cancers, including Glioblastoma Multiforme (GBM). STAT-3 is a promising target for GBM therapy because it is a convergence point for several signaling pathways that promote glioma growth and maintenance, and because aberrant STAT-3 activation results from upstream dysregulation, not constitutively active STAT-3 mutations. STAT-3 inhibition has been achieved through inhibition of upstream kinases, including the JAKs and receptor tyrosine kinases, such as EGFR and VEGFR. Pharmacologic inhibitors including AG490, WP1066, cucurbitacin I, and gefitinib have shown promising results in glioblastoma cells in vitro and are in early stages of clinical trials. PIAS3, a negative regulator of STAT-3, is a protein whose function is of great importance in understanding the regulation of STAT-3 signal transduction in vivo. The work presented here provides evidence of PIAS3 dysregulation in GBM, and the promotion of STAT-3 transcriptional activity in human GBM. The presence or absence of PIAS3 may determine GBM patients that would be responsive to STAT-3 inhibitors in future clinical trials.
Acknowledgments
This work was supported in part by National Institutes of Health grants P50 CA-097247 (to E.N.B., L.B.N., C.A.P., and G.Y.G.), CA-112397 (to L.B.N.) and NS-54158 (to E.N.B.). K.R. was supported by R25 CA-47888, UAB Cancer Prevention and Control Training Program.
The authors thank Dr. Susan Nozell for the PIAS3-pcDNA3 construct and technical assistance, Dr. Hongwei Qin for the SOCS-3 promoter construct and helpful advice, and Dr. Sun-Jung Lee for assistance in manuscript preparation. We acknowledge Dr. Kevin Roth and Cecelia Latham of the UAB Neuroscience Core Facility (P30 NS-47466) for technical assistance and discussions. We also acknowledge Drs. Hirano, Hibi, and Nakajima (Osaka University Medical School, Osaka, Japan) for kindly providing the STAT-3F construct and Dr. D. Boyd (MD Anderson Cancer Center, Houston, TX) for the MMP-9 luciferase promoter construct.
References
- 1.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol (Berl) 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 2.Rao RD, James CD. Altered molecular pathways in gliomas: an overview of clinically relevant issues. Semin Oncol. 2004;31:595–604. doi: 10.1053/j.seminoncol.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Brantley EC, Benveniste EN. STAT-3: A molecular hub for signal transduction pathways in GBM. Molecular Cancer Research. 2008 doi: 10.1158/1541-7786.MCR-07-2180. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–11. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 5.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedranzini L, Leitch A, Bromberg J. Stat3 is required for the development of skin cancer. J Clin Invest. 2004;114:619–22. doi: 10.1172/JCI22800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 8.Repovic P, Fears CY, Gladson CL, Benveniste EN. Oncostatin-M induction of vascular endothelial growth factor expression in astroglioma cells. Oncogene. 2003;22:8117–24. doi: 10.1038/sj.onc.1206922. [DOI] [PubMed] [Google Scholar]
- 9.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–42. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–13. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 11.Schaefer LK, Ren Z, Fuller GN, Schaefer TS. Constitutive activation of Stat3alpha in brain tumors: localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2) Oncogene. 2002;21:2058–65. doi: 10.1038/sj.onc.1205263. [DOI] [PubMed] [Google Scholar]
- 12.Mizoguchi M, Betensky RA, Batchelor TT, Bernay DC, Louis DN, Nutt CL. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol. 2006;65:1181–8. doi: 10.1097/01.jnen.0000248549.14962.b2. [DOI] [PubMed] [Google Scholar]
- 13.Weissenberger J, Loeffler S, Kappeler A, et al. IL-6 is required for glioma development in a mouse model. Oncogene. 2004;23:3308–16. doi: 10.1038/sj.onc.1207455. [DOI] [PubMed] [Google Scholar]
- 14.Chen SH, Benveniste EN. Oncostatin M: a pleiotropic cytokine in the central nervous system. Cytokine Growth Factor Rev. 2004;15:379–91. doi: 10.1016/j.cytogfr.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Loeffler S, Fayard B, Weis J, Weissenberger J. Interleukin-6 induces transcriptional activation of vascular endothelial growth factor (VEGF) in astrocytes in vivo and regulates VEGF promoter activity in glioblastoma cells via direct interaction between STAT3 and Sp1. Int J Cancer. 2005;115:202–13. doi: 10.1002/ijc.20871. [DOI] [PubMed] [Google Scholar]
- 16.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep. 2004;11:1319–24. [PubMed] [Google Scholar]
- 18.Zhang Q, Raghunath PN, Xue L, et al. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive T/null-cell lymphoma. J Immunol. 2002;168:466–74. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- 19.Wible BA, Wang L, Kuryshev YA, Basu A, Haldar S, Brown AM. Increased K+ efflux and apoptosis induced by the potassium channel modulatory protein KChAP/PIAS3beta in prostate cancer cells. J Biol Chem. 2002;277:17852–62. doi: 10.1074/jbc.M201689200. [DOI] [PubMed] [Google Scholar]
- 20.Ogata Y, Osaki T, Naka T, et al. Overexpression of PIAS3 suppresses cell growth and restores the drug sensitivity of human lung cancer cells in association with PI3-K/Akt inactivation. Neoplasia. 2006;8:817–25. doi: 10.1593/neo.06409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross M, Liu B, Tan J, French FS, Carey M, Shuai K. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene. 2001;20:3880–7. doi: 10.1038/sj.onc.1204489. [DOI] [PubMed] [Google Scholar]
- 22.Shrikant P, Weber E, Jilling T, Benveniste EN. Intercellular adhesion molecule-1 gene expression by glial cells. Differential mechanisms of inhibition by IL-10 and IL-6. J Immunol. 1995;155:1489–501. [PubMed] [Google Scholar]
- 23.MacCumber MW, Ross CA, Snyder SH. Endothelin in brain: receptors, mitogenesis, and biosynthesis in glial cells. Proc Natl Acad Sci U S A. 1990;87:2359–63. doi: 10.1073/pnas.87.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen SH, Gillespie GY, Benveniste EN. Divergent effects of oncostatin M on astroglioma cells: influence on cell proliferation, invasion, and expression of matrix metalloproteinases. Glia. 2006;53:191–200. doi: 10.1002/glia.20264. [DOI] [PubMed] [Google Scholar]
- 25.Qin H, Wilson CA, Roberts KL, Baker BJ, Zhao X, Benveniste EN. IL-10 inhibits lipopolysaccharide-induced CD40 gene expression through induction of suppressor of cytokine signaling-3. J Immunol. 2006;177:7761–71. doi: 10.4049/jimmunol.177.11.7761. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima K, Yamanaka Y, Nakae K, et al. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. Embo J. 1996;15:3651–8. [PMC free article] [PubMed] [Google Scholar]
- 27.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 28.Qin H, Roberts KL, Niyongere SA, Cong Y, Elson CO, Benveniste EN. Molecular mechanism of lipopolysaccharide-induced SOCS-3 gene expression in macrophages and microglia. J Immunol. 2007;179:5966–76. doi: 10.4049/jimmunol.179.9.5966. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest. 2004;84:941–51. doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 31.Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23–31. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–65. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 33.Komyod W, Bohm M, Metze D, Heinrich PC, Behrmann I. Constitutive suppressor of cytokine signaling 3 expression confers a growth advantage to a human melanoma cell line. Mol Cancer Res. 2007;5:271–81. doi: 10.1158/1541-7786.MCR-06-0274. [DOI] [PubMed] [Google Scholar]
- 34.Zhou H, Miki R, Eeva M, et al. Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res. 2007;13:2344–53. doi: 10.1158/1078-0432.CCR-06-2303. [DOI] [PubMed] [Google Scholar]
- 35.Ma Z, Shah RC, Chang MJ, Benveniste EN. Coordination of cell signaling, chromatin remodeling, histone modifications, and regulator recruitment in human matrix metalloproteinase 9 gene transcription. Mol Cell Biol. 2004;24:5496–509. doi: 10.1128/MCB.24.12.5496-5509.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHale K, Tomaszewski JE, Puthiyaveettil R, Livolsi VA, Clevenger CV. Altered expression of prolactin receptor-associated signaling proteins in human breast carcinoma. Mod Pathol. 2008 doi: 10.1038/modpathol.2008.7. Advance Online Publication. [DOI] [PubMed] [Google Scholar]
- 37.Herrmann A, Vogt M, Monnigmann M, et al. Nucleocytoplasmic shuttling of persistently activated STAT3. J Cell Sci. 2007;120:3249–61. doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- 38.Qu J, Liu GH, Wu K, et al. Nitric oxide destabilizes pias3 and regulates sumoylation. PLoS ONE. 2007;2:e1085. doi: 10.1371/journal.pone.0001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam-Himlin D, Espey MG, Perry G, Smith MA, Castellani RJ. Malignant glioma progression and nitric oxide. Neurochem Int. 2006;49:764–8. doi: 10.1016/j.neuint.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–89. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 41.Rao RD, Mladek AC, Lamont JD, et al. Disruption of parallel and converging signaling pathways contributes to the synergistic antitumor effects of simultaneous mTOR and EGFR inhibition in GBM cells. Neoplasia. 2005;7:921–29. doi: 10.1593/neo.05361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herrmann A, Vogt M, Monnigmann M, et al. Nucleocytoplasmic shuttling of persistently activated STAT3. J Cell Sci. 2007 doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland KD, Lindeman GJ, Choong DY, et al. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004;23:7726–33. doi: 10.1038/sj.onc.1207787. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi K, Sakai I, Narumi H, et al. Expression of SOCS3 mRNA in bone marrow cells from CML patients associated with cytogenetic response to IFN-alpha. Leuk Res. 2005;29:173–8. doi: 10.1016/j.leukres.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Fojtova M, Boudny V, Kovarik A, et al. Development of IFN-γ resistance is associated with attenuation of SOCS genes induction and constitutive expression of SOCS3 in melanoma cells. Br J Cancer. 2007;97:231–37. doi: 10.1038/sj.bjc.6603849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho-Vega JH, Rassidakis GZ, Amin HM, et al. Suppressor of cytokine signaling 3 expression in anaplastic large cell lymphoma. Leukemia. 2004;18:1872–8. doi: 10.1038/sj.leu.2403495. [DOI] [PubMed] [Google Scholar]
- 47.de la Iglesia N, Konopka G, Puram SV, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–62. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwamaru A, Szymanski S, Iwado E, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–44. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 49.Jang HD, Yoon K, Shin YJ, Kim J, Lee SY. PIAS3 suppresses NF-kappaB-mediated transcription by interacting with the p65/RelA subunit. J Biol Chem. 2004;279:24873–80. doi: 10.1074/jbc.M313018200. [DOI] [PubMed] [Google Scholar]
- 50.Bruna A, Darken RS, Rojo F, et al. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–60. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.