Abstract
Background
Human ascending thoracic aortic aneurysms (ATAAs) are life threatening and constitute a leading cause of mortality in the United States. Previously, we demonstrated that collagens α2(V) and α1(XI) mRNA and protein expression levels are significantly increased in ATAAs.
Methods and Results
In this report, the authors extended these preliminary studies using high‐throughput proteomic analysis to identify additional biomarkers for use in whole blood real‐time RT‐PCR analysis to allow for the identification of ATAAs before dissection or rupture. Human ATAA samples were obtained from male and female patients aged 65±14 years. Both bicuspid and tricuspid aortic valve patients were included and compared with nonaneurysmal aortas (mean diameter 2.3 cm). Five biomarkers were identified as being suitable for detection and identification of ATAAs using qRT‐PCR analysis of whole blood. Analysis of 41 samples (19 small, 13 medium‐sized, and 9 large ATAAs) demonstrated the overexpression of 3 of these transcript biomarkers correctly identified 79.4% of patients with ATAA of ≥4.0 cm (P<0.001, sensitivity 0.79, CI=0.62 to 0.91; specificity 1.00, 95% CI=0.42 to 1.00).
Conclusion
A preliminary transcript biomarker panel for the identification of ATAAs using whole blood qRT‐PCR analysis in men and women is presented.
Keywords: ascending thoracic aortic aneurysm, collagen, FHL1
Introduction
Ascending thoracic aortic aneurysms (ATAAs) are a major cause of morbidity and mortality worldwide, especially in those older than 65 years.1–3 Both men and women are affected, with a considerably increased risk in men compared with women.4–5 While ATAAs can occur as a result of genetic diseases such as Marfan, Ehlers‐Danlös and Loeyes‐Dietz syndromes, the most common cause of ATAA is through cardiovascular malformation of the aortic valve. This malformation resulting in a bicuspid aortic valve (BAV), has a heritability of 89%, and has been shown to affect 1% to 2% of the population, and it is known to be associated with the development of ATAA .5 The vast majority of ATAAs, however, are unrelated to aortic valve malformation or heritable degenerative disease and occur in patients with a normal cardiovascular formation, with a normal tricuspid aortic valve (TAV).
The presentation of ATAAs is varied and often silent. Thus, ATAA detection is often fortuitous, with identification occurring during a routine physical examination or during an unrelated medical evaluation. Once suspected, confirmation by radiography, magnetic resonance imaging, computed tomography scanning, or ultrasound is needed to allow for elective surgery or endovascular repair before dissection or rupture.
At present, there are no biomarkers available to identify ATAAs before visible symptoms, dissection, or rupture. The development of a simple and rapid detection methodology to identify ATAAs that can be performed at major centers and at centers lacking sophisticated visual technologies is therefore needed. In this report, we use proteomic analysis of ATAA tissue to first identify proteins altered in ATAA patients compared with controls. These proteins allowed for the identification of annotation clusters and canonical pathways associated with ATAA in tissue biopsies. This discovery phase, or “sieving approach,” allowed for the identification of putative genetic markers of interest.
This approach has been previously used by Liew and colleagues, who have demonstrated that whole blood provides a useful complement for tissue biopsy.6–7 Their work and that of others have demonstrated that gene profiling in the blood identifies secreted protein transcripts at the tissue level.8–9 The mechanism for these alterations is the result of the close association of blood with the tissue. When tissue is damaged, the genomic pattern in blood is altered to reflect that in the affected organ. Thus, the blood acts as a bioinformational scout to determine the disease state in the body.10 This approach has been successfully used for the diagnosis of heart failure,11 Chagas' disease cardiomyopathy,12 cancer,10,13 autoimmune diseases,14 osteoarthritis, rheumatoid arthritis, and Crohn's disease.15 Using this approach, we have identified a preliminary cohort of transcript biomarkers allowing for the rapid identification of ATAA in whole blood using real‐time RT‐PCR analysis.
Methods
Study Design
Aortic specimens were collected from male and female control and ATAA patients. Both TAV and BAV ATAA specimens were obtained. Protein was isolated from 6 control and 18 ATAA samples and used for proteomic analysis to identify possible biomarkers. Proteomic analysis was confirmed by Western blot analysis. Transcripts of identified proteins up‐ or down‐regulated in ATAA were evaluated as possible biomarkers (circulating transcript biomarkers). Following identification of putative transcript biomarkers, qRT‐PCR analysis with whole blood was performed on 6 age‐matched healthy volunteers (3 men [aged 63±13 years] and 3 women [aged 61±15 years]) and 41 ATAA patients (aged 65.3±13.9 years) consisting of 13 of the 18 ATAA samples used for proteomic analysis and 28 ATAA patients. All samples were unique to this study. Statistical analysis was performed on each group and on the overall group in comparison with controls.
Aortic Specimens and Patient Characteristics
Aortic specimens were collected during a 2‐year period (October 2009 to September 2011) from patients with non‐AATAs undergoing heart transplantation and patients undergoing replacement of ATAA. Six specimens were obtained from patients (3 men and 3 women, mean age 62.5±7.8 years) who underwent heart transplantation procedures with normal ascending aortic diameters (mean diameter 2.3 cm) and were used as controls. ATAA samples (n=41; aged 65.3±13.9 years; mean diameter 5.3±0.8 cm, P<0.001 versus controls) were obtained at the time of surgery. The ATAA patients included 35 men (aged 64.0±14.2 years) and 6 women (aged 73.0±10.1 years). Patients had either bicuspid aortic valves (BAVs) (n=17; aged 55.6±12.9 years) or tricuspid aortic valves (TAVs) (n=24; aged 71.8±10.8 years) and were subdivided into 3 groups according to the maximal diameter of the aneurysm: small, with diameters between 4.0 and 4.9 cm (n=19; 3 woman and 16 men); medium sized, between 5.0 and 5.5 cm (n=13; 1 woman and 12 men); and large, with diameters >5.5 cm (n=9; 2 women and 7 men).
All patients with ATAA underwent elective surgery and required graft replacement of the ascending aorta. Aortic samples from patients with aneurysms secondary to genetic syndromes such as Marfan, Ehlers‐Danlös, and Loeys‐Dietz syndromes or with other cardiovascular diseases, cancer, or chronic disease were excluded from this study. Aortic replacement was indicated when the size of the aorta was 5.5 cm for TAV or 5.0 cm for BAV. If valve surgery was indicated, the standard size for aortic replacement was decreased dependent on tricuspid valve, younger age, or thin aortic wall (4 to 4.5 cm). None of the specimens analyzed were atherosclerotic and none were lined with thrombus. Control specimens were taken from donors to ensure that these aortas would have minimal alterations in matrix composition.
Aortic Specimens
Research protocols were approved by the Institutional Review Board at Beth Israel Deaconess Medical Center, and informed consents were obtained under Institutional Review Board No. 2007P‐000252. Full‐thickness biopsy samples containing all 3 layers of the aorta (tunica adventitia, tunica media, and tunica intima) were collected from the right‐lateral aspect of the ascending aorta (the greater curvature, roughly in line with the commissure between the right and noncoronary sinuses) in the operating room, fresh frozen, and stored at −80°C until analysis.
Proteomic Analysis
Proteomic analysis was performed on 6 control patients (3 men, 3 women; mean age 62.5±7.8 years) who underwent heart transplantation procedures with normal ascending aortic diameters (mean diameter 2.3 cm). ATAA samples were obtained from 18 patients and sorted by size. Each group contained 6 samples consisting of 3 men and 3 women. Each group consisted of 3 TAV and 3 BAV specimens. Mean age was 65±14 years in controls, 60±16 years in patients with small ATAAs (4.0 to 4.9 cm), 59±8 years in patients with medium‐sized ATAAs (5.0 to 5.5 cm), and 60±16 years in patients with large ATAAs (<5.5 cm). Total aortic tissue protein was isolated, and quality and purity were assessed by SDS‐PAGE, as previously described.1 High‐throughput profiling of protein samples was performed at the Genomics and Proteomics Center Core facility of the Beth Israel Deaconess Medical Center using the 8‐plex iTRAQ (AB Sciex, Foster City, CA) labeling protocol and standard MudPIT methodology coupled with the 4800 MALDI TOF/TOF Plus instrument.16
Unsupervised and Quality Control Analysis of Proteomic Data
Raw data were analyzed by using ProteinPilot v3.0 software with the Paragon algorithm (AB Sciex). Searches were performed against the latest available FASTA format fully annotated SwissProt human protein database (UniProtKB/Swiss‐Prot; release 2010–12 [January 18, 2011 to February 7, 2011] downloaded from the website: http://www.uniprot.org/downloads). Proteins with confidence score >90% and with at least 2 peptides of 95% identification confidence were used for quality control and differential expression analysis.
Supervised Analysis
To identify the differentially expressed proteins, the relative protein expression values were compared between groups (small ATAAs versus controls, medium‐sized ATAAs versus controls, large ATAAs versus controls). Control samples were matched for age and sex and were used to identify constitutive protein expression. Proteins were considered overexpressed in ATAAs relative to control if the iTRAQ ratio of ATAA to control was >2.0 and if the corresponding maximum control‐to‐control ratio was less than the ATAA‐to‐control ratio. Similarly, proteins were considered underexpressed in ATAA relative to control if the iTRAQ ratio of ATAA to control was <0.5 and if the corresponding minimum control‐to‐control ratio was higher than the ATAA‐to‐control ratio.
Western Blotting
Western blotting was used to confirm proteomic results. Protein samples (25 μg) from full‐thickness aortic tissue from the site of the maximal diameter containing all 3 layers were fractionated on 10% Novex Tris‐glycine gels (Invitrogen) and then electroblotted onto nitrocellulose membranes (Invitrogen). Protein equivalency, transfer efficiency, membrane blocking, immunoblotting, detection, and densitometry analysis (Image J analysis software; http://rsbweb.nih.gov/ij/) were performed as previously described.1
RNA Isolation
Whole blood was collected into PAXgene Blood RNA Tubes and stored at 4°C until use. RNA was isolated from whole blood within 5 days of collection. Total RNA was isolated using the PAXgene Blood RNA Kit (PreAnalytiX, A Qiagen/BD Company; http://www.preanalytix.com/).
Real‐Time RT‐PCR
qRT‐PCR analysis was performed on 13 of the ATAA samples used for proteomic analysis and an additional 28 ATAA samples (see Aortic Specimens and Patient Characteristics). qRT‐PCR analysis was performed using an Eppendorf Realplex2 Mastercycler and software package (Eppendorf North America). The iScript One‐Step RT‐PCR Kit with SYBR Green solution (Bio‐Rad) was used for the qRT‐PCRs. In brief, 100 ng of total RNA and 600 nmol/L of both forward and reverse primers were added to each reaction. No threshold cycle for any probe was >32. Control and ATAA samples were run for each primer set in triplicates. Control reactions without reverse transcriptase were also performed for each reaction.16
Oligonucleotide primers (Table 1) were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and were synthesized by Oligos ETC, Inc.
Table 1.
Oligonucleotide Primers Used for Quantitative Real‐Time RT‐PCR
| mRNA Target | Forward Primer | Reverse Primer |
|---|---|---|
| 18s | 5′‐ATGGCCGTTCTTAGTTGGTG‐3′ | 5′‐CGCTGAGCCAGTCAGTGTAG‐3′ |
| FHL1 | 5′‐CCTGGTCTAGGCCATCACAT‐3′ | 5′‐TCTTGCATCCAGCACACTTC‐3′ |
| ENOA1 | 5′‐GGAGCTTGGCAGAAGTTCAC‐3′ | 5′‐TGATTCACAAGCCGTAGCTG‐3′ |
| CSRP1 | 5′‐AAGTCCTGCTACGGCAAGAA‐3′ | 5′‐CACCAATCTTCTGGGCAAAT‐3′ |
| CRP2 | 5′‐CCCACCTGCCAGTGTTATTT‐3′ | 5′‐TTGACAGCACAAGGCTCAAC‐3′ |
| PPIA | 5′‐TGTTTGTGGTTGCCAGTCAT‐3′ | 5′‐TCGAGTTGTCCACAGTCAGC‐3′ |
| Col α1(I) | 5′‐CTCTGACTGGAAGAGTGGAGAGTA‐3′ | 5′‐TTGGTGGTTTTGTATTCAATCACT‐3′ |
| Col α1(III) | 5′‐AGTGACCGACAAAATTCCAGTTAT‐3′ | 5′‐CTTTTACTGGTGAGCACAGTCATT‐3′ |
| Col α2(V) | 5′‐TGAGTTGTGGAGCTGACTCTAATC‐3′ | 5′‐TAACAGAAGCATAGCACCTTTCAG‐3′ |
| Col α1(XI) | 5′GAAATTGTACCTTGGTGCCACCAAC‐3′ | 5′‐GGATGGATGAGAATGAGCACCATAT‐3′ |
Col α1(I) indicates collagen α1(I); Col α1(III), collagen α1(III); Col α2(V), collagen α2(V); Col α1(XI), collagen α1(XI); CRP2, cysteine‐rich protein 2; CSRP1, cysteine‐ and glycine‐rich protein 1; ENOA1, α‐enolase 1; FHL1, four and a half LIM domains protein 1; PIPA, peptidyl‐prolyl cis‐trans isomerase A; 18s, ribosomal 18s.
Reaction kinetics were optimized for each primer set. RT of RNA template occurred at 50°C for 10 minutes with inactivation at 95°C for 5 minutes. Amplification and detection occurred over 40 cycles (denaturing 95°C; 10 seconds, annealing 60°C; 10 seconds, extension 72°C; 20 seconds, with plate read at 78°C). Melting curves were performed for each reaction at the conclusion of the cycling parameters from 60° to 95°C. Fold changes in gene expression were calculated using the ΔΔCT (ddCT) method.17 Fold change was determined using the formula: fold change=2−[(CT target gene−CT 18s) sample−(CT target gene−CT 18s) control].
Statistical Analysis
Statistical analysis was performed using SAS (version 6.12) software package (SAS Institute). The mean±SEM for all data was calculated for all variables. Statistical differences between groups were evaluated by 1‐way ANOVA. One‐way ANOVA was used for comparison of patient's demographics. Dunnett's test was used for comparisons between control and other groups to adjust for the multiplicity of tests. Fisher's exact test was used for comparisons of transcript biomarkers and ATAA prevalence compared with control. Power analysis was calculated with G*Power 3.1.5 (www.psycho.uni-duesseldorf.de/abteilungen/app/gpower3/download-and-register) (at test positive rate of control 0.1, test positive rate of ATAA 0.8, α error 0.05, β error 0.05, number needed for control 6, number needed for ATAA 36). Sensitivity and specificity of qRT‐PCR test for each gene or for combination of genes were calculated with 95% CIs. Sensitivity/specificity results were measured as up‐regulated transcript biomarker within diseased patients (sensitivity) and down‐regulated transcript biomarker within control patients (specificity) compared with controls.
Results
Proteomic Analysis Sample Groups
Samples for proteomic analysis were obtained from 6 control patients (3 men and 3 women, mean age 62.5±7.8 years) who underwent heart transplantation procedures with normal ascending aortic diameters (mean diameter 2.3 cm). ATAA samples were obtained from 18 patients and sorted by size. Each group contained 6 samples, from 3 men and 3 women. Each group consisted of 3 TAV and 3 BAV specimens. Mean age was 65±14 years in controls, 60±16 years in patients with small ATAAs (4.0 to 4.9 cm), 59±8 years in patients with medium‐sized ATAAs (5.0 to 5.5 cm), and 60±16 years in patients with large ATAAs (<5.5 cm).
Proteomic Analysis
A total of 3396 proteins were identified with valid iTRAQ labeling, of which 369 were identified with >95% confidence (P<0.05). From these, 82 proteins were found to be commonly expressed in all ATAA size groups and controls.
Principal component analysis (PCA) is shown in Figure 1A. PCA projects multivariate data objects onto a lower‐dimensional space while retaining as much of the original variance as possible. This is necessary because in analyzing proteomic data; due to a dimensionality problem, the number of proteins always considerably exceeds the number of samples. Each principal component is associated with an eigenvalue, which corresponds to the amount of variability explained by the corresponding principal component. An eigenvalue is a measure of strength of each component.
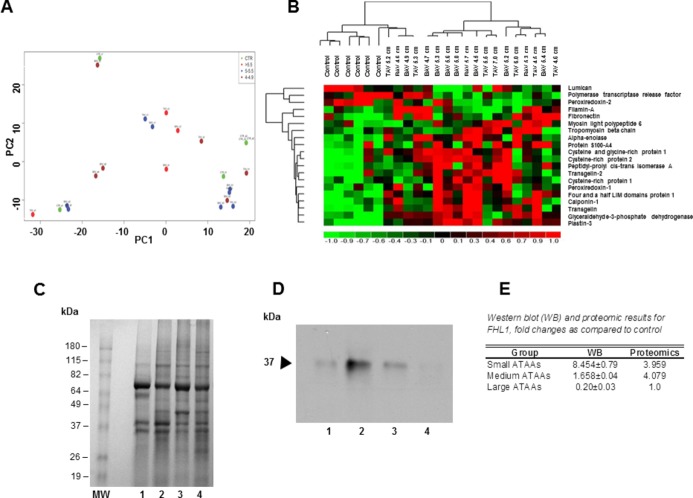
Figure 1.
A, Principal component analysis. B, Hierarchical cluster analysis of differentially expressed proteins. C, Western blot analysis. D, Immunoblot analysis. E, Coomassie gel. A, Principal component analysis (PCA) indicated no difference in differential protein expression patterns between BAV vs TAV or male vs female (identified with gender icon). B, Differentially expressed proteins compared with control were identified by supervised analysis on the basis of P value <0.01 in each group. The differentially expressed proteins are identified with short descriptions obtained from the SWISS‐PROT database. The log‐fold change (LFC) in protein expression is shown with pseudocolor scale (−1 to 1) with red denoting up‐regulation and green denoting down‐regulation. The columns represent LFC comparisons and the rows represent the proteins. Dendrograms are found on the left side, experimental groups are found on the top, and protein names are found on the right side of the figure. C, Representative 10% Novex Tris‐glycine gel. Protein samples (25 μg) from full‐thickness aortic tissue from the site of the maximal diameter containing all 3 layers were fractionated on 10% Novex Tris‐glycine gel, Validation of proteomic results via Western blot (WB) analysis. Results displayed as fold changes compared with controls. 1, Control; 2, small ATAAs; 3, medium‐sized ATAAs; 4, large ATAAs. D, Image of immunoblot, performed using mouse monoclonal four and a half LIM domains protein 1 (FHL1) antibody (1:2000 dilution, Abcam, Inc). 1, Control; 2, small ATAAs; 3, medium‐sized ATAAs; 4, large ATAAs. E, Image of Coomassie blue–stained gel. MW, Molecular weight marker. 1, Control; 2, small ATAAs; 3, medium‐sized ATAAs; 4, large ATAAs. ATAA indicates ascending thoracic aortic aneurysm; BAV, bicuspid aortic valve; PC1, principal component 1; TAV, tricuspid aortic valve.
PCA is used to convert a set of observations of possibly correlated variables to a set of uncorrelated variables called principal components. The number of principal components is less than or equal to the number of original variables. Principal component 1 has the highest variance possible and therefore accounts for as much variability in the data as feasible. Principal component 2 has the second highest variance of the data.
Principal component analysis indicated there was no difference in differential protein expression patterns between BAV versus TAV or male versus female (Figure 1A).
However, comparison of all ATAA size groups (small, medium sized, and large) versus controls identified 26 unique iTRAQ validated proteins as being up‐ or down‐regulated (Figure 1B). Four of these proteins (calponin‐1, plastin‐3, and peptidyl‐prolyl cis‐trans isomerase) were commonly expressed and up‐regulated in all ATAA size groups (Figure 1B).
Hierarchical cluster analysis showed distinct protein expression patterns among small, medium‐sized, and large ATAA size groups compared with controls (Figure 1B). Comparison between small ATAAs versus controls, medium‐sized ATAAs versus controls, and large ATAAs versus controls identified 13, 17, and 12 proteins, respectively, that were differentially expressed (Figure 1A).
Analysis of up‐ and down‐regulated proteins in ATAA size groups indicated that there were 2 unique proteins altered in small ATAAs (filamin‐A and G protein–coupled receptor 98), 7 unique proteins altered in medium‐sized ATAAs (cysteine‐rich protein [CRP] 1, cysteine‐ and glycine‐rich protein [CSRP] 2, transgelin‐2, peroxiredoxin‐1, tropomyosin β chain, protein disulfide‐isomerase, and lumican), and 5 unique proteins altered in large ATAAs (vimentin, polymerase 1 and transcript release factor, peroxiredoxin‐2, prolargin, and α2‐HS‐glycoprotein) (Table 2). Proteomic results were confirmed by Western blot analysis (Figure 1C through 1E).
Table 2.
Protein Expression by ATAA Size
| Protein Name | t‐Statistic | P Value | Control Mean | Group Mean | No. of Peptides | Control SE | GRP2 SE |
|---|---|---|---|---|---|---|---|
| ATAA 4.0 to 4.9 cm | |||||||
| Glyceraldehyde‐3‐phosphate dehydrogenase | 5.954 | 0.001 | 0.807 | 2.446 | 6 | 0.15 | 0.09 |
| G protein–coupled receptor 98* | 5.332 | 0.001 | 0.657 | 1.753 | 2 | 0.09 | 0.04 |
| Plastin‐3 | 5.232 | 0.001 | 1.553 | 4.527 | 2 | 0.38 | 0.39 |
| Transgelin | 3.977 | 0.004 | 1.547 | 4.417 | 13 | 0.35 | 0.65 |
| Four and a half LIM domains protein 1 | 3.959 | 0.004 | 1.627 | 5.113 | 2 | 0.46 | 0.81 |
| Fibronectin | 3.231 | 0.009 | 0.680 | 2.163 | 8 | 0.26 | 0.31 |
| Prelamin‐A/C | 3.003 | 0.013 | 1.017 | 1.752 | 6 | 0.13 | 0.12 |
| Filamin‐A* | 2.904 | 0.023 | 0.467 | 1.626 | 31 | 0.13 | 0.3 |
| Cysteine‐rich protein 2 | 2.691 | 0.025 | 7.833 | 15.130 | 2 | 2.88 | 1.96 |
| Cysteine‐ and glycine‐rich protein 1 | 2.607 | 0.026 | 1.852 | 3.795 | 2 | 0.59 | 0.57 |
| Calponin‐1 | 2.581 | 0.041 | 2.658 | 7.530 | 6 | 0.68 | 2.1 |
| α‐Enolase 1 | 2.533 | 0.030 | 0.873 | 1.610 | 5 | 0.15 | 0.17 |
| Peptidyl‐prolyl cis‐trans isomerase A | 2.474 | 0.044 | 1.000 | 1.685 | 3 | 0.09 | 0.21 |
| ATAA 5.0 to 5.5 cm | |||||||
| Plastin‐3 | 4.668 | 0.001 | 1.553 | 4.240 | 2 | 0.38 | 0.44 |
| Calponin‐1 | 4.595 | 0.001 | 2.658 | 7.471 | 6 | 0.68 | 0.79 |
| Cysteine‐ and glycine‐rich protein 1 | 4.215 | 0.003 | 1.852 | 4.664 | 2 | 0.59 | 0.31 |
| Glyceraldehyde‐3‐phosphate dehydrogenase | 4.154 | 0.001 | 0.807 | 1.788 | 6 | 0.15 | 0.18 |
| Four and a half LIM domains protein 1 | 4.079 | 0.002 | 1.627 | 5.619 | 2 | 0.46 | 0.86 |
| Transgelin | 3.843 | 0.003 | 1.547 | 4.731 | 13 | 0.35 | 0.75 |
| α‐Enolase 1 | 3.509 | 0.005 | 0.873 | 1.604 | 5 | 0.15 | 0.14 |
| Cysteine‐rich protein 2 | 3.42 | 0.006 | 7.833 | 20.816 | 2 | 2.88 | 2.48 |
| Transgelin‐2 | 3.345 | 0.009 | 4.030 | 12.031 | 5 | 2.04 | 1.24 |
| Peroxiredoxin‐1* | 3.327 | 0.006 | 1.020 | 1.586 | 3 | 0.11 | 0.13 |
| Cysteine‐rich protein 1* | 2.706 | 0.019 | 0.710 | 1.434 | 3 | 0.17 | 0.21 |
| Cysteine‐ and glycine‐rich protein 2* | 2.584 | 0.024 | 6.757 | 13.735 | 2 | 1.65 | 2.14 |
| Tropomyosin β chain* | 2.548 | 0.029 | 0.940 | 1.463 | 22 | 0.09 | 0.18 |
| Protein S100‐A4 | 2.283 | 0.048 | 0.847 | 1.166 | 2 | 0.12 | 0.08 |
| Peptidyl‐prolyl cis‐trans isomerase A | 2.259 | 0.050 | 1.000 | 1.536 | 3 | 0.09 | 0.22 |
| Protein disulfide‐isomerase* | −3.564 | 0.004 | 1.242 | 0.820 | 2 | 0.06 | 0.1 |
| Lumican* | −3.718 | 0.004 | 0.730 | 0.276 | 11 | 0.1 | 0.07 |
| ATAA >5.5 | |||||||
| Plastin‐3 | 4.429 | 0.009 | 1.553 | 5.285 | 2 | 0.38 | 0.75 |
| Peptidyl‐prolyl cis‐trans isomerase A | 3.265 | 0.051 | 0.457 | 0.020 | 3 | 0.09 | 0.23 |
| Protein S100‐A4 | 3.201 | 0.013 | 0.847 | 1.275 | 2 | 0.12 | 0.07 |
| Prelamin‐A/C | 2.828 | 0.024 | 1.017 | 1.428 | 6 | 0.13 | 0.08 |
| Calponin‐1 | 2.73 | 0.032 | 2.658 | 5.670 | 6 | 0.68 | 0.87 |
| Fibronectin | 2.642 | 0.041 | 0.680 | 1.913 | 8 | 0.26 | 0.39 |
| α‐Enolase 1 | 2.602 | 0.032 | 0.873 | 1.388 | 5 | 0.15 | 0.12 |
| Vimentin* | −2.381 | 0.045 | 1.950 | 0.940 | 23 | 0.32 | 0.28 |
| Prolargin* | −2.454 | 0.041 | 0.792 | 0.395 | 6 | 0.14 | 0.09 |
| Peroxiredoxin‐2* | −2.491 | 0.039 | 0.822 | 0.450 | 4 | 0.11 | 0.1 |
| α2‐HS‐glycoprotein* | −2.552 | 0.051 | 0.457 | 0.020 | 2 | 0.17 | 0 |
| Polymerase 1 and transcript release factor* | −3.137 | 0.015 | 1.508 | 0.673 | 2 | 0.23 | 0.14 |
t‐Statistic indicates fold change compared with control; ATAA, ascending thoracic aortic aneurysm.
Unique protein expression in ATAA size group.
Functional Enrichment Analysis
Functional enrichment analysis revealed 11 functional annotation clusters in small ATAAs, 7 annotation clusters in medium‐sized ATAAs, and 6 annotation clusters in large ATAAs (P<0.05, enrichment score >2.0) were significantly up‐regulated (Table 3).
Table 3.
Protein Functional Annotation Clusters for Ascending Thoracic Aortic Aneurysms (ATAAs) Based on Size
| Annotation Cluster | Count | P Value |
|---|---|---|
| Functional annotation clusters for ATAAs 4.0 to 4.9 cm | ||
| Cytoskeletal protein binding | 5 | 0.001 |
| Actin binding | 4 | 0.003 |
| Muscle organ development | 3 | 0.012 |
| Actin cytoskeleton organization | 3 | 0.014 |
| Actin filament–based process | 3 | 0.016 |
| Glycolysis pathway | 2 | 0.023 |
| Cytosol | 4 | 0.029 |
| Glycolysis | 2 | 0.038 |
| Glucose catabolic process | 2 | 0.046 |
| Glycolysis/gluconeogenesis | 2 | 0.046 |
| Cytoskeleton organization | 3 | 0.047 |
| Functional annotation clusters for ATAAs 5.0 to 5.5 cm | ||
| Muscle organ development | 4 | 0.001 |
| Actin binding | 4 | 0.006 |
| Cytoskeletal protein binding | 4 | 0.019 |
| Regulation of cell size | 3 | 0.021 |
| Glycolysis pathway | 2 | 0.023 |
| Glycolysis/gluconeogenesis | 2 | 0.023 |
| Regulation of cellular component size | 3 | 0.035 |
| Functional annotation clusters for ATAAs >5.5 cm | ||
| Acute inflammatory response | 3 | 0.003 |
| Structural molecule activity | 4 | 0.014 |
| Extracellular matrix | 3 | 0.018 |
| Inflammatory response | 3 | 0.027 |
| Acute‐phase response | 2 | 0.032 |
| Cytosol | 4 | 0.042 |
Count, number of proteins in each functional annotation cluster. P value was determined by Fisher exact test. A P value ≤0.01 indicates the annotation cluster is specifically associated (enriched) in the ATAA size group pathway rather than by random chance.
In small and medium‐sized ATAAs (4.0 to 5.5 cm), functional annotation clusters for cytoskeletal protein binding, actin binding, muscle organ development, glycolysis pathway, and glycolysis/gluconeogenesis were significantly up‐regulated (Table 3). In the large ATAAs (>5.5 cm), functional annotation clusters for acute inflammatory response, structural molecule activity, extracellular matrix, and acute‐phase response were significantly up‐regulated. All annotation clusters were unique to ATAA size with no common functional annotation clusters found in ATAAs 4.0 to 5.5 cm compared with ATAAs >5.5 cm. No down‐regulated annotation clusters were identified in any ATAA size groups (Table 3).
Pathway Analysis
Pathway analysis of all ATAA size groups showed a significant up‐regulation of 11 pathways. In small ATAAs (4.0 to 4.9 cm), 4 pathways were significantly up‐regulated (Figure 2A), while in medium‐sized ATAAs (5.0 to 5.5 cm), 6 pathways were significantly up‐regulated (Figure 2B). In large ATAAs, 6 pathways were significantly up‐regulated (Figure 2C). One pathway—phenylalanine, tyrosine, and tryptophan biosynthesis—was commonly up‐regulated in all ATAAs. There were 2 pathways common in small and medium‐sized ATAAs and 3 pathways common between medium‐sized and large ATAAs. Only 1 pathway was common between small and large ATAAs (ILK signaling).
Figure 2.
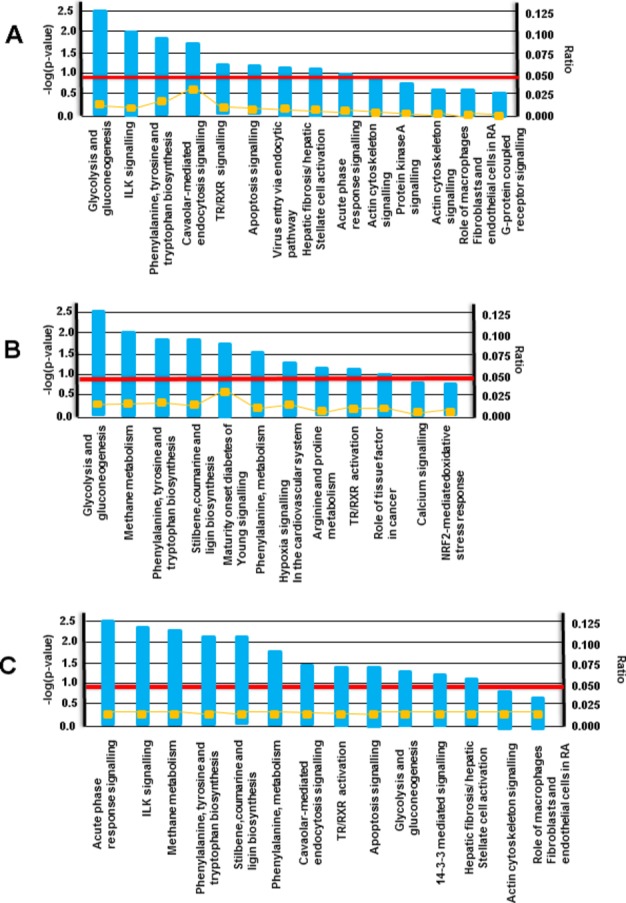
Pathway analysis based on size. A, Small aneurysms (4.0 to 4.9 cm); B, medium‐sized aneurysms (5.0 to 5.5 cm); C, large aneurysms (>5.5 cm). Pathway analysis was performed by identifying the over‐represented GO categories in differentially expressed proteins: this was done using the Biological Processes and Molecular Functions Enrichment Analysis available from the Database for Annotation, Visualization and Integrated Discovery (DAVID). Functional pathways are labeled on the abscissa and the −log (P value) is on the ordinate. On the right side is the ratio of proteins in pathway over total proteins and the yellow line shows the ratio of each pathway. The red line is the threshold at P=0.05. Any pathway that passes the red line is significantly enriched. ILK indicates integrin linked kinase; NRF, nuclear respiratory factor; RA, rheumatoid arthritis; TR/RXR, thyroid hormone receptor/retinoid X receptor.
Real‐Time RT‐PCR Analysis
For the identification of ATAAs in whole blood, RNA biomarkers were chosen as determined from proteomic analysis. qRT‐PCR analysis of whole blood demonstrated that transcripts for α‐enolase 1 and peptidyl‐prolyl cis‐trans isomerase A were up‐regulated in all ATAA size groups compared with nonaneurysm controls and four and a half LIM domains protein 1 (FHL1), CSRP1, and CRP2 transcripts were up‐regulated in small and medium‐sized ATAA groups compared with nonaneurysm controls.
qRT‐PCR analysis of whole blood demonstrated elevated expression of transcripts for collagen α1(I), collagen α1(III), collagen α2(V), collagen α1(XI), and FHL1 based in ATAAs compared with nonaneurysm controls (Table 4). Our results show that collagen α1(III), collagen α1(XI), and FHL1 were down‐regulated in all control samples, whereas collagen α1(I) and collagen α2(V) were uniquely up‐regulated in 3 individual control samples.
Table 4.
Whole Blood ATAA Biomarker qRT‐PCR Fold Changes in Control and ATAA roups
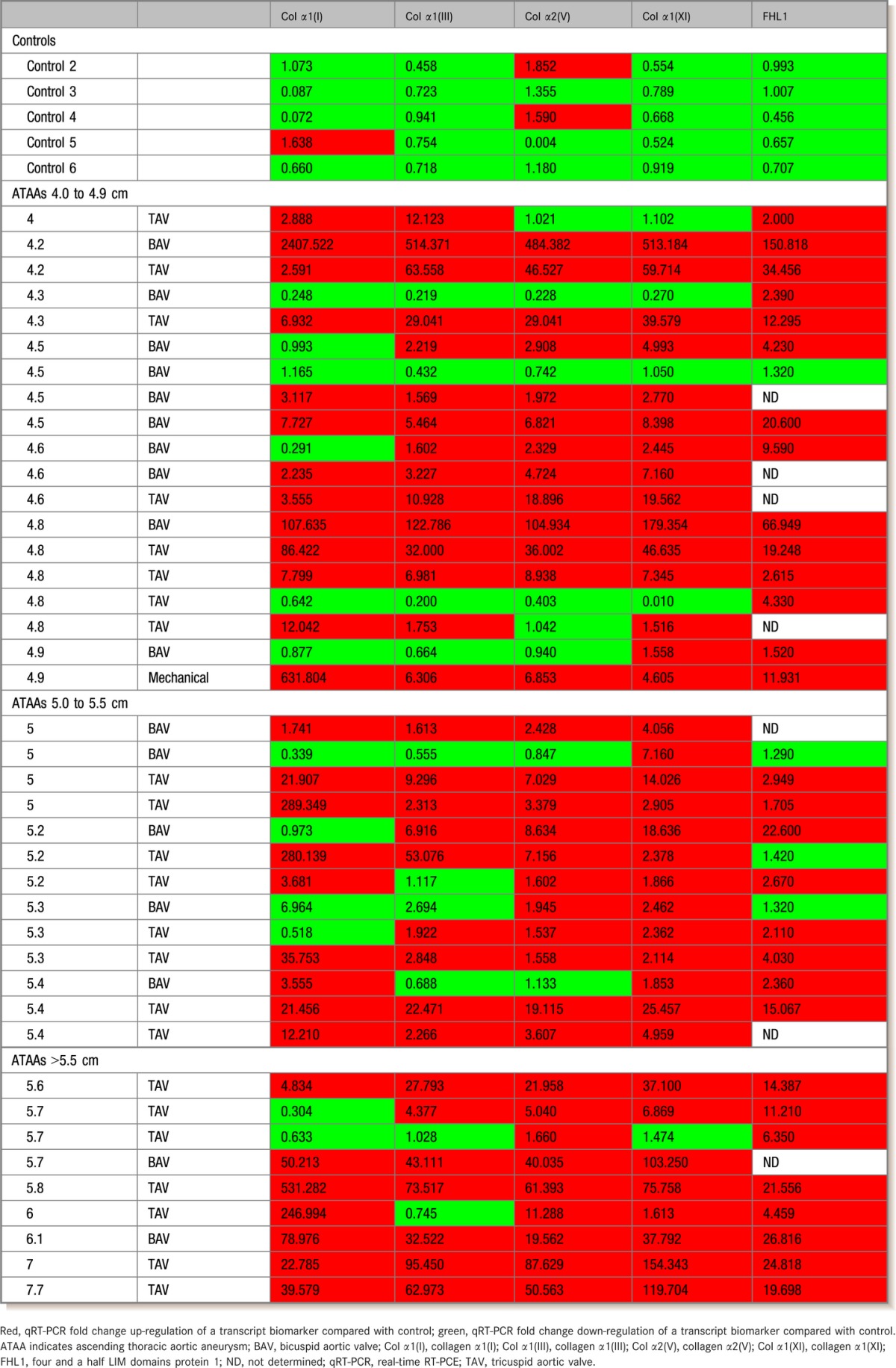
Based on these results, we have used collagen α1(I), collagen α1(III), collagen α2(V), collagen α1(XI), and FHL1 for biomarker analysis of ATAAs in whole blood samples. Using these 5 biomarkers for whole blood qRT‐PCR analysis, we observed that 3 of these 5 markers were significantly increased in 15 of 19 ATAA samples of 4.0 to 4.9 cm (79.0% positive) and in 19 of 21 ATAA samples >5.0 cm (90.5% positive) (Table 4).
Comparison between TAV versus control and BAV versus control demonstrated that these 5 biomarkers were significantly increased in 21 of 23 TAV (91.3% positive) and 12 of 17 BAV (70.6% positive) ATAA samples >4.0 cm (Table 4). Five ATAA samples were excluded from analysis due to insufficient sample volume: 4.2 BAV, 4.5 TAV, 5.0 BAV, 5.3 BAV, and 7.0 TAV. These samples were from 3 women and 2 men.
Analysis of biomarkers showed that identification of ATAAs in whole blood samples was possible having only 3 of 5 biomarkers over‐expressed compared with control values (Table 4). Sensitivity, specificity, probability, and CIs for ATAA identification using 1 to 3 biomarkers for qRT‐PCR in whole blood showed significantly higher rates compared with control blood with varying sensitivity of 0.53 to 0.97 (Table 5).
Table 5.
Sensitivity, Specificity, Probability, and CIs for ATAA Identification Using 1 to 3 Biomarkers for qRT‐PCR in Whole Blood
| Positive ATAA Patients | % of ATAA Patients | P Value | Sensitivity | 95% CI | Specificity | 95% CI | |
|---|---|---|---|---|---|---|---|
| 1 Up‐regulated transcript biomarker compared with control | |||||||
| FHL1 | 30 | 88.2 | <0.0001 | 0.88 | 0.73 to 0.97 | 1.00 | 0.42 to 1.00 |
| Col α1(XI) | 36 | 87.8 | <0.0001 | 0.88 | 0.74 to 0.96 | 1.00 | 0.42 to 1.00 |
| Col α2(V) | 33 | 80.5 | 0.03 | 0.81 | 0.65 to 0.91 | 0.67 | 0.22 to 0.96 |
| Col α1(III) | 31 | 75.6 | <0.001 | 0.76 | 0.60 to 0.88 | 1.00 | 0.42 to 1.00 |
| Col α1(I) | 29 | 70.7 | 0.02 | 0.71 | 0.55 to 0.84 | 0.83 | 0.36 to 1.00 |
| ATAA patients identified for any 1 transcript biomarker being up‐regulated compared with control | 33 | 97.1 | <0.01 | 0.97 | 0.85 to 1.00 | 0.50 | 0.12 to 0.88 |
| 2 Up‐regulated transcript biomarkers compared with control | |||||||
| Col α2(V)+Col α1(XI) | 32 | 78.0 | <0.001 | 0.78 | 0.63 to 0.89 | 1.00 | 0.42 to 1.00 |
| Col α1(XI)+FHL1 | 26 | 76.5 | <0.001 | 0.77 | 0.59 to 0.89 | 1.00 | 0.42 to 1.00 |
| Col α2(V)+FHL1 | 25 | 73.5 | <0.01 | 0.74 | 0.56 to 0.87 | 1.00 | 0.42 to 1.00 |
| Col α1(III)+Col α1(XI) | 30 | 73.2 | <0.01 | 0.73 | 0.57 to 0.86 | 1.00 | 0.42 to 1.00 |
| Col α1(III)+Col α2(V) | 29 | 70.7 | <0.01 | 0.71 | 0.55 to 0.84 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α1(XI) | 28 | 68.3 | <0.01 | 0.68 | 0.52 to 0.82 | 1.00 | 0.42 to 1.00 |
| Col α1(III)+FHL1 | 23 | 67.6 | <0.01 | 0.68 | 0.50 to 0.83 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α1(III) | 26 | 63.4 | <0.01 | 0.64 | 0.47 to 0.78 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α2(V) | 26 | 63.4 | <0.01 | 0.64 | 0.47 to 0.78 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+FHL1 | 21 | 61.8 | <0.01 | 0.62 | 0.44 to 0.78 | 1.00 | 0.42 to 1.00 |
| ATAA patients identified for any 2 transcript biomarkers being up‐regulated compared with control | 30 | 88.2 | <0.0001 | 0.88 | 0.73 to 0.97 | 1.00 | 0.42 to 1.00 |
| 3 Up‐regulated transcript biomarkers compared with control | |||||||
| Col α1(III)+Col α2(V)+Col α1(XI) | 29 | 70.7 | <0.01 | 0.71 | 0.55 to 0.84 | 1.00 | 0.42 to 1.00 |
| Col α2(V)+Col α1(XI)+FHL1 | 24 | 70.6 | <0.01 | 0.71 | 0.53 to 0.85 | 1.00 | 0.42 to 1.00 |
| Col α1(III)+Col α2(V)+FHL1 | 22 | 64.7 | <0.01 | 0.65 | 0.47 to 0.80 | 1.00 | 0.42 to 1.00 |
| Col α1(III)+Col α1(XI)+FHL1 | 22 | 64.7 | <0.01 | 0.65 | 0.47 to 0.80 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α2(V)+Col α1(XI) | 26 | 63.4 | <0.01 | 0.63 | 0.47 to 0.78 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α1(III)+Col α1(XI) | 25 | 61.0 | <0.01 | 0.61 | 0.45 to 0.76 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α1(XI)+FHL1 | 20 | 58.8 | 0.02 | 0.59 | 0.41 to 0.75 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α1(III)+Col α2(V) | 24 | 58.5 | <0.01 | 0.59 | 0.42 to 0.74 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α2(V)+FHL1 | 19 | 55.9 | 0.02 | 0.56 | 0.38 to 0.73 | 1.00 | 0.42 to 1.00 |
| Col α1(I)+Col α1(III)+FHL1 | 18 | 52.9 | 0.02 | 0.53 | 0.35 to 0.70 | 1.00 | 0.42 to 1.00 |
| ATAA patients correctly identified for any 3 transcript biomarkers being up‐regulated compared with control | 27 | 79.4 | <0.001 | 0.79 | 0.62 to 0.91 | 1.00 | 0.42 to 1.00 |
P values were given by Fisher's exact tests comparing the proportions among variables of each screening test and ATAA prevalence. Seven samples in which FHL1 was not determined due to sample deficiency were not included in the analysis. Note that for specificity, the percentage of control subjects with up‐regulated biomarker values was 0% for all except for Col α2(V) and Col α1(I). ATAA indicates ascending thoracic aortic aneurysm; Col α1(I), collagen α1(I); Col α1(III), collagen α1(III); Col α2(V), collagen α2(V); Col α1(XI), collagen α1(XI); FHL1, four and a half LIM domains protein 1; qRT‐PCR, real‐time RT‐PCR.
Discussion
We present a preliminary biomarker panel for the identification of ATAAs in male and female patients with BAVs or TAVs in whole blood. We have used both BAVs and TAVs and both male and female patient samples for biomarker identification as we reasoned that to provide clinical efficacy, both BAV and TAV needed to be included.18
Proteomic studies were performed to identify likely candidate biomarkers to use in whole blood analysis by real‐time RT‐PCR. Our proteomic studies show significant differences in differential protein expression between ATAA and controls. However, we found no difference in differential protein expression patterns between BAV versus TAV or male versus female. These findings agree with Fedak et al,19 who demonstrated no difference in differential genomic expression patterns between BAV versus TAV, and are in agreement with current proposed mechanisms leading to aortic rupture.20–22
In both BAV ATAAs and TAV ATAAs, our studies show that enriched pathways for actin band, cytoskeletal, and LIM domain proteins were differentially expressed in small and medium‐size ATAA samples, while the pathways for acute inflammatory response and extracellular matrix proteins were differentially expressed in large ATAA samples.
From these studies, we identified 8 proteins found in aortic tissue samples regardless of sex and aortic valve type: α‐enolase 1, peptidyl‐prolyl cis‐trans isomerase A, plastin‐3, calponin‐1, S100‐A4, FHL1, CSRP1, and CRP2. These proteins have been shown to have important roles in modulating ATAA development through the modulation of matrix metalloproteinases (MMPs).
Peptidyl‐prolyl cis‐trans isomerase A encodes for cyclophilin A, which has been shown to and have been associated with extracellular matrix metalloproteinase activation.23–24 Calponin 1 is a regulator of vascular smooth muscle tone and has been shown to be a target of MMP2,25 whereas S100‐A4 is a calcium signal transducer that has been shown to positively regulate the secretion of MMPs from endothelial cells and fibroblasts and to regulate the transcriptional activation of collagenase 3 (MMP‐13) mRNA followed by subsequent release of MMP protein and the proteolytic degradation of extracellular matrix.26–28 α‐Enolase is a multifunctional protein and is involved in the synthesis of pyruvate but also acts as a plasminogen receptor, where it regulates the activation of plasmin leading to MMP activation and extracellular matrix degradation.20 CSRP1 is expressed in vascular smooth muscle, and CRP2 is a cofactor for smooth muscle cell differentiation.29 Recent knock‐out studies have shown that while CSRP1 and CRP2 are not essential for normal neointimal formation, they act antagonistically to modulate the smooth muscle response to pathophysiological stress.30 Plastin 3 or fimbrin is an actin binding protein containing a calcium‐binding domain and is dispersed throughout the lamella. Overall, these proteins and their effects on MMPs and aneurysm formation agree with clinical data demonstrating that ascending aortic aneurysms exhibit increased MMP expression in the affected tissue.21–22
In initial studies, we tested the genetic sequences for all of these proteins for preliminary diagnostic efficacy using qRT‐PCR analysis of whole blood. Our analysis showed that only FHL1, a unique protein that has been shown to play an important role in vascular smooth muscle cell differentiation, was useful as a transcript biomarker in whole blood analysis for ATAA identification by qRT‐PCR analysis.31 α‐Enolase 1, CSRP1, CRP2, and peptidyl‐prolyl cis‐trans isomerase A did not provide general transcript biomarker identification in whole blood samples. Collagen α1(I), collagen α1(III), collagen α2(V), and collagen α1(XI) were used for qRT‐PCR analysis of whole blood based on our previous studies.1
Our data demonstrate that the identification of patients with ATAA using whole blood analysis is possible and that, by using only the collagen transcript biomarker expression, levels we were able to correctly identify 30 of 41 patients with ATAA. We also show that in 30 of 41 ATAA patients, at least 4 of the 5 biomarkers are up‐regulated compared with controls. We suggest that to properly identify an aneurysm of ≥4.0 cm, at least 3 of 5 biomarkers must be elevated >1.5‐fold (Tables 4 and 5). Using this criterion, 79.4% of ATAA patients were correctly identified (P<0.001; sensitivity 0.79, specificity 1.00).
The use of whole blood analysis for identification of ATAAs of ≥4.0 cm can be performed rapidly and provides a cost‐effective method for screening before the use of expensive magnetic resonance imaging, computed tomography scanning, or ultrasound evaluation.32 The ability to provide for rapid evaluation of potential ATAA in patients using whole blood strengthens the armamentarium currently available to clinicians and surgeons and should significantly decrease morbidity and mortality in asymptomatic ATAAs. The use of whole blood allows for a simple approach that can be used at major hospital centers and at clinics lacking sophisticated imaging technology.32
It must be noted that while the present study provides initial preliminary results based on sample size, the sample population does not include ethnic diversity as all subjects were white and therefore applicability to patients of varied racial heritage cannot be confirmed. The ages of the patients are representative of our clinical population and may not reflect those of other areas. It is also important to note that qRT‐PCR did not allow for direct replication of the tissue finding. The data presented do, however, provide preliminary data and evidence that ATAAs can be readily identified in whole blood samples using qRT‐PCR analysis.
Sources of Funding
This study was supported by National Institutes of Health grants HL029077 and HL103542.
Disclosures
None.
References
- 1.Toumpoulis IK, Oxford JT, Cowan DB, Anagnostopoulos CE, Rokkas CK, Chamogeorgakis TP, Angouras DC, Shemin RJ, Navab M, Ericsson M, Federman M, Levitsky S, McCully JD. Differential expression of collagen type V and XI α‐1 in human ascending thoracic aortic aneurysms. Ann Thorac Surg. 2009; 88:506-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Upadhye S, Schiff K. Acute aortic dissection in the emergency department: diagnostic challenges and evidence‐based management. Emerg Med Clin North Am. 2012; 30:307-327 [DOI] [PubMed] [Google Scholar]
- 3.Wittels K. Aortic emergencies. Emerg Med Clin North Am. 2011; 29:789-800 [DOI] [PubMed] [Google Scholar]
- 4.Elefteriades JA. Natural history of thoracic aortic aneurysms: indications for surgery, and surgical versus nonsurgical risks. Ann Thorac Surg. 2002; 74:S1877-S1880 [DOI] [PubMed] [Google Scholar]
- 5.Minino AM, Heron MP, Murphy SL, Kochanek KD. Deaths: final data for 2004. Natl Vital Stat Rep. 2007; 55:1-119 [PubMed] [Google Scholar]
- 6.Ma J, Liew CC. Gene profiling identifies secreted protein transcripts from peripheral blood cells in coronary artery disease. J Mol Cell Cardiol. 2003; 35:993-998 [DOI] [PubMed] [Google Scholar]
- 7.Liew CC. Expressed genome molecular signatures of heart failure. Clin Chem Lab Med. 2005; 43:462-469 [DOI] [PubMed] [Google Scholar]
- 8.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006; 147:126-132 [DOI] [PubMed] [Google Scholar]
- 9.Burczynski ME, Dorner AJ. Transcriptional profiling of peripheral blood cells in clinical pharmacogenomic studies. Pharmacogenomics. 2006; 7:187-202 [DOI] [PubMed] [Google Scholar]
- 10.Xi L, Nicastri DG, El‐Hefnawy T, Hughes SJ, Luketich JD, Godfrey TE. Optimal markers for real‐time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin Chem. 2007; 53:1206-1215 [DOI] [PubMed] [Google Scholar]
- 11.Vanburen P, Ma J, Chao S, Mueller E, Schneider DJ, Liew CC. Blood gene expression signatures associate with heart failure outcomes. Physiol Genomics. 2011; 43:392-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunha‐Neto E, Dzau VJ, Allen PD, Stamatiou D, Benvenutti L, Higuchi ML, Koyama NS, Silva JS, Kalil J, Liew CC. Cardiac gene expression profiling provides evidence for cytokinopathy as a molecular mechanism in Chagas' disease cardiomyopathy. Am J Pathol. 2005; 167:305-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao S, Ying J, Liew G, Marshall W, Liew CC, Burakoff R. Blood RNA biomarker panel detects both left‐ and right‐sided colorectal neoplasms: a case‐control study. J Exp Clin Cancer Res. 2013; 32:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesko B, Poliska S, Nagy L. Gene expression profiles in peripheral blood for the diagnosis of autoimmune diseases. Trends Mol Med. 2011; 17:223-233 [DOI] [PubMed] [Google Scholar]
- 15.Marshall KW, Zhang H, Yager TD, Nossova N, Dempsey A, Zheng R, Han M, Tang H, Chao S, Liew CC. Blood‐based biomarkers for detecting mild osteoarthritis in the human knee. Osteoarthritis Cartilage. 2005; 13:861-871 [DOI] [PubMed] [Google Scholar]
- 16.Black KM, Barnett R, Bhasin MK, Daly C, Dillon ST, Libermann TA, Levitsky S, McCully JD. Microarray and proteomic analysis of cardioprotection in the mature and aged male and female. Physiol Genomics. 2012; 44:1027-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods. 2001; 25:402-408 [DOI] [PubMed] [Google Scholar]
- 18.Pisano C, Maresi E, Balistreri CR, Candore G, Merlo D, Fattouch K, Bianco G, Ruvolo G. Histological and genetic studies in patients with bicuspid aortic valve and ascending aorta complications. Interact Cardiovasc Thorac Surg. 2012; 14:300-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedak PW, Verma S, David TE, Leask RL, Weisel RD, Butany J. Clinical and pathophysiological implications of bicuspid aortic valve. Circulation. 2002; 106:900-904 [DOI] [PubMed] [Google Scholar]
- 20.Capello M, Ferri‐Borgogno S, Cappello P, Novelli F. α‐Enolase: a promising therapeutic and diagnostic tumor target. FEBS J. 2011; 278:1064-1074 [DOI] [PubMed] [Google Scholar]
- 21.Jackson V, Olsson T, Kurtovic S, Folkersen L, Paloschi V, Wågsäter D, Franco‐Cereceda A, Eriksson P. Matrix metalloproteinase 14 and 19 expression is associated with thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 2012; 144:459-466 [DOI] [PubMed] [Google Scholar]
- 22.Theruvath TP, Jones JA, Ikonomidis JS. Matrix metalloproteinases and descending aortic aneurysms: parity, disparity, and switch. J Card Surg. 2012; 27:81-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satoh K, Nigro P, Matoba T, O'Dell MR, Cui Z, Shi X, Mohan A, Yan C, Abe J, Illig KA, Berk BC. Cyclophilin A enhances vascular oxidative stress and the development of angiotensin II‐induced aortic aneurysms. Nat Med. 2009; 15:649-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weintraub NL. Understanding abdominal aortic aneurysm. N Engl J Med. 2009; 361:1114-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro MM, Cena J, Cho WJ, Walsh MP, Schulz R. Matrix metalloproteinase‐2 proteolysis of calponin‐1 contributes to vascular hypocontractility in endotoxemic rats. Arterioscler Thromb Vasc Biol. 2012; 32:662-668 [DOI] [PubMed] [Google Scholar]
- 26.Lases EC, Schepens MA, Haas FJ, Aarts LP, ter Beek HT, van Dongen EP, Siegers HP, van der Tweel I, Boezeman EH. Clinical prospective study of biochemical markers and evoked potentials for identifying adverse neurological outcome after thoracic and thoracoabdominal aortic aneurysm surgery. Br J Anaesth. 2005; 95:651-661 [DOI] [PubMed] [Google Scholar]
- 27.Schmidt‐Hansen B, Ornås D, Grigorian M, Klingelhöfer J, Tulchinsky E, Lukanidin E, Ambartsumian N. Extracellular S100A4(mts1) stimulates invasive growth of mouse endothelial cells and modulates MMP‐13 matrix metalloproteinase activity. Oncogene. 2004; 23:5487-5495 [DOI] [PubMed] [Google Scholar]
- 28.Zhang HY, Zheng XZ, Wang XH, Xuan XY, Wang F, Li SS. S100A4 mediated cell invasion and metastasis of esophageal squamous cell carcinoma via the regulation of MMP‐2 and E‐cadherin activity. Mol Biol Rep. 2012; 39:199-208 [DOI] [PubMed] [Google Scholar]
- 29.Kihara T, Shinohara S, Fujikawa R, Sugimoto Y, Murata M, Miyake J. Regulation of cysteine‐rich protein 2 localization by the development of actin fibers during smooth muscle cell differentiation. Biochem Biophys Res Commun. 2011; 411:96-101 [DOI] [PubMed] [Google Scholar]
- 30.Lilly B, Clark KA, Yoshigi M, Pronovost S, Wu ML, Periasamy M, Chi M, Paul RJ, Yet SF, Beckerle MC. Loss of the serum response factor cofactor, cysteine‐rich protein 1, attenuates neointima formation in the mouse. Arterioscler Thromb Vasc Biol. 2010; 30:694-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan MJ, Madgwick AJ, Charleston B, Pell JM, Loughna PT. The developmental regulation of a novel muscle LIM‐protein. Biochem Biophys Res Commun. 1995; 212:840-846 [DOI] [PubMed] [Google Scholar]
- 32.Booher AM, Eagle KA. Diagnosis and management issues in thoracic aortic aneurysm. Am Heart J. 2011; 162:38-46 [DOI] [PubMed] [Google Scholar]