Abstract
Background
The cardiac conduction system consists of the sinus node, nodal extensions, atrioventricular (AV) node, penetrating bundle, bundle branches, and Purkinje fibers. Node‐like AV ring tissue also exists at the AV junctions, and the right and left rings unite at the retroaortic node. The study aims were to (1) construct a 3‐dimensional anatomical model of the AV rings and retroaortic node, (2) map electrical activation in the right ring and study its action potential characteristics, and (3) examine gene expression in the right ring and retroaortic node.
Methods and Results
Three‐dimensional reconstruction (based on magnetic resonance imaging, histology, and immunohistochemistry) showed the extent and organization of the specialized tissues (eg, how the AV rings form the right and left nodal extensions into the AV node). Multiextracellular electrode array and microelectrode mapping of isolated right ring preparations revealed robust spontaneous activity with characteristic diastolic depolarization. Using laser microdissection gene expression measured at the mRNA level (using quantitative PCR) and protein level (using immunohistochemistry and Western blotting) showed that the right ring and retroaortic node, like the sinus node and AV node but, unlike ventricular muscle, had statistically significant higher expression of key transcription factors (including Tbx3, Msx2, and Id2) and ion channels (including HCN4, Cav3.1, Cav3.2, Kv1.5, SK1, Kir3.1, and Kir3.4) and lower expression of other key ion channels (Nav1.5 and Kir2.1).
Conclusions
The AV rings and retroaortic node possess gene expression profiles similar to that of the AV node. Ion channel expression and electrophysiological recordings show the AV rings could act as ectopic pacemakers and a source of atrial tachycardia.
Keywords: action potential, arrhythmogenesis, atrioventricular ring tissues, cardiac conduction system, ion channels
Introduction
The cardiac conduction system (CCS), generally considered to consist of the sinus node (SN) and the atrioventricular (AV) conduction axis, is responsible for the initiation and conduction of the action potential (AP) throughout the heart, leading to coordinated contraction of the heart.1 The AV conduction axis is known to be made up of nodal extensions, AV node (AVN), penetrating or His bundle, bundle branches, and Purkinje fibers. Further areas of histologically specialized tissue, distinct from these well‐recognized areas of the CCS, have been shown to exist within the muscular AV junctions, initially being illustrated by Kent in 1893, who incorrectly suggested that they provided multiple pathways for normal AV conduction. It was subsequently established, however, that the AV conduction axis provides the only pathway for conduction across the AV junctions in structurally normal hearts and that the nodes identified by Kent were part of so‐called AV rings, which are rings of node‐like myocytes occupying the vestibules of the tricuspid and mitral valvar orifices and which unite in the retroaortic node.2 The AV rings are also anatomically continuous with the CCS,3 although they are not thought to contribute to the normal functioning of the heart. Their location suggests that they could be a source of atrial tachycardia.4
For most of the preceding 100 years, the components of the CCS have been identified on the basis of the histological criteria established by Aschoff and Monckeberg. With the emergence of new techniques, it is now possible to identify transcription factors involved in the development of the CCS, these factors then serving to identify the conduction tissues. Work by Hoogaars et al5 showed that the transcription factor Tbx3 is expressed in the entire CCS, including the previously described AV rings.2 Recently, Aanhaanen et al6 demonstrated that the AV rings are formed from the embryonic AV canal musculature, which also contributes to the retroaortic node, nodal extensions, and compact AVN. AV rings have been demonstrated using immunohistochemistry in many species: human,7 rabbit, guinea pig, rat,3 and mouse.3,6 The fact that the AV rings share common developmental origins with the AVN suggests that they may have similar gene expression profiles and possess some subsidiary pacemaker functionality. HCN4, the principal ion channel responsible for the funny current (If), an important pacemaker current in the SN and AVN, is known to be expressed in the AV rings.3 We showed previously that the AV rings have high expression of connexin45 (Cx45) and low expression of connexin43 (Cx43), characteristics shared with the SN and AVN.3,8 Electrophysiological investigations have shown node‐type APs in the vestibule of the tricuspid valve in pig and dog hearts.9–10 The functionality of the AV rings in the adult heart, however, is unknown. The aims of this study were 3‐fold: (1) to develop a 3‐dimensional (3D) model of the anatomy of the AV rings and retroaortic node to show their extent and location; (2) to map electrical activation in the right AV ring and study its AP characteristics; and (3) to examine the expression of functionally important molecules (including transcription factors, ion channels, Ca2+‐handling proteins, and gap junction channels) in the right ring.
Methods
Animals
All animals were humanely killed via cervical dislocation in accordance with the United Kingdom Animals (Scientific Procedures) Act, 1986. Male Wistar‐Hanover rats weighing 250 to 400 g were used throughout.
Cryosectioning for Histology, Immunohistochemistry, and Quantitative PCR
Eighteen hearts were excised and flash frozen in cold isopentane and stored at −80°C until use. Hearts were serially sectioned at 25 μm (6 hearts for 3D reconstruction and immunohistochemistry) and 50 μm (12 hearts for quantitative PCR [qPCR]) thickness in the plane of the long‐axis (coronal plane) from posterior to anterior sides avoiding nonlinear deformation. Sections were collected on either Superfrost plus slides (VWR; for 3D reconstruction and protein expression) or ultraviolet light treated PEN‐membrane slides (Leica Microsystems; for qPCR study) and stored at −80°C until use.
Histology
During 3D reconstruction, to show tissue morphology, Masson's trichrome staining was performed on either immunolabeled sections or sections adjacent to such sections. When Masson's trichrome staining was performed on sections that had previously been immunolabeled, coverslips were removed. All sections were washed with PBS 3 times (10 minutes each wash). Sections were fixed in Bouin's fluid (Sigma Aldrich) overnight. Next, sections were washed in 70% ethanol 3 times (10 minutes each wash), stained in celestine blue for 5 minutes, and rinsed in distilled water. Sections were then stained in Mayer's alum hematoxylin for 10 minutes, washed in tap water for 15 minutes, stained in acid fuchsin for 3 minutes, and rinsed in distilled water. Next, sections were treated with phosphomolybdic acid for 5 minutes, drained, stained in methyl blue for 5 minutes, rinsed in distilled water, and treated with 1% acetic acid for 2 minutes, followed by dehydration through alcohols (1 minute 70% ethanol, 1 minute 90% ethanol, 2×2 minutes 100% ethanol). Finally, sections were placed in Histoclear (2×5 minutes) and mounted in DPX. Images of sections were taken using light microscopy (Carl Zeiss Microscopy) using Axiovision software (Carl Zeiss Microscopy). With this technique, connective tissue was stained royal blue, cardiac myocytes were stained pink, and nuclei were stained dark blue.
Immunohistochemistry
Tissue sections at 50‐ to 100‐μm intervals from each heart were fixed in 10% neutral buffered formalin (Sigma Aldrich) for 30 minutes and then washed 3 times (10 minutes each) in 0.01 mol/L PBS containing 0.138 mol/L NaCl and 0.027 mol/L KCl (pH 7.4; Sigma Aldrich). The sections were permeabilized by treatment with 0.1% Triton X‐100 (Sigma Aldrich) in PBS for 30 minutes followed by 3 PBS washes (10 minutes each). Sections were blocked using 1% bovine serum albumin (BSA) in PBS (Sigma Aldrich) for 60 minutes. The sections were incubated in primary antibodies diluted in 1% BSA (Table 1) overnight. The following day, the sections were incubated in secondary antibodies diluted in 1% BSA (Table 2) for 2 hours at room temperature and then washed 3 times in PBS (10 minutes each) and mounted in Vectashield mounting medium (Vector Laboratories). Sections were imaged using a Zeiss LSM5 laser scanning confocal microscope (Carl Zeiss Microscopy) using Pascal software (Carl Zeiss Microscopy).
Table 1.
List of Primary Antibodies Used
| Protein | Labelling Type/Use | Host | Type | Concentration | Manufacturer | |
|---|---|---|---|---|---|---|
| Nav1.5 | Single | Rabbit | Polyclonal | IgG | 1:20 | Alomone Labs |
| Cav3.1 | Single | Rabbit | Polyclonal | IgG | 1:20 | Alomone Labs |
| HCN4 | Double | Rabbit | Polyclonal | IgG | 1:20 | Alomone Labs |
| HCN4 | Western blot | Rabbit | Polyclonal | IgG | 1:200 | Alomone Labs |
| SK1 | Single | Goat | Polyclonal | IgG | 1:20 | Santa Cruz Biotechnology |
| Kir2.1 | Single | Rabbit | Polyclonal | IgG | 1:20 | Alomone Labs |
| Kir3.1 | Single | Rabbit | Polyclonal | IgG | 1:20 | Alomone Labs |
| Kir3.4 | Single | Rabbit | Polyclonal | IgG | 1:50 | Alomone Labs |
| NCX1 | Single | Mouse | Monoclonal | IgM | 1:20 | Thermo Scientific |
| RYR2 | Single | Mouse | Monoclonal | IgG | 1:20 | Thermo Scientific |
| Cx40 | Double | Goat | Polyclonal | IgG | 1:50 | Santa Cruz Biotechnology |
| Cx43 | Double | Mouse | Monoclonal | IgG | 1:50 | Millipore |
| Cx43 | Western blot | Rabbit | Polyclonal | IgG | 1:1000 | Sigma Aldrich |
| Caveolin 3 | Double | Mouse | Monoclonal | IgG | 1:100 | BD Biosciences |
| A‐Tubulin | Western blot | Mouse | Monoclonal | IgG | 1:2000 | Abcam |
Table 2.
List of Secondary Antibodies Used
| Host | Type | Conjugate | Concentration | Manufacturer | |
|---|---|---|---|---|---|
| Goat | Rabbit | IgG | FITC | 1:100 | Millipore |
| Donkey | Rabbit | IgG | FITC | 1:100 | Millipore |
| Goat | Mouse | IgG | Cy3 | 1:100 | Millipore |
| Donkey | Mouse | IgG | Cy3 | 1:100 | Millipore |
| Goat | Mouse | IgM | Cy3 | 1:100 | Millipore |
| Donkey | Goat | IgG | Alexa Fluor 488 | 1:100 | Invitrogen |
| Donkey | Goat | IgG | Alexa Fluor 546 | 1:100 | Invitrogen |
| Goat | Rabbit | IgG | HRP | 1:2000 | Cell Signaling Technology |
Western Blotting
Right ring tissue, sinus node, and atrial and ventricular myocardium were dissected from 6 hearts and flash frozen in liquid N2 and stored at −80°C. Protein was isolated by homogenizing tissue in RIPA buffer (10 mL PBS, 10 mL 10% IGEPAL CA‐630 [Sigma Aldrich], 5 mL 10% Na deoxycholate [Sigma Aldrich], 1 mL 10% SDS [BDH], 0.5 mL 100 mmol/L PMSF [Sigma Aldrich], 50 μL 1 mg/mL leupeptin [Sigma Aldrich], 50 μL 2 mg/mL aprotinin [Sigma Aldrich], 0.25 μL 1 mg/mL pepstatin [Sigma Aldrich], and 73.15 mL deionized water). Homogenate was then centrifuged at 3000 rpm for 5 minutes at 4°C, and the supernatant was collected. Protein yield was quantified using Qubit protein assay (Life Technologies). Eighteen micrograms of protein was used for each sample, and volume was increased to 7.5 μL with deionized water. To each sample, 2.5 μL NuPAGE LDS sample buffer was added. Samples were loaded onto NuPAGE 4% to 12% bis‐tris gels (Life Technologies). NuPAGE MOPS SDS running buffer (Life Technologies) was used, and electrophoresis was run at 200 V constant for 50 minutes. Western blot onto Immobilon‐P PVDF membrane (Millipore) was performed using a transfer buffer consisting of 100 mL 10× running buffer (30 g Tris base [144 g glycine in 1 L deionized water], 200 mL methanol, 700 mL deionized water) and run at 30 V constant for 2 hours. Transfer was checked by staining membranes with Ponceau red solution for 5 minutes, followed by rinsing in deionized water. After staining, membranes were cleared by washing for 30 seconds in 0.1 mol/L NaOH solution. Membranes were blocked in 3% BSA (Sigma Aldrich) in TBS‐T (100 mL TBS 10× [24.23 g Trizma HCl, 80.06 g NaCl in 1 L deionized water, pH 7.6], 900 mL deionized water, 1 mL Tween20 [Sigma Aldrich]) overnight at 4°C. Membranes were washed 3 times in TBS‐T followed by staining with primary antibody diluted in 1% TBS‐T to the appropriate concentration (Table 1) for 2 hours at room temperature. Membranes were washed 3 times in TBS‐T. Appropriate HRP‐conjugated secondary antibody was diluted to the required concentration (Table 2) in 1% BSA in TBS‐T applied to membranes for 2 hours at room temperature. Membranes were washed 3 times in TBS‐T. Amersham ECL detection reagents (GE Healthcare) used to visualize signal using ChemiDoc MP imaging system (Bio‐Rad). Band density was quantified using Image Studio Lite (LI‐COR) and normalized to the expression of α‐tubulin to correct for variation in protein loading. Outliers were determined using a robust statistical modified z‐score method based on the median of absolute deviation and were excluded from the statistical analysis. One‐way ANOVA was performed to find statistical differences in expression. P<0.05 was taken to be significant.
Magnetic Resonance Imaging
Three hearts were rapidly removed and placed in Krebs–Ringer solution (in mmol/L: NaCl 120, KCl 4, CaCl2 1.2, MgSO4 1.3, NaHCO3 25.2, glucose 5.8) equilibrated with 95% O2 and 5% CO2 at 4°C. After removal of excessive connective tissue, the heart was immobilized in Fomblin, and then magnetic resonance (MR) images were acquired with use of a 9.4‐T spectrometer (Bruker BioSpin) using a standard 3D fast low‐angle shot MR sequence with an echo time of 7.9 ms, a repetition time of 50 ms, and 20 averages. Field of view was 12.8×25.6×12.8 mm with a matrix size of 128×256×128, to give a voxel resolution of 0.1 mm isotropic.
3D Reconstruction
The process of reconstruction is illustrated in Figure 1 and consisted of 7 steps:
MR images and equivalent histological and immunohistochemical sections at 100‐μm intervals were selected.
Based on histological and the immunohistochemical staining, the SN, right and left rings, retroaortic node, inferior nodal extensions, AVN, penetrating bundle, and right and left bundle branches were delineated on the sections.
The delineated structures were outlined on the MR images.
The MR images with the delineated structures were read into MATLAB (The MathWorks, Inc) to build a 3D mathematical array in the Workspace of MATLAB.
The voxel size in the mathematical array was interpolated as 100×100×100 μm using bicubic interpolation. The array consisted of 1 489 839 nonzero nodes.
Different structures in the array were indexed as different integer numbers in previously described custom‐developed software.11 The atria and ventricles of the heart were also segmented and indexed separately. In the array, the atria consisted of 284 094 nodes (indexed as 1), the ventricles consisted of 1 180 021 nodes (indexed as 2), the SN consisted of 3316 nodes (indexed as 3), the AVN consisted of 1326 nodes (indexed as 4), the AV rings consisted of 3013 nodes (indexed as 5), and the Purkinje fibers consisted of 18 069 nodes (indexed as 6).
The isosurface method was used for the visualization of the resulting 3D anatomical model.
Figure 1.

Flow diagram showing the steps used for 3‐dimensional (3D) reconstruction. A, Whole heart was sectioned and immunolabeled. B, Based on immunolabeling, the SN, AV rings, retroaortic node, AVN, penetrating or His bundle, bundle branches, and Purkinje fibers were delineated. C, Delineated structures were transferred onto the histological images. D, Delineated structures were transferred onto corresponding MR images. E, MR images with the delineated structures were imported in MATLAB and a 3D mathematical model constructed. AVN indicates atrioventricular node; MR, magnetic resonance; SN, sinus node.
Tissue Preparation for Electrophysiology
Nineteen hearts were used in electrophysiology experiments. The chest was opened, and each heart along with the aortic arch was removed. A stainless steel cannula was inserted into the aorta, and the heart was Langendorff perfused with oxygenated (95% O2 and 5% CO2) Krebs–Ringer solution (in mmol/L: NaCl 120.3, KCl 4.0, CaCl2 1.2, MgSO4 1.3, NaH2PO4 1.2, NaHCO3 25.2, glucose 11, pH 7.4) for 10 minutes to wash out blood within the coronary arteries and veins. Thereafter, a longitudinal incision was made along the atrial septum, and the right atrium with part of right ventricle was separated from the rest of the heart. Tissue preparations with an intact SN, part of the right ring, and part of the right atrium were pinned to the floor of a chamber with the endocardial surface facing up. The chamber was superfused with oxygenated (95% O2 and 5% CO2) Krebs–Ringer solution at 10 mL/min, 37°C and tissue was allowed to equilibrate for 20 minutes before recordings were made. The bath temperature was monitored using a miniature thermistor. To study the isolated right ring, the SN was detached along with part of the right atrium.
Multiextracellular Electrode Mapping
Experiments were carried out using a custom‐designed multiextracellular electrode array mapping system. Tissue was mapped with a rectangular (4.5 mm×3.7 mm) 8×8 electrode array (0.2‐mm electrode diameter; ≈0.6‐mm interelectrode distance). For isolated right ring preparations, a smaller (4.5 mm×1.5 mm) 6×10‐electrode array (0.25‐mm electrode diameter; ≈0.2‐mm interelectrode distance) was used. Signals were acquired at 1.5 kHz, amplified (100 times), and digitized with 4 PXI‐6031E cards (National Instrument Inc) and stored on the hard disk of a computer for offline analysis. Data were acquired and analyzed using custom‐written software. The moment of maximal negative rate of change of potential in the extracellular electrograms was automatically detected and saved as the time of local activation. The local activation time data were then used to draw activation maps in MATLAB. Signals were not filtered.
Action Potential Recording
Intracellular AP recordings were obtained using the sharp microelectrode technique. Glass microelectrodes (20 to 40 MΩ resistance) filled with 3 mol/L KCl were used. Data acquired at intervals of 0.005 ms were passed through a 10‐kHz low‐pass Bessel filter and amplified 10 times by an Axon Instruments GeneClamp 500 (Molecular Devices Inc) amplifier, digitized with an Axon Instruments Digidata 1440A (Molecular Devices Inc), and stored on a computer for later analysis. Action potential recordings were analyzed using the software programs WinEDR V3.3.6 and WinWCP V4.6.1 (Dr J. Dempster, University of Strathclyde, Glasgow, UK). Action potentials were detected using the WinEDR program after rejecting any occasional signals arising due to electrical interference. Frequency was calculated by counting the number of APs detected in 10 seconds, and the APs were then exported to WinWCP, in which the maximum diastolic potential (MDP) and the following AP parameters were measured: AP amplitude (APA), peak APA measured relative to the MDP; overshoot, amplitude of the positive/overshoot phase of the AP; dV/dtmax, the maximum slope of the AP upstroke; T50, time taken for the AP to fall from peak amplitude to 50% of peak amplitude; T90, time taken for the AP to fall from peak amplitude to 90% of peak amplitude, and APD30, AP duration at −30 mV.
All results are expressed as mean±SEM, and n refers to the number of tissue preparations. Sigma Plot 12 (Systat Software Inc) was used to perform a‐way ANOVA with Tukey's multiple comparisons posttest to check the statistical significance of the AP data.
Overview of qPCR
Tissue samples were collected using laser microdissection (LMD), RNA was isolated, the RNA was reverse transcribed to produce cDNA, and the abundance of transcripts was measured using qPCR.
LMD
The frozen sections for LMD (from 12 hearts) were selected at 400‐μm intervals from each heart, and immunohistochemistry was performed as described earlier using anti‐HCN4 and anti–Cx43 as well as anti‐Cx40 and anti‐caveolin3 IgGs (Table 1), to determine the extent of each specialized structure to be investigated (ie, right ring, retroaortic node, SN, and AVN). The LMD procedure was performed using a Leica LMD6000 system (Leica Microsystems). Based on immunohistochemical images, sections selected for microdissection were stained with a quick hematoxylin and eosin stain to enable the tissue to be visualized under the light microscope (Figure 2). Sections were placed in 100% ethanol (30 seconds) and then stained with Mayers's hematoxylin solution (Sigma Aldrich) for 1 minute. Next, the sections were washed in 70% ethanol (30 seconds) and 95% ethanol (30 seconds). The sections were stained with alcoholic eosin solution (Sigma Aldrich) for 1 minute and placed into 70% ethanol (30 seconds), 95% ethanol (30 seconds), and 100% ethanol (30 seconds). The slides were kept in 100% ethanol until ready to be used for LMD. LMD samples were collected from the right ring, retroaortic node, SN, AVN, right atrium, and right ventricle from ≈40 sections per region from each heart. The immunolabeled images for each heart were used to guide the LMD from the quick hematoxylin and eosin–stained sections (Figure 2). There was no variation in the extent of the structures to be investigated among the 12 hearts used. Tissues collected from 2 hearts were combined during the LMD procedure in order to increase the yield of total RNA from each region.
Figure 2.

Laser microdissection (LMD). A, HCN4 (green signal) immunolabeling to identify the atrioventricular node (AVN). The AVN corresponds to the crescent‐shaped area of bright green signal. B, Quick hematoxylin and eosin–stained adjacent section prior to LMD. C, Same section as in B after LMD of the AVN.
RNA Isolation
Total RNA was isolated using an RNAqueous‐Micro kit (Applied Biosystems) according to the manufacturer's standard protocol for LMD samples and DNase I treatment. Samples were homogenized for 1 minute with an Ika T10 homogeniser (Ika Werke). The concentration of total RNA obtained from each sample was measured using a Nanodrop ND1000 spectrophotometer (Thermo Scientific), and the RNA integrity was analyzed using an Agilent Bioanalyzer (Agilent Technologies).
Reverse Transcription
Single‐stranded cDNA was synthesized from 150 ng total RNA using High Capacity RNA‐to‐cDNA Master Mix (Applied Biosystems) in a 20 μL reaction according to the manufacturer's protocol. Samples were run on a PCR Express thermal cycler (Thermo Hybaid).
qPCR
The gene targets were chosen after analyzing data from previous studies into cardiac gene and protein expression related to cardiac function, pacemaking, and development. qPCR was performed using 2 methods:
Taqman low density array (TLDA) microfluidic cards with 96 targets per sample (96a format) (Applied Biosystems) used with an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Total RNA 67.5 ng converted to cDNA in 50 μL water was loaded into each sample reservoir with 50 μL Taqman Universal Master Mix (Applied Biosystems).
QuantiTect primer assays (Qiagen) with Power SYBR green fluorescent reporter (Applied Biosystems). This method was used to look at a number of extra targets (Table 3) in 10 μL reactions using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems).
Table 3.
QuantiTect Assays Used
| Target | Assay Number |
|---|---|
| Tbx3 | QT00195608 |
| Nav1.5 | QT00182663 |
| Cav1.2 | QT01571822 |
| Cav1.3 | QT00194306 |
| Cav3.1 | QT00193669 |
| HCN1 | QT00184674 |
| HCN4 | QT00191387 |
| Kv1.4 | QT00376349 |
| Kv1.5 | QT01827245 |
| Kv4.2 | QT01080744 |
| KvLQT1 (Kv7.1) | QT00184387 |
| ERG (Kv11.1) | QT00184534 |
| Kir2.1 | QT00411292 |
| Kir3.1 | QT01694728 |
| Cx40 | QT00194782 |
| Cx43 | QT00177737 |
| Cx45 | QT01864390 |
PCR cycle parameters were: 2 minutes at 50°C and 10 minutes at 95°C initial incubation followed by 15 seconds at 95°C and 1 minute at 60°C cycles. Each run consisted of 40 cycles.
qPCR Data Analysis
TLDA cards were analyzed using RQ manager (Applied Biosystems) and StatMiner (Integromics). Average threshold cycle (Ct) values were obtained using RQ manager, and amplification curves were analyzed to check for experimental errors. Reactions that failed quality analysis were removed from further analysis. Expression levels were calculated using the ΔCt method with 18S used as a reference transcript. 18S was shown to be more stable than GADPH using the minimum variance median stability scoring method within StatMiner. Biological replicate outliers were removed using StatMiner via the use of MAD unit comparison, with samples with >3 flags being removed from the analysis. Graphs show mean 2−ΔCt+SEM. Data obtained using QuantiTect primer assays were analyzed using SDS 2.4 (Applied Biosystems) to obtain Ct values and the efficiency (E) of PCRs. 28S was used as a reference gene transcript for ΔCt expression level calculation. Outliers were determined using a robust statistical modified z‐score method based on the median of absolute deviation and were excluded from the statistical analysis. Data were calculated as E−ΔCt. Similar results were obtained with both methods, but only data obtained using the TLDA cards are shown.
Statistics—TLDA Cards
A moderated t test (limma parametric) was performed on ΔΔCt values using StatMiner. A Benjamini–Hochberg adjustment for false discovery rate (FDR) was also calculated as a correction for multiple comparisons. We follow the example of Myers et al12 and show both uncorrected t test and FDR results for all comparisons. FDR <0.2 was the primary method used for identifying significance, but targets with P<0.05 (limma) were also considered as being significant. Correlation of mean ΔCt values between regions for all targets was calculated using Spearman Rank Order (Sigma Plot 12, Systat Software Inc).
Statistics—QuantiTect Assays
One‐way ANOVA of mean E−ΔCt values was used to identify significant mRNA expression differences between tissues. Normality of data was tested using a Shapiro–Wilk test, and Holm–Sidak post hoc tests were used for comparison between tissues using Sigma Plot software (Sigma Plot 12, Systat Software Inc). P<0.05 was taken to be significant. A Benjamini–Hochberg adjustment for FDR was also calculated as a correction for multiple comparisons. We have used http://strimmerlab.org/notes/fdr.html for FDR analysis as recommended by a statistician.
Cluster Analysis
Two‐way hierarchical analysis was performed using StatMiner on ΔCt values using Ward's clustering method and Euclidean distance similarity measure.
Results
Location of AV Rings and Retroaortic Node and Relationship to CCS
By using histology and immunohistochemistry, we were able to localize all components of the CCS of the rat heart including the AV rings and retroaortic node: the SN, right and left rings, retroaortic node, nodal extensions, AVN His or penetrating bundle, and right and left bundle branches were HCN4+ and Cx43– (Figure 3). Immunodetection using HCN4 and Cx43 antibodies performed on Western blot demonstrated specific bands at the expected molecular weights (≈150 and ≈43 kDa, respectively) (Figure 4). Cx43 expression was shown to be significantly higher in the ventricular muscle than in the right ring tissue and atrial muscle (Figure 4, top), while HCN4 expression was significantly higher in SN than in all the other tissues (Figure 4, middle). Immunolabeling of HCN4 and Cx43 at various levels through the heart is shown in Figures 5 through 7. HCN4 is a positive marker for the CCS, while Cx43 is a positive marker for working myocardium.13 Although the Purkinje fibers did not show discernible labeling of HCN4, they could be localized because they were positive for Cx40; Cx40 was not discernibly expressed in the SN, AV rings, and AVN (retroaortic node not tested), whereas it was expressed in the penetrating bundle, bundle branches, and Purkinje fibers (Figures 7 and 8). Cx40 is a positive marker for the ventricular conduction system. The identified components occupied the same location in all 6 investigated hearts. MRI data for one of the hearts were used as a template onto which we mapped the location of the AV rings, retroaortic node, and the remainder of the CCS (Figures 1 and 9; Video S1). The 3D reconstruction or model shows the relation between the AV rings, retroaortic node, and principal parts of the CCS. The SN (Figure 9, yellow) is an extensive structure found at the junction of the superior vena cava with the right atrium. Based on HCN4 immunofluorescence, we found no evidence for any discrete specialized connections within the atrial walls between the SN and the AVN. The AVN (Figure 9, red) lies in the atrial septal component of the AV junctional area, with a prominent tongue of HCN4+ tissue extending from the AVN into the atrial septum (Figure 6). The right and left rings (Figure 9, green) occupy the vestibules of the tricuspid and mitral valves, respectively. They are seen as small HCN4+ regions at the insertion of the atrial myocardium into the AV junctions (Figure 5). When traced within the tricuspid vestibule, the right ring is seen to be continuous with the right nodal extension (Figure 5). The left ring is less well defined than the right ring, being no more than a thin strip of HCN4+ tissue within the mitral valvar vestibule at the level of the left AV junction (Figure 5). The retroaortic node is situated at the base of the atrial septum superior to the penetrating portion of the AV conduction axis (Figures 7 and 9, dark green). It is an extended structure, with connections to both the right and left rings. Expression of HCN4, as seen on immunofluorescence, was less in the AV rings than in the SN and AVN and less in the left ring than in the right ring. Tracing the HCN4+ signal through the heart shows how the ring tissues form a figure‐of‐eight configuration around the tricuspid and mitral valvar orifices, with the central components occupying the base of the atrial septum. As shown in Figure 9, the right and left rings are also continuous with both right and left nodal extensions from the AVN, the AV rings then uniting ventrally in the retroaortic node as they cross cranially over the penetrating portion of the AV conduction axis. The transition of the AVN into the penetrating bundle is marked by the insulation of the AV conduction axis from the atrial myocardium (Figure 6). The Purkinje fibers ramify within the ventral aspect of the ventricular musculature (Figures 7 through 9, blue).
Figure 3.
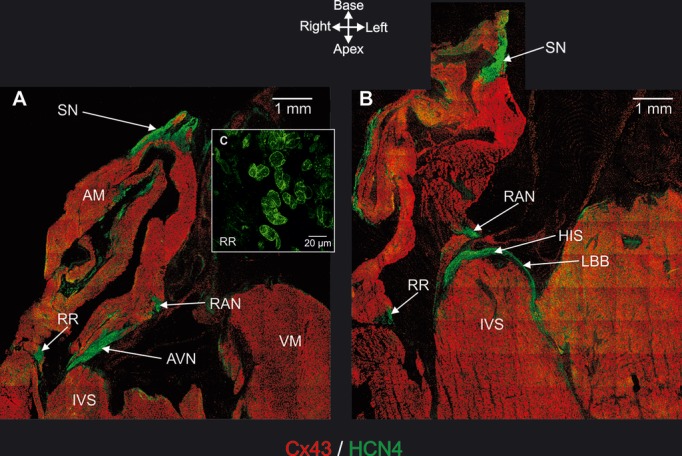
Immunohistochemical detection of AV rings and retroaortic node (and also principal tissues making up CCS). A and B, Long‐axis tissue sections immunolabeled for HCN4 (green signal) and Cx43 (red signal). Sections taken at level of AVN (A) and His bundle (B). C, High‐magnification image of HCN4 labeling in right ring. AM indicates atrial muscle; AVN, atrioventricular node; HIS, penetrating or His bundle; IVS, interventricular septum; LBB, left bundle branch; RAN, retroaortic node; RR, right ring; SN, sinus node; VM, ventricular muscle.
Figure 4.
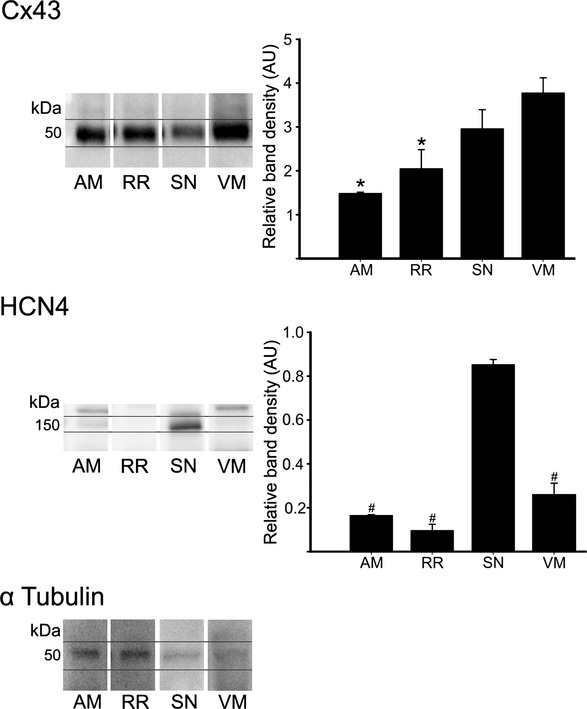
Western blot showing protein expression of Cx43 (top), HCN4 (middle), and α‐tubulin (bottom). Cx43 and HCN4 band density normalized to α‐tubulin presented in graphs. *P<0.05 vs VM; #P<0.05 vs SN. AM indicates atrial muscle; RR, right ring; SN, sinus node; VM, ventricular muscle.
Figure 5.
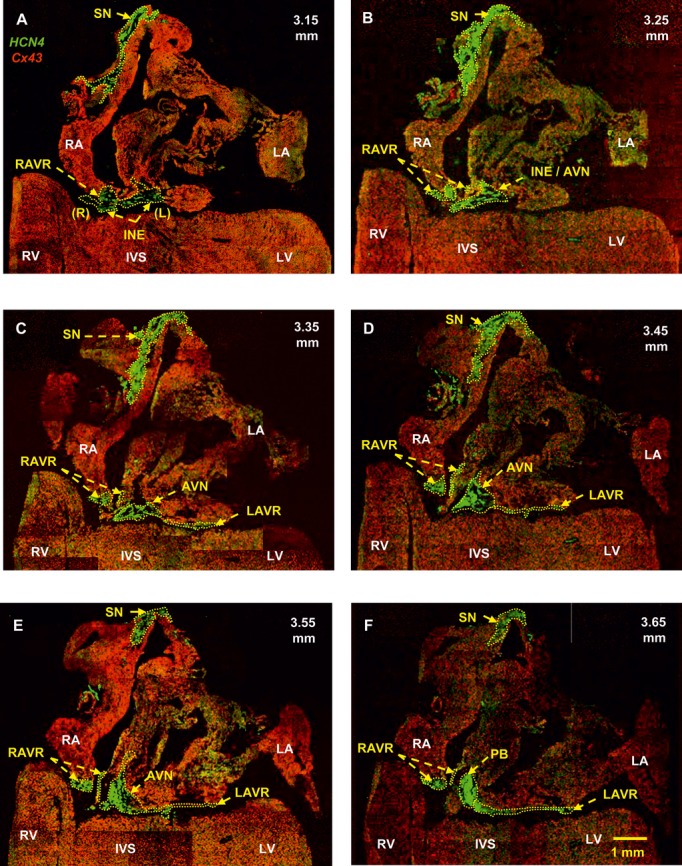
Identification of the SN, AV rings, inferior nodal extensions, AVN, and penetrating bundle. Double labeling of HCN4 (green signal) and Cx43 (red signal) proteins in long‐axis sections at different levels through the heart is shown (distance in mm from the back to the front of the heart shown). A, Section at 3.15 mm showing that the right and left rings are continuous with the right and left nodal extensions. B, Section at 3.25 mm showing the transitional area between the nodal extension and the AVN. C, Section at 3.35 mm showing the SN, right and left rings, and AVN. D, Section at 3.45 mm showing the atrial component of the AVN. The left ring is continuous with the AVN. E, Section at 3.55 mm showing again that the left ring is continuous with the AVN. F, Section at 3.65 mm showing that the left ring is continuous with the penetrating bundle. AVN indicates atrioventricular node; INE, inferior nodal extension (right and left); IVS, interventricular septum; LA, left atrium; LAVR, left atrioventricular ring; LV, left ventricle; PB, penetrating bundle; RA, right atrium; RAVR, right atrioventricular ring; RV, right ventricle; SN, sinus node.
Figure 7.
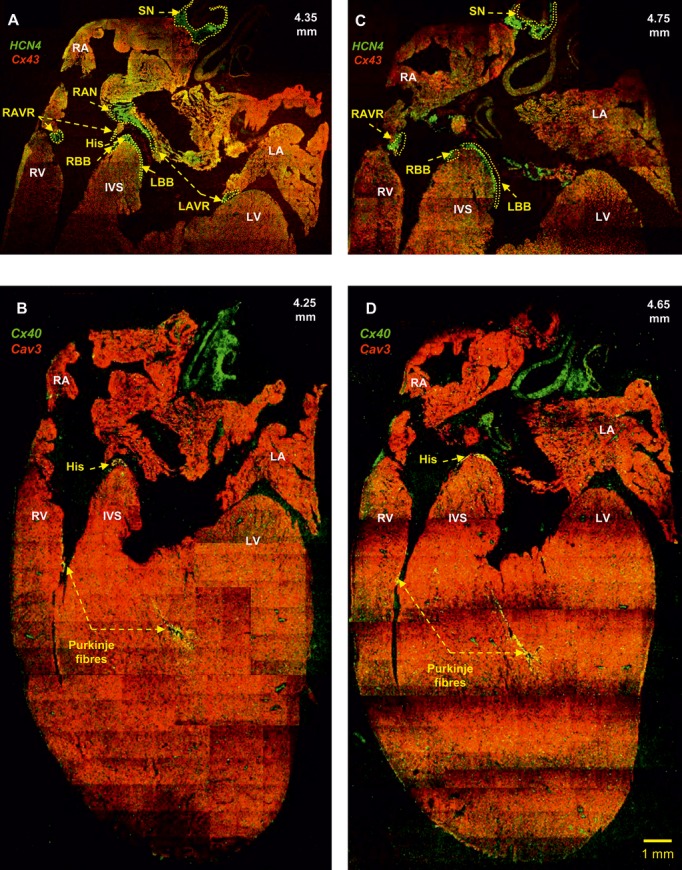
Identification of the AV rings, retroaortic node, right and left bundle branches, and Purkinje fibers. A and C, Double labeling of HCN4 (green signal) and Cx43 (red signal) proteins in long‐axis sections at 4.35 and 4.75 mm (from the back to the front of the heart). The right and left rings can be seen to unite within the retroaortic node. B and D, Double labeling of Cx40 (green signal) and caveolin3 (red signal) proteins in long‐axis sections at 4.25 and 4.65 mm (from the back to the front of the heart) showing the penetrating bundle and the Purkinje fibers. Same heart as shown in Figure 5. His indicates bundle of His; IVS, interventricular septum; LA, left atrium; LAVR, left atrioventricular ring; LBB, left bundle branch; LV, left ventricle; RA, right atrium; RAVR, right atrioventricular ring; RAN, retroaortic node; RBB, right bundle branch; RV, right ventricle; SN, sinus node.
Figure 8.
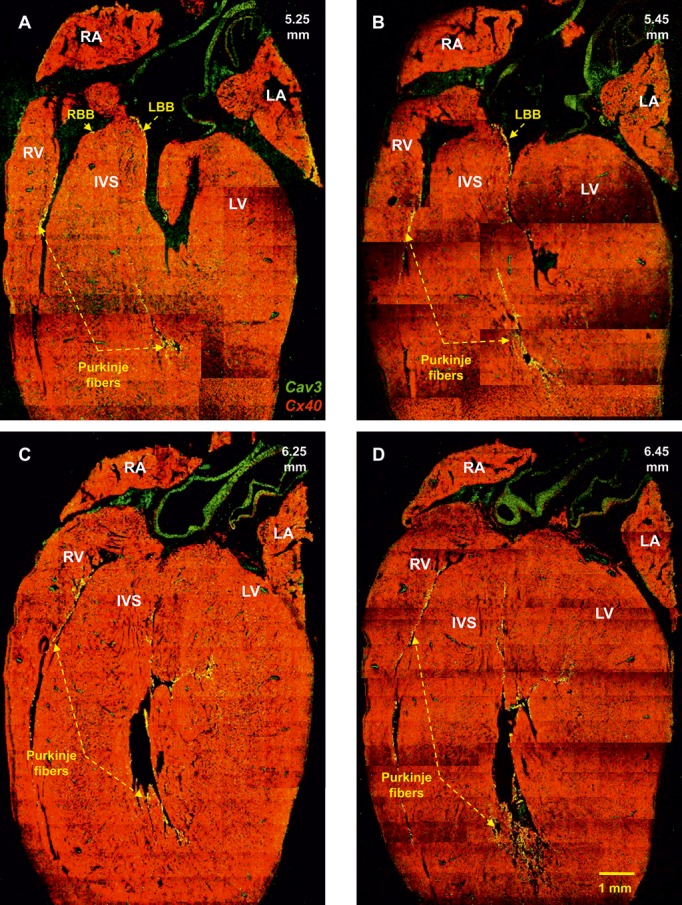
Identification of the right and left bundle branches and Purkinje fibres. Double labeling of Cx40 (green signal) and caveolin3 (red signal) proteins in long‐axis sections at 5.25 to 6.45 mm (from the back to the front of the heart) is shown. Same heart as shown in Figure 5. IVS indicates interventricular septum; LA, left atrium; LBB, left bundle branch; LV, left ventricle; RA, right atrium; RBB, right bundle branch; RV, right ventricle.
Figure 9.
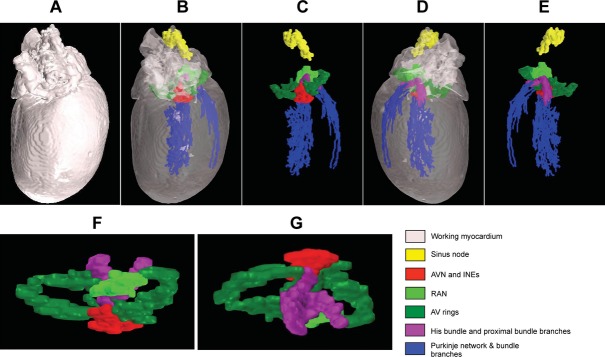
A three‐dimensional (3D) model of extended CCS. A, 3D model of heart (with opaque myocardium) viewed from dorsal surface. B, 3D model of heart (with transparent myocardium) viewed from dorsal surface. C, 3D model of extended CCS only (myocardium removed) viewed from dorsal surface. D, 3D model of heart (with transparent myocardium) viewed from ventral surface. E, 3D model of extended CCS only (myocardium removed) viewed from ventral surface. F and G, Top (F) and bottom (G) views of 3D model of AV rings, retroaortic node, and AV conduction axis. AVN indicates atrioventricular node; INEs, inferior nodal extensions; RAN, retroaortic node.
Figure 6.
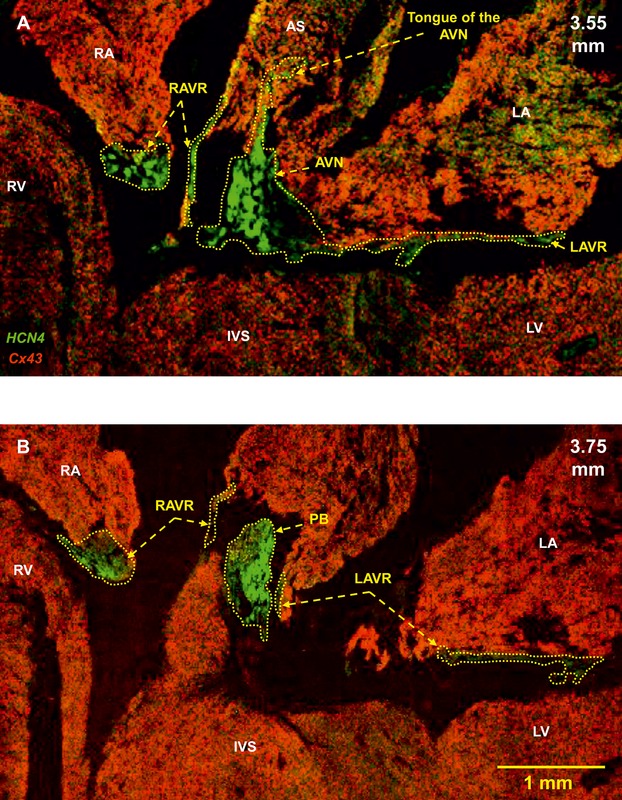
High‐magnification images of the AV rings, AVN, and penetrating bundle. Double labeling of HCN4 (green signal) and Cx43 (red signal) proteins in long‐axis sections at different levels through the heart is shown (distance in mm from the back to the front of the heart shown). A, Section at 3.55 mm showing the AVN with a tongue extending into the atrial septum. B, Section at 3.75 mm showing the penetrating bundle. Same heart as shown in Figure 5. AS indicates atrial septum; AVN, atrioventricular node; IVS, interventricular septum; LA, left atrium; LAVR, left atrioventricular ring; LV, left ventricle; PB, penetrating bundle; RA, right atrium; RAVR, right atrioventricular ring; RV, right ventricle.
Electrophysiology
Multiextracellular electrode mapping in the isolated rear wall of the right atrium (Figure 10A) revealed the site of earliest activation at the level of the SN. From here, electrical activity propagated to the rest of the right atrium and farther onto the right ring. A representative activation map is shown in Figure 10A. After detaching the SN, all 19 preparations showed spontaneous activation, and the site of earliest activation was located in the right ring (Figure 10B) (the valve leaflets were removed; therefore, this spontaneous activity was from the right ring rather than the tricuspid valve). Electrical excitation occurred at irregular intervals, and activation maps revealed multiple pacemaker sites within the right ring. In 5 of 19 preparations, exit blocks occurred; an example is shown in Figure 10B. Microelectrode recordings from the sites of earliest activation in isolated right ring preparations revealed spontaneous APs with characteristic diastolic depolarization (Figure 10C). The frequency of spontaneous APs varied from 0.9 to 4.4 Hz, with a mean value of 2.1±0.3 Hz (n=4 preparations; 13 impalements; Table 4). The upstroke of the AP (phase 0) was slow, with a maximum upstroke velocity (dV/dtmax) of 7±1 V/s (Table 4). The APA was 49±3 mV, and there was little or no overshoot (Table 4). The repolarization phase lacked a prominent plateau, and the duration of the AP (at −30 mV) varied from 35 to 144 ms, with an average value of 78±11 ms (Table 4). The maximum diastolic potential (MDP) reached after the AP was −50±3 mV (Table 4). The characteristics of right AV ring pacemaker APs are summarized in Table 4, along with corresponding measurements for APs recorded from the center of the SN and the terminal crest of the right atrium.
Figure 10.
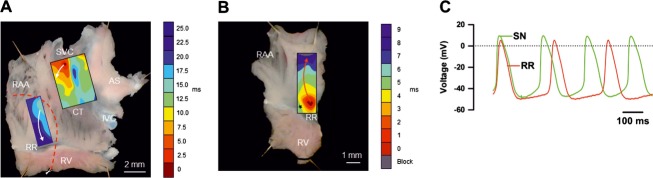
Pacemaker activity in right ring. A, Preparation of rear wall of right atrium containing SN and right ring. Boxes shows position of mapping arrays covering area of SN (8×8 extracellular electrodes) and right ring (6×10 extracellular electrodes). Leading pacemaker site (identified as site of earliest activation) located in SN, and anterograde conduction in right atrial free wall. Arrows show direction of AP conduction from SN to rest of preparation. To isolate right ring (as shown in B), tissue was cut along dashed line. B, Preparation containing right ring only (SN and AVN removed). Box shows position of mapping array (6×10 extracellular electrodes). Stars denotes pacemaker site in right ring and arrows show retrograde conduction from pacemaker site to remainder of right atrium. C, Typical intracellular action potentials recorded from pacemaker sites in right ring and SN. AS indicates atrial septum; CT, crista terminalis; IVC, inferior vena cava; RAA, right atrial appendage; RR, right ring; RV, right ventricle; SN, sinus node; SVC, superior vena cava.
Table 4.
Characteristics of Action Potentials
| Frequency, Hz | MDP, mV | APA, mV | Overshoot, mV | dV/dt, mV/ms | T50, ms | T90, ms | APD30, ms | |
|---|---|---|---|---|---|---|---|---|
| RR (n=4) | 2.1±0.3*† | −50.0±2.5* | 48.8±2.9* | −1.21±0.6* | 7.0±0.7* | 45.9±4.8* | 86.8±7.5* | 78.4±11.3* |
| SN (n=4) | 4.5±0.2‡ | −46.4±2.9* | 49.3±4.3* | 2.9±2.7* | 12.8±4.6* | 47.4±2.7* | 74.8±3.6* | 74.5±3.0* |
| RA (n=4) | 4.7±0.1†‡ | −78.1±0.8†‡ | 94.3±2.0†‡ | 16.2±2.4†‡ | 180±21.1†‡ | 13.3±1.2†‡ | 45.2±3.4†‡ | 13.2±0.8†‡ |
Frequency, maximum diastolic potential (MDP), action potential amplitude (APA), overshoot, maximum upstroke velocity (dV/dtmax), time taken for 50% repolarization (T50), time taken for 90% repolarization (T90), and action potential duration at −30 mV (APD30) in right ring (RR), sinus node (SN), and atrial muscle of terminal crest in right atrium (RA) are shown as mean±SEM values. Total of 13, 6, and 13 impalements were made from right ring, SN, and atrial muscle, respectively, from 4 preparations.
*P<0.05 vs atrial muscle; †P<0.05 vs SN; ‡P<0.05 vs right ring.
qPCR
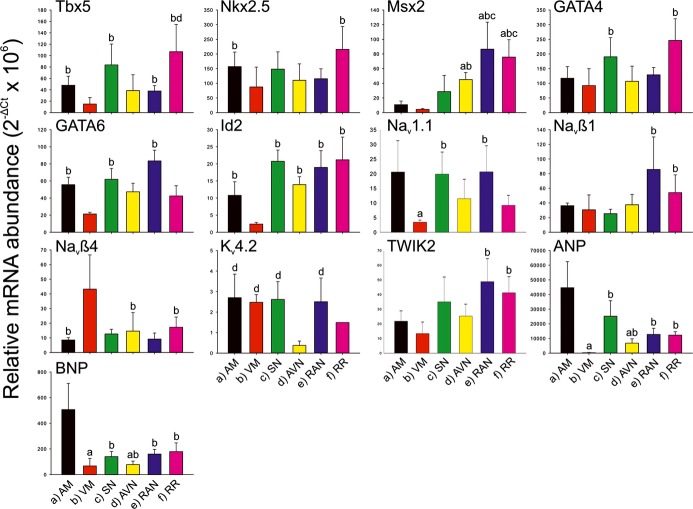
In the right ring and retroaortic node (as well as SN and AVN and atrial and ventricular muscle for comparison), qPCR was used to measure the expression (at mRNA level) of 79 targets: transcription factors, ion channels, Na+–K+ pump, exchangers, Ca2+‐handling proteins, connexins, extracellular matrix components, and a few miscellaneous targets. For information on the expression of all targets, see Tables S1 and S2. The expression of 2 atrial markers (ANP and BNP) was, as expected, high in atrial muscle and low in ventricular muscle and intermediate in the specialized tissues (Figure 11). Generally, there was significantly higher expression of extracellular matrix components (eg, collagens 1 and 3, vimentin, and elastin) in the right ring and retroaortic node than in the ventricular muscle (and, in some cases, atrial muscle); the SN and AVN tended to show the same expression pattern (Figure 12). This suggests that the right ring and retroaortic node contain more fibrous tissue (well‐known properties of SN and AVN). The expression pattern of transcription factors (responsible for tissue phenotype) and ion channels, Na+–K+ pump, exchangers, Ca2+‐handling proteins, and connexins (collectively responsible for electrical activity) are described in detail later. An overview of differences in the expression of these targets between regions is shown in Table 5.
Figure 11.
Relative abundance of mRNA for markers, transcription factors and ion channels. Means of 2−∆Ct (+SEM) shown (AM, n=4; VM, n=4; SN, n=5; AVN, n=4; RAN, n=5; RR, n=4). a through f, Significant difference (FDR <0.2, P<0.05) from corresponding bar. AM indicates atrial muscle; AVN, atrioventricular node; FDR, false discovery rate; FITC, fluorescein isothiocyanate; HRP, horseradish peroxidase; RAN, retroaortic node; RR, right ring tissue; SN, sinus node; VM, ventricular muscle.
Figure 12.

Relative abundance of mRNA for components of the extracellular matrix. Means of 2−∆Ct (+SEM) shown (AM, n=4; VM, n=4; SN, n=5; AVN, n=4; RAN, n=5; RR, n=4). A through F, Significant difference (FDR<0.2, P<0.05) from corresponding bar. AM indicates atrial muscle; ANP, atrial natriuretic peptide; AVN, atrioventricular node; BNP, brain natriuretic peptide; FDR, false discovery rate; RAN, retroaortic node; RR, right ring tissue; SN, sinus node; VM, ventricular muscle.
Table 5.
Comparison of Expression of Transcription Factors, Ion Channels, Na+–K+ Pump, Exchangers, Ca2+‐Handling Proteins, and Connexins Between Regions of Interest
| Transcript | Retroaortic Node vs Ventricular Muscle | Right AV Ring vs Ventricular Muscle | Retroaortic Node vs Atrial Muscle | Right AV Ring vs Atrial Muscle | Retroaortic Node vs SN | Right AV Ring vs SN | Retroaortic Node vs AV Node | Right AV Ring vs AV Node | Retroaortic Node vs Right AV Ring |
|---|---|---|---|---|---|---|---|---|---|
| Transcription factors | |||||||||
| Id2 | ↑ | ↑ | = | = | = | = | = | = | = |
| Msx1 | = | = | = | = | = | = | = | = | = |
| Msx2 | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | = | = | = |
| Tbx3 | ↑ | ↑ | ↑ | ↑ | = | = | = | = | = |
| Tbx5 | ↑ | ↑ | = | = | = | = | = | ↑ | = |
| Nkx2.5 | = | ↑ | = | = | = | = | = | = | = |
| GATA4 | = | ↑ | = | = | = | = | = | = | = |
| GATA6 | ↑ | = | = | = | = | = | = | = | = |
| Inward current carrying ion channels | |||||||||
| Nav1.1 | ↑ | = | = | = | = | = | = | = | = |
| Nav1.5 | = | = | ↓ | = | ↓ | = | = | = | = |
| Navβ1 | ↑ | ↑ | = | = | = | = | = | = | = |
| Navβ4 | ↓ | = | = | = | = | = | = | = | = |
| Cav1.2 | = | = | = | = | = | = | = | = | = |
| Cav1.3 | = | = | = | = | = | = | = | ↓ | = |
| Cav3.1 | ↑ | ↑ | = | = | = | = | = | = | = |
| Cav3.2 | ↑ | ↑ | = | = | = | = | = | = | = |
| HCN1 | ↑ | ↑ | ↑ | ↑ | ↓ | = | = | = | = |
| HCN2 | ↓ | ↓ | = | = | = | = | = | = | = |
| HCN4 | ↑ | ↑ | ↑ | ↑ | ↓ | = | = | = | = |
| Outward current carrying K+ channels | |||||||||
| Kv1.2 | = | = | ↓ | = | = | = | = | = | = |
| Kv1.4 | = | = | = | = | = | = | = | = | = |
| Kv1.5 | ↑ | ↑ | ↑ | ↑ | = | = | = | = | = |
| Kv2.1 | = | = | = | = | = | = | = | = | = |
| Kv4.2 | = | = | = | = | = | = | ↑ | = | = |
| Kv4.3 | = | = | = | = | = | = | = | ↑ | = |
| KvLQT1 (Kv7.1) | = | = | = | = | = | = | = | = | = |
| ERG (Kv11.1) | = | ↑ | = | = | = | = | = | = | = |
| KChIP2 | = | = | = | = | = | = | = | ↑ | = |
| Kvβ1 | = | ↑ | = | ↑ | = | = | ↑ | ↑ | = |
| SK1 (KCa2.1) | ↑ | ↑ | = | ↑ | = | = | = | = | = |
| SK2 (KCa2.2) | = | = | = | = | = | = | = | = | = |
| SK3 (KCa2.3) | = | ↑ | = | ↑ | = | = | = | = | = |
| Kir2.1 | ↓ | ↓ | = | = | = | = | = | = | = |
| Kir2.2 | ↑ | ↑ | = | = | = | = | = | = | = |
| Kir3.1 | ↑ | ↑ | ↓ | ↓ | = | = | = | = | = |
| Kir3.4 | ↑ | ↑ | = | = | = | = | = | = | = |
| Kir6.1 | = | = | = | = | = | = | = | = | = |
| Kir6.2 | = | = | = | = | = | = | = | = | = |
| SUR1 | = | ↑ | = | = | = | = | = | ↑ | = |
| SUR2 | ↓ | = | ↓ | = | = | = | = | = | = |
| TREK1 (K2P2.1) | = | = | = | = | = | = | = | = | = |
| TASK1 (K2P3.1) | = | = | = | ↓ | = | = | = | = | = |
| TASK2 (K2P5.1) | ↑ | ↑ | = | = | = | = | = | = | = |
| TWIK2 (K2P6.1) | ↑ | ↑ | = | = | = | = | = | = | = |
| Na+–K+ pump and exchangers | |||||||||
| Na+–K+ pump α3 | ↑ | ↑ | ↑ | ↑ | = | = | = | = | = |
| Na+–H+ exchanger | ↑ | ↑ | = | = | = | = | = | = | = |
| NCX1 | ↑ | ↑ | = | = | = | = | = | = | = |
| Ca2+‐handling proteins | |||||||||
| SERCA2a | = | ↑ | = | = | = | = | = | = | = |
| Phospholamban | = | = | = | = | = | = | = | = | = |
| Sarcolipin | ↑ | ↑ | = | = | = | = | = | ↑ | = |
| RYR2 | = | = | = | = | = | = | = | = | = |
| RYR3 | = | ↑ | = | = | = | = | = | = | = |
| Gap junction channels | |||||||||
| Gja5/Cx40 | = | ↑ | = | = | = | = | = | = | = |
| Gja1/Cx43 | = | = | ↓ | = | = | = | = | = | = |
| Gja7/Cx45 | ↑ | ↑ | = | = | = | = | = | = | = |
AV indicates atrioventricular; SN, sinus node; ↑, significantly upregulated; ↓, significantly downregulated; =, no difference.
Transcription Factors
The right ring and retroaortic node (and, in general, SN and AVN) had significantly higher expression of Tbx3 (Figure 13), Tbx5, Id2, and Msx2 (Figure 11) than the ventricular (and, in general, atrial) muscle. The right ring also had significantly higher expression of Nkx2.5 and GATA4 than the ventricular muscle (Figure 11); the retroaortic node had significantly higher expression of GATA6 than the ventricular muscle (Figure 11). There were no significant regional differences in Msx1 expression (Table 5).
Figure 13.
Relative abundance of mRNA for Tbx3 and ion channels underlying cardiac action potential. Means of 2−∆Ct (+SEM) shown (AM, n=4; VM, n=4; SN, n=5; AVN, n=4; RAN, n=5; RR, n=4). A through F, Significant difference (P<0.05, FDR<0.2) from corresponding bar. AM indicates atrial muscle; AVN, atrioventricular node; FDR, false discovery rate; MMP, matrix metalloproteinase; RAN, retroaortic node; RR, right ring; SN, sinus node; TGF, transforming growth factor; TIMP, tissue inhibitor of metalloproteinase; VM, ventricular muscle.
Channels Carrying Inward Current
HCN channels are important pacemaker channels and, as expected, HCN4 was the most abundantly expressed HCN channel (Figure 13). Expression of HCN4 was significantly higher in the right ring and retroaortic node than in the working myocardium—expression was similar to that in the AVN but not as high as that in the SN (Figure 13). The expression pattern of HCN1 was similar to that of HCN4, whereas HCN2 was most abundant in the ventricular muscle (Figure 13). A minor Na+ channel in the heart, Nav1.1, tended to be evenly expressed in the tissues investigated (Figure 11). However, expression of the dominant Na+ channel, Nav1.5 (responsible for AP upstroke in working myocardium), tended to be lower in the right ring and retroaortic node (and in AVN, but unexpectedly not in SN) than in the working myocardium (Figure 13). Two Na+ channel β subunits were differentially expressed in the right ring and retroaortic node (Figure 11). Whereas there were no significant differences in expression of the major L‐type Ca2+ channel, Cav1.2, between tissues, expression of 2 T‐type Ca2+ channels, Cav3.1 and Cav3.2 (important for pacemaker activity), was higher in the right ring and retroaortic node (and SN and AVN) than in the ventricular muscle (Figure 13).
K+ Channels Carrying Outward Current
Voltage‐gated and Ca2+‐activated K+ channels control APD. The expression of some of these (Kv1.5, ERG, Kvβ1, SK1/KCa2.1, and SK3/KCa2.3) tended to be higher in the right ring and retroaortic node (and SN and AVN) than in the working myocardium (Figure 13). In contrast, expression of Kv1.2, tended to be lower (Figure 13) and others (Kv1.4, Kv2.1, Kv4.2 [Figure 11], Kv4.3, KvLQT1, and SK2) unchanged (Table 5). In rat atrium, Kv1.2, Kv1.5, and Kv4.2 are reported to be functionally important channels.14 Kir2.1 is the major inward rectifier K+ channel responsible for the resting potential in the working myocardium. Expression of Kir2.1 was significantly lower in the right ring and retroaortic node (and SN and AVN) than in the ventricular muscle (Figure 13). Kir2.1 was more highly expressed in the ventricle than the atrium, but the reverse was true for Kir2.2; expression of Kir2.2 tended to be less in the right ring and retroaortic node (and AVN) than in the atrial muscle (Figure 13; Table 5). Expression of Kir3.1 and Kir3.4 (responsible for acetylcholine‐activated K+ channel) was significantly higher in all tissues including the right ring and retroaortic node than in the ventricular muscle (Figure 13). Twin pore K+ channels potentially could play a role in the resting potential; interestingly, TASK2 (K2P5.1) and TWIK2 (K2P6.1) expression tended to be higher in the right ring and retroaortic node (and SN and AVN) than in the working myocardium (Figure 11 and 13).
Na+–K+ Pump, Transporters, and Ca2+‐Handling Proteins
Expression of the α3 isoform of the Na+–K+ pump (Na+–K+,ATPase α3), the Na+–H+ exchanger, and the Na+–Ca2+ exchanger (NCX1) was significantly higher in the right ring and retroaortic node than in the ventricular muscle (and, in 1 case, atrial muscle); the SN and AVN showed a similar expression profile (Figure 14). Of the 2 ryanodine receptors (sarcoplasmic reticulum Ca2+ release channels), RYR2 was more highly expressed in all tissues (Figure 14). In terms of 4 Ca2+‐handling molecules (RYR2, RYR3, SERCA2, and sarcolipin), expression in the right ring was similar to that in the SN, whereas expression in the retroaortic node was similar to that in the AVN; in the right ring (and SN), expression of RYR3, SERCA2, and sarcolipin was significantly higher than in the ventricular muscle (Figure 14). Interestingly, sarcolipin was not detected in ventricular muscle (Figure 14).
Figure 14.

Relative abundance of mRNA for Na+–K+ pump, exchangers, Ca2+‐handling proteins, and connexins. Means of 2−∆Ct (+SEM) shown (AM, n=4; VM, n=4; SN, n=5; AVN, n=4; RAN, n=5; RR, n=4). A through F, Significant difference (P<0.05, FDR<0.2) from corresponding bar. AM indicates atrial muscle; AVN, atrioventricular node; FDR, false discovery rate; NCX1, Na+–Ca2+ exchanger; RAN, retroaortic node; RR, right ring; RYR, ryanodine receptor; SN, sinus node; VM, ventricular muscle.
Connexins
Of the 3 connexins measured (Cx40, Cx43, and Cx45), Cx43 was the most abundant in all tissues (Figure 14). There was a trend of lower expression of Cx43 in the right ring and retroaortic node (and SN and AVN) compared with the working myocardium (Figure 14). Cx40 expression was higher in the right ring (and in SN and AVN, but not in retroaortic node) than in the working myocardium (Figure 14, Table 5). Cx45 was uniformly expressed in all tissues, except for the ventricular muscle, in which Cx45 expression was significantly lower (Figure 14).
Cluster Analysis and Correlation
Based on the expression of 79 transcripts measured using TLDA cards, 2‐way hierarchical cluster analysis separated the samples into 7 clusters (Figure 15A). The samples fell into 4 unique sets, with the right ring, retroaortic node, and AVN samples clustering together (clusters 3 and 7), the SN and atrial muscle samples clustering together (clusters 1 and 6), and the ventricular muscle samples clustering separately (clusters 2 and 5). The only exception to this pattern was the grouping of the retroaortic node with the SN in cluster 4. Spearman rank order correlation shows that the right ring has a greater similarity to the retroaortic node and the remainder of the CCS than the working myocardium—the order of correlation for the right ring was retroaortic node>AVN>SN>atrial muscle>ventricular muscle (Figure 15B and 15C). The retroaortic node also had greatest similarity to the right ring and the remainder of the CCS:AVN=right ring>SN>atrial muscle>ventricular muscle (Figure 15C).
Figure 15.
Hierarchical clustering. A, Two‐way hierarchical cluster analysis applied to 79 genes and to samples of right ring (n=4), retroaortic node (n=5), sinus node (n=5), atrioventricular node (n=4), atrial muscle (n=4), and ventricular muscle (n=4). B, Plot of ∆Ct values of all transcripts for ventricular muscle vs those for right ring and plot of ∆Ct values of all transcripts for retroaortic node vs those for right ring. Line represents y=x. C, Spearman rank order correlation data for same samples used for cluster analysis. AM indicates atrial muscle; AVN, atrioventricular node; RAN, retroaortic node; RR, right ring; SN, sinus node; VM, ventricular muscle.
Protein Expression
Expression of some targets was also investigated at the protein level in the right ring, dependent on the availability of suitable antibodies. Expression of HCN4, Cx40, and Cx43 at the protein level has already been described (Figures 3 through 8) and is consistent with the mRNA expression (Figures 13 and 14). Figure 16A shows that Nav1.5 protein labeling was not found (above background fluorescence) within the right ring (and AVN) compared with the working myocardium. In the working myocardium, Nav1.5 protein was expressed at the cell membrane. Consistent with mRNA expression (Figure 13), expression of Cav3.1 protein was observed in the right AV ring (Figure 16A). High SK1 (KCa2.1) protein expression (at cell membrane) was observed in the penetrating bundle (Figures 16B and 17G). Expression of SK1 was also observed in the right ring (Figure 16C), while no expression was observed in the working myocardium (Figures 16D and 17F). This is generally consistent with SK1 mRNA expression (Figure 13). Kir2.1 protein expression (at cell membrane) was lower in the right ring than in the ventricular muscle (Figure 18A and 18B), consistent with mRNA expression (Figure 13). There was no detectable labeling of Kir3.1 in the right ring, while there was in the atrial muscle (Figure 17D and 17E). In contrast, Kir3.4 protein expression was observed in the right ring (at cell membrane), while there was weaker expression in the ventricular muscle (Figure 18C and 18D). The expression pattern of Kir3.1 and Kir3.4 proteins is consistent with the mRNA expression (Figure 13). Expression of RYR2 (Figure 18E and 18F) and NCX1 (Figure 17B and 17C) proteins was similar in the right ring and ventricular muscle, consistent with the mRNA expression (Figure 14).
Figure 16.

Expression of Nav1.5 and SK1 proteins in AV junctional area. A, Immunolabeling of Nav1.5 (bright green signal) in right ring and AVN in AV junctional area. Long‐axis section. B, Immunolabeling of SK1 (bright red signal) in penetrating bundle. C, High magnification image of immunolabeling of SK1 in right ring. D, High magnification image of immunolabeling of SK1 in right ventricle. AVN indicates atrioventricular node; PB, penetrating bundle; RA, right atrium; RR, right ring; RV, right ventricle; VS, ventricular septum.
Figure 17.
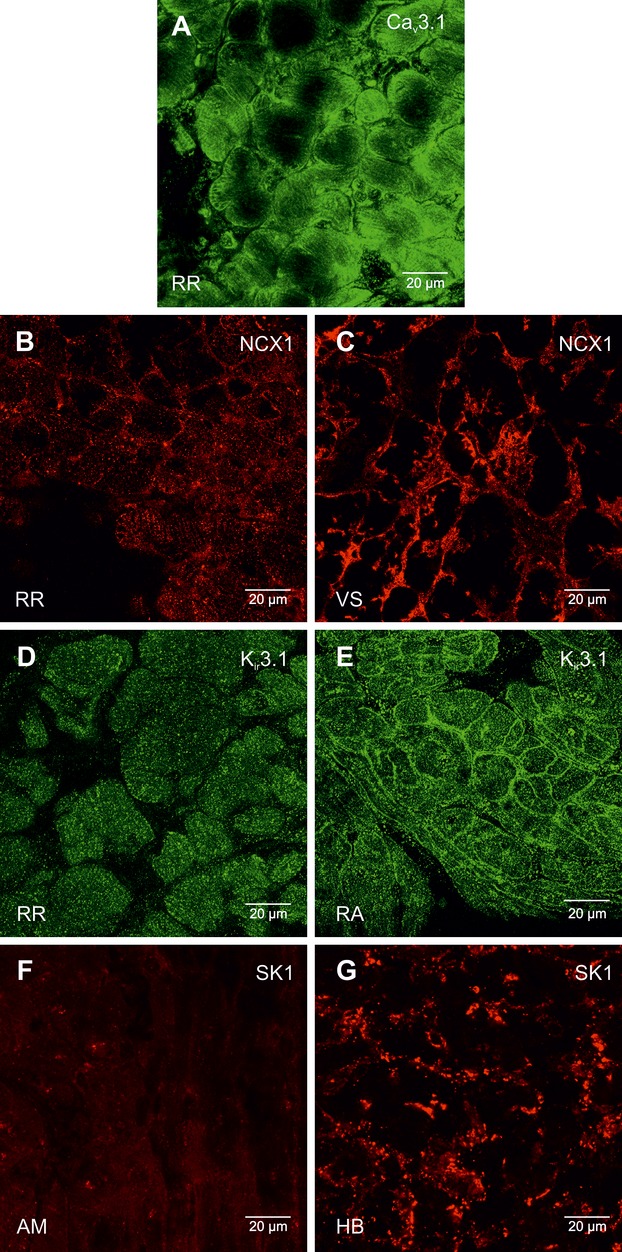
High magnification images of Cav3.1, NCX1, Kir3.1, and SK1 protein labeling. A, Cav3.1 protein labeling (green signal) in the right ring. B and C, NCX1 protein labeling (red signal) in the right ring (B) and ventricular septum (C). D and E, Kir3.1 protein labeling (green signal) in the right ring (D) and atrial muscle (E) RA. F and G, SK1 protein labeling (red signal) in atrial muscle (F) AM and the His bundle (G). AM indicates atrial muscle; HB, bundle of His; NCX1, Na+–Ca2+ exchanger; RA, right atrium; RR, right ring tissue; VS, ventricular septum.
Figure 18.

Expression of Kir2.1, Kir3.4, and RYR2 proteins in right ring and right ventricle. High magnification images of immunolabeling (bright green signal) shown. RR indicates right ring; RV, right ventricle; RYR, ryanodine receptor; VS, ventricular septum.
Discussion
This study (1) presents the first 3D reconstruction/model of the entirety of the CCS including the AV rings and retroaortic node, (2) demonstrates the pacemaker potentiality of the right ring, and (3) shows that the expression profile of key transcription factors, ion channels, Na+‐K+ pump, exchangers, Ca2+‐handling proteins, connexins, and extracellular matrix components of the right ring and retroaortic node is similar to that of the SN and AVN. Together, these data show that the right ring and retroaortic node have characteristics that could result in the generation of ectopic beats and thus underlie atrial tachycardias known to originate within the vestibules of the AV valves.4
Anatomy of Extended CCS
The rival notions of multiple muscular pathways for normal AV conduction, as proposed by Kent (1893), and a solitary pathway, as proposed by His (1893), existed until the work of Tawara15 conclusively proved that there was but a solitary pathway connecting (anatomically and electrically) the atrial and ventricular muscular masses. The node‐like structures illustrated by Kent, however, do exist and have been shown to be constant features within the muscular vestibule of the tricuspid valve in structurally normal human hearts, albeit insulated from the ventricular myocardium.16 There is little doubt that these structures, as seen in the human heart, are analogous to the more complete rings of specialized tissue identified in the hearts of rabbits, guinea pigs, rats, and mice.2–3 The morphology of the AV rings was misinterpreted in early studies, with 2 separate rings being shown surrounding the AV valves.2 Our current study (Figure 9; Video S1) shows unequivocally that the AV rings and retroaortic node are continuous with the AV conduction axis. There is strong evidence showing that the AV rings are remnants of the embryonic AV canal.6 Our 3D model demonstrates how the AV rings cross over the penetrating part of the AV conduction axis, forming the extensive retroaortic node recognized both histologically and immunohistochemically (Figure 9; Video S1). This “third” node is anatomically discrete from the AVN3 and not continuous with it, as suggested by Blom et al.17 We presume that, initially, the retroaortic node is also in continuity with the ventral continuation of the AV conduction axis, because in the developing heart a ring is known to encircle the aortic root and return to the right atrial vestibule5,18; we were unable to demonstrate this pathway in the current investigation. Based on its extent and its continuity with the AV rings, we also presume that the retroaortic node must have a functional role, although as yet this is unknown.
The 3D model (Figure 9; Video S1) also provides insights into the rest of the CCS. The 3D model shows the sinus node in situ (Figure 9; Video S1). The SN is an extensive inverted U‐shaped structure in the rat, endorsing the previous description of Yamamoto et al8 and in agreement with observations in the human and rabbit.13,19 The 3D model shows that the bulk of the SN lies in the region of the superior caval vein at the crest of the right atrial appendage, with a tail of nodal tissue extending down both the terminal crest and the interatrial groove, as observed in some human hearts.20 In the 3D model (Figure 9; Video S1), the SN is more extensive than is generally depicted in anatomical textbooks.21
In the developing heart, the nodal primordia are extensive and interconnected. Thus, in the mouse, as identified by marker genes such as the transcription factor Tbx3, a continuous tract of primary myocardium, the future nodal tissue, extends from within the confines of the systemic venous sinus, becoming continuous with the primary myocardium of the AV canal, from which will develop the AVN.22–23 The continuous tract of primary myocardium in the developing heart is suggestive of an “internodal tract,” such as that proposed by James24 to provide preferential conduction from the SN to the AVN. The 3D model (Figure 9; Video S1), however, reveals there is a gap between the SN and the AVN in the adult heart. Moreover, use of standard histological techniques in the postnatal heart, basing the findings on the criterions proposed by Aschoff and Monkeberg, has consistently failed to identify any insulated tracts within the atrial myocardium.25 It follows, therefore, that much of the primary myocardium must lose the histological features of nodal tissue as the embryo matures.23 Furthermore, in the postnatal heart, preferential conduction from the SN to the AVN, as well as into the left atrium, can adequately be explained on the basis of the orderly longitudinal packing of atrial myocytes in prominent muscular bundles, such as the terminal crest and Bachmann bundle.26–27
Transcription Factors
The establishment of the pacemaking phenotype in specific regions of the heart is achieved during the embryonic development of the heart, with expression of transcription factors being critical in enhancing or repressing the expression of certain genes during development.28 Tbx3 has been shown to repress chamber (ie, working myocardium)‐specific genes such as Nav1.5, Cx40, and Cx45.5–6 Our results show Tbx3 expression in the right ring and retroaortic node to be comparable to that in other components of the CCS (Figure 13). Expression of Tbx5 and Nkx2.5 was also high in the right ring (Figure 11). Nkx2.5 expression in the nodal regions is reported to be low (it is thought to act as a repressor of HCN4,29 although some evidence exists for HCN4 expression in Nkx2.5‐positive regions in humans).30 Msx2 has been found in the AV canal, particularly in the cushions, and in close association with the CCS,31 where it is thought to interact with the Tbx transcription factors to regulate connexin expression.31–32 Msx2 knockout mice have been shown to develop abnormal AV valves, along with the expression of chamber‐specific markers in the cardiomyocytes of the AV junction.31–32 The finding of a high level of Msx2 in the right ring (Figure 11) supports the hypothesis that AV rings are developmentally similar to the main components of the CCS. The GATA4 and GATA6 transcription factors are known to affect the expression of genes within the heart, with GATA binding motifs identified on many genes, including NCX1.33 We observed high GATA4 expression in the right ring and high GATA6 expression in the retroaortic node (Figure 11), although the functional consequence of these differences is unclear. Id2 is a promoter of conduction system gene expression and has been shown to have functional binding sites for Tbx5 and is cooperatively regulated by Tbx5 and Nkx2.5.34–35 Expression of Id2 was high in the right ring and retroaortic node as well as the SN and AVN (Figure 11). The complex interactions between the transcription factors, the high degree of redundancy, and multiple levels of regulation including gene expression levels and posttranscriptional modification are thought to provide a mechanism ensuring tight regulation of the patterning and functioning of the CCS.36 Our qPCR data show that the right ring and retroaortic node share an expression profile of transcription factors closer to that of the CCS than the working myocardium (Table 5), again supporting the developmental evidence that the right ring and retroaortic node originate from the same primordium as the AVN.
Pacemaking in Right Ring
Using surface electrode mapping, we demonstrated that the right ring is capable of pacemaking and propagating electrical activity to the atria, albeit only when the dominant pacemaking tissue of the SN and AVN were detached (Figure 10). Microelectrode recordings revealed a less negative MDP in the right ring, comparable to the MDP in the SN (Figure 10C; Table 4). Consistent with this, we observed low mRNA and protein levels in the right ring and SN for Kir2.1 (Figures 13 and 18), a channel that contributes to IK,1, which is responsible for the resting membrane potential in the working myocardium.37 Expression of Kir2.2 (another channel contributing to IK,1) in the right ring was also relatively low (Table 5). A less negative MDP in the right ring, like in the SN, is expected to favor pacemaking. At the site of earliest activation in the right ring, APs showed spontaneous diastolic depolarizations, akin to that in the SN. Our results clearly support the pacemaking potentiality of the right AV ring and are consistent with the observation of APs with diastolic depolarizations recorded from the AV ring region of other species.10,38–39 Although the MDP (and dV/dtmax, APA, and APD in the right ring was similar to that in the SN, the spontaneous rate in the right ring was slower (Figure 10; Table 4). While the mechanisms underlying the diastolic depolarization in the right ring have yet to be fully investigated, the expression data from this study suggest involvement of both membrane‐ and Ca2+‐clock mechanisms. HCN channels and If are major determinants of the diastolic depolarization in the SN.40 In the right ring and retroaortic node, we observed expression of HCN1 and HCN4 channels at mRNA and protein levels comparable to that in the AVN (Figures 3, 5 through 7, and 13). Cav3.1 and Cav3.2 are responsible for the T‐type Ca2+ current (ICa,T), which is also thought to contribute to the diastolic depolarization in the SN.41 High HCN, Cav3.1, and Cav3.2 expression in the right ring and retroaortic node supports the operation of the membrane‐clock pacemaking mechanism. According to the Ca2+‐clock mechanism of pacemaking, spontaneous Ca2+ release (via ryanodine receptor RYR2) from the sarcoplasmic reticulum initiates an inward NCX1 current (INaCa) that contributes to the diastolic depolarization.42 The data show high mRNA and protein expression of RYR2 and NCX1 in the right ring and possibly retroaortic node (Figures 14, 16, and 18). Thus, our data suggest that a number of mechanisms contribute to pacemaking in the right ring, and this is not unlike pacemaking in the SN. Nav1.5 is responsible for the large and fast Na+ current, INa, which in turn is responsible for the AP upstroke in working myocardium. Because of the lower expression of Nav1.5 in nodal tissues (Figures 13 and 17A), Cav1.2 (and Cav1.3) and the smaller and slower L‐type Ca2+ current, ICa,L, are responsible for the AP upstroke in nodal tissues.43 This is why dV/dtmax is high in the working myocardium and low in nodal tissues (Table 4). In the right ring, dV/dtmax was ≈7 V/s, comparable to that in the SN (≈12 V/s), as opposed to 180 V/s in the right atrium (Table 4). This is likely to be the result of the low expression of Nav1.5 in the right ring (Figures 13 and 17A), and it presumably indicates that ICa,L is responsible for the AP upstroke in the right AV ring. The AP in the right ring, like that in the SN, was longer than that in the atrial muscle (Table 4). This could be the result of a decrease in expression of Kv1.2 in the right ring and SN (Figure 13). There was high expression of SK1 (KCa2.1) (Figures 13 and 17) in the right ring. This channel is involved in phase 3 repolarization during the AP44 and has been linked to spontaneous activity and arrhythmia.45–46 In patients with focal atrial tachycardia, ectopic foci are often found in the tricuspid and mitral valve annuli.4,47 It is likely that the AV rings are responsible.
Conduction Velocity and Connexin Expression
Connexin expression determines the degree of electrical coupling of myocytes, which in turn is an important determinant of the conduction velocity of the AP. The lowest expression of the principal connexin, Cx43, at the mRNA level, was found in the right ring, retroaortic node and AVN (Figure 14). Figures 3 and 5 through 7 show that there was no discernible labeling of Cx43 protein in the AV rings and retroaortic node as well as the SN and AVN. It is well known that the conduction velocity of the SN and AVN is low, and this suggests that the conduction velocity of the AV rings is also low; this may be important for arrhythmogenesis. In the rabbit, De Carvalho et al38 reported a low conduction velocity in the expected location of the right ring. We observed high Cx45 (small conductance connexin) expression in the right ring (and retroaortic node, SN, and AVN; Figure 14), which is consistent with the findings of Coppen et al,48 who demonstrated the presence of Cx45 expression at the AV junction at a location that matches the anatomy of the AV rings.
Similarities and Differences Among AV Rings, Retroaortic Node, and AV Node
We have shown that, anatomically, the AV rings, retroaortic node, and AVN form a continuous structure surrounding the AV valvar orifices. It is known that, developmentally, the ring tissues are derived from the AV canal, implying that they would possess some of the same functionality as the AVNs.6 In our study, of the 79 transcripts studied using qPCR, we observed no significant differences between the right ring and retroaortic node, 10 significant differences between the right ring and the AVN, and 3 significant differences between the retroaortic node and the AVN (Table S1). This suggests that the different tissues have similar profiles of expression of the genes investigated, albeit with some differences between the right ring/retroaortic node and the AVN, which presumably could be critical in preventing the ring tissues from acting as primary pacemakers under normal conditions. In general, the gene expression profile of the right ring and retroaortic node is that of a pacemaker tissue, and it is likely that both tissues could act as ectopic pacemakers.
Sources of Funding
This study was supported by the British Heart Foundation (PG/08/055/25310).
Disclosures
None.
References
- 1.Anderson RH, Yanni J, Boyett MR, Chandler NJ, Dobrzynski H. The anatomy of the cardiac conduction system. Clin Anat. 2009; 22:99-113 [DOI] [PubMed] [Google Scholar]
- 2.Anderson RH. The disposition and innervation of atrioventricular ring specialized tissue in rats and rabbits. J Anat. 1972; 113:197-211 [PMC free article] [PubMed] [Google Scholar]
- 3.Yanni J, Boyett MR, Anderson RH, Dobrzynski H. The extent of the specialized atrioventricular ring tissues. Heart Rhythm. 2009; 6:672-680 [DOI] [PubMed] [Google Scholar]
- 4.Kistler PM, Roberts‐Thomson KC, Haqqani HM, Fynn SP, Singarayar S, Vohra JK, Morton JB, Sparks PB, Kalman JM. P‐wave morphology in focal atrial tachycardia: development of an algorithm to predict the anatomic site of origin. J Am Coll Cardiol. 2006; 48:1010-1017 [DOI] [PubMed] [Google Scholar]
- 5.Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004; 62:489-499 [DOI] [PubMed] [Google Scholar]
- 6.Aanhaanen WT, Mommersteeg MT, Norden J, Wakker V, de Gier‐de Vries C, Anderson RH, Kispert A, Moorman AF, Christoffels VM. Developmental origin, growth, and three‐dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ Res. 2010; 107:728-736 [DOI] [PubMed] [Google Scholar]
- 7.Anderson RH, Taylor IM. Development of atrioventricular specialized tissue in human heart. Br Heart J. 1972; 34:1205-1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto M, Dobrzynski H, Tellez J, Niwa R, Billeter R, Honjo H, Kodama I, Boyett MR. Extended atrial conduction system characterised by the expression of the HCN4 channel and connexin45. Cardiovasc Res. 2006; 72:271-281 [DOI] [PubMed] [Google Scholar]
- 9.McGuire MA, de Bakker JM, Vermeulen JT, Opthof T, Becker AE, Janse MJ. Origin and significance of double potentials near the atrioventricular node. Correlation of extracellular potentials, intracellular potentials, and histology. Circulation. 1994; 89:2351-2360 [DOI] [PubMed] [Google Scholar]
- 10.McGuire MA, de Bakker JM, Vermeulen JT, Moorman AF, Loh P, Thibault B, Vermeulen JL, Becker AE, Janse MJ. Atrioventricular junctional tissue. Discrepancy between histological and electrophysiological characteristics. Circulation. 1996; 94:571-577 [DOI] [PubMed] [Google Scholar]
- 11.Li J, Greener ID, Inada S, Nikolski VP, Yamamoto M, Hancox JC, Zhang H, Billeter R, Efimov IR, Dobrzynski H, Boyett MR. Computer three‐dimensional reconstruction of the atrioventricular node. Circ Res. 2008; 102:975-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers SA, Eriksson N, Burow R, Wang SC, Muscat GE. Beta‐adrenergic signaling regulates NR4A nuclear receptor and metabolic gene expression in multiple tissues. Mol Cell Endocrinol. 2009; 309:101-108 [DOI] [PubMed] [Google Scholar]
- 13.Chandler N, Aslanidi O, Buckley D, Inada S, Birchall S, Atkinson A, Kirk D, Monfredi O, Molenaar P, Anderson R, Sharma V, Sigg D, Zhang H, Boyett M, Dobrzynski H. Computer three‐dimensional anatomical reconstruction of the human sinus node and a novel paranodal area. Anat Rec (Hoboken). 2011; 294:970-979 [DOI] [PubMed] [Google Scholar]
- 14.Bou‐Abboud E, Nerbonne JM. Molecular correlates of the calcium‐independent, depolarization‐activated K+ currents in rat atrial myocytes. J Physiol. 1999; 517:407-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tawara S. Das Reizleitungssystem des Säugetierherzens. Eine Anatomisch Histologische Studie Über das Atrioventrikularbündel und die Purkinjeschen Fäden. 1906Jena: Gustav Fischer [Google Scholar]
- 16.Anderson RH, Becker AE, Arnold R, Wilkinson JL. The conduction tissues in congenitally corrected transposition. Circulation. 1974; 50:911-923 [DOI] [PubMed] [Google Scholar]
- 17.Blom NA, Gittenberger‐de Groot AC, DeRuiter MC, Poelmann RE, Mentink MM, Ottenkamp J. Development of the cardiac conduction tissue in human embryos using HNK‐1 antigen expression: possible relevance for understanding of abnormal atrial automaticity. Circulation. 1999; 99:800-806 [DOI] [PubMed] [Google Scholar]
- 18.Lamers WH, Wessels A, Verbeek FJ, Moorman AF, Virágh S, Wenink AC, Gittenberger‐de Groot AC, Anderson RH. New findings concerning ventricular septation in the human heart. Implication for maldevelopment. Circulation. 1992; 86:1194-1205 [DOI] [PubMed] [Google Scholar]
- 19.Dobrzynski H, Li J, Tellez J, Greener ID, Nikolski VP, Wright SE, Parson SH, Jones SA, Lancaster MK, Yamamoto M, Honjo H, Takagishi Y, Kodama I, Efimov IR, Billeter R, Boyett MR. Computer three‐dimensional reconstruction of the sinoatrial node. Circulation. 2005; 111:846-854 [DOI] [PubMed] [Google Scholar]
- 20.Anderson RH, Ho SY. The architecture of the sinus node, the atrioventricular conduction axis, and the internodal atrial myocardium. J Cardiovasc Electrophysiol. 1998; 9:1233-1248 [DOI] [PubMed] [Google Scholar]
- 21.Iaizzo PA. Handbook of Cardiac Anatomy, Physiology, and Devices. 20092nd edTotowa, NJ: Humana Press [Google Scholar]
- 22.Rentschler S, Vaidya DM, Tamaddon H, Degenhardt K, Sassoon D, Morley GE, Jalife J, Fishman GI. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001; 128:1785-1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moorman AF, Christoffels VM, Anderson RH. Anatomic substrates for cardiac conduction. Heart Rhythm. 2005; 2:875-886 [DOI] [PubMed] [Google Scholar]
- 24.James TN. The connecting pathways between the sinus node and A‐V node and between the right and the left atrium in the human heart. Am Heart J. 1963; 66:498-508 [DOI] [PubMed] [Google Scholar]
- 25.Janse MJ, Anderson RH. Specialized internodal atrial pathways: fact or fiction? Eur J Cardiol. 1974; 2:117-136 [Google Scholar]
- 26.Spach MS, Kootsey JM. The nature of electrical propagation in cardiac muscle. Am J Physiol. 1983; 244:H3-H22 [DOI] [PubMed] [Google Scholar]
- 27.Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity. Promoting understanding of sick sinus syndrome. Circulation. 2007; 115:1921-1932 [DOI] [PubMed] [Google Scholar]
- 28.Hatcher CJ, Basson CT. Specification of the cardiac conduction system by transcription factors. Circ Res. 2009; 105:620-630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mommersteeg MT, Hoogaars WM, Prall OW, De Gier‐de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, Christoffels VM. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007; 100:354-362 [DOI] [PubMed] [Google Scholar]
- 30.Sizarov A, Devalla HD, Anderson RH, Passier R, Christoffels VM, Moorman AF. Molecular analysis of patterning of conduction tissues in the developing human heart. Circ Arrhythm Electrophysiol. 2011; 4:532-542 [DOI] [PubMed] [Google Scholar]
- 31.Boogerd CJ, Moorman AF, Barnett P. Expression of muscle segment homeobox genes in the developing myocardium. Anat Rec (Hoboken). 2010; 293:998-1001 [DOI] [PubMed] [Google Scholar]
- 32.Boogerd CJ, Moorman AF, Barnett P. Protein interactions at the heart of cardiac chamber formation. Ann Anat. 2009; 191:505-517 [DOI] [PubMed] [Google Scholar]
- 33.Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004; 63:196-207 [DOI] [PubMed] [Google Scholar]
- 34.Lim JY, Kim WH, Kim J, Park SI. Induction of Id2 expression by cardiac transcription factors GATA4 and Nkx2.5. J Cell Biochem. 2008; 103:182-194 [DOI] [PubMed] [Google Scholar]
- 35.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including Id2, Tbx5, and Nk2‐5 required for cardiac conduction system development. Cell. 2007; 129:1365-1376 [DOI] [PubMed] [Google Scholar]
- 36.Brewer A, Pizzey J. GATA factors in vertebrate heart development and disease. Expert Rev Mol Med. 2006; 8:1-20 [DOI] [PubMed] [Google Scholar]
- 37.Noma A, Nakayama T, Kurachi Y, Irisawa H. Resting K+ conductances in pacemaker and non‐pacemaker heart cells of the rabbit. Jpn J Physiol. 1984; 34:245-254 [DOI] [PubMed] [Google Scholar]
- 38.De Carvalho AP, deMello WC, Hoffman BF. Electrophysiological evidence for specialized fiber types in rabbit atrium. Am J Physiol. 1959; 196:483-488 [DOI] [PubMed] [Google Scholar]
- 39.Rozanski GJ, Jalife J. Automaticity in atrioventricular valve leaflets of rabbit heart. Am J Physiol. 1986; 250:H397-H406 [DOI] [PubMed] [Google Scholar]
- 40.DiFrancesco D, Ducouret P, Robinson RB. Muscarinic modulation of cardiac rate at low acetylcholine concentrations. Science. 1989; 243:669-671 [DOI] [PubMed] [Google Scholar]
- 41.Mangoni ME, Couette B, Bourinet E, Platzer J, Reimer D, Striessnig J, Nargeot J. Functional role of L‐type Cav1.3 Ca2+ channels in cardiac pacemaker activity. Proc Natl Acad Sci USA. 2003; 100:5543-5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart's pacemaker. Circ Res. 2010; 106:659-673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schram G, Pourrier M, Melnyk P, Nattel S. Differential distribution of cardiac ion channel expression as a basis for regional specialization in electrical function. Circ Res. 2002; 90:939-950 [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Tuteja D, Zhang Z, Xu D, Zhang Y, Rodriguez J, Nie L, Tuxson HR, Young JN, Glatter KA, Vázquez AE, Yamoah EN, Chiamvimonvat N. Molecular identification and functional roles of a Ca2+‐activated K+ channel in human and mouse hearts. J Biol Chem. 2003; 278:49085-49094 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q, Timofeyev V, Lu L, Li N, Singapuri A, Long MK, Bond CT, Adelman JP, Chiamvimonvat N. Functional roles of a Ca2+‐activated K+ channel in atrioventricular nodes. Circ Res. 2008; 102:465-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diness JG, Sørensen US, Nissen JD, Al‐Shahib B, Jespersen T, Grunnet M, Hansen RS. Inhibition of small‐conductance Ca2+‐activated K+ channels terminates and protects against atrial fibrillation. Circ Arrhythm Electrophysiol. 2010; 3:380-390 [DOI] [PubMed] [Google Scholar]
- 47.Morton JB, Sanders P, Das A, Vohra JK, Sparks PB, Kalman JM. Focal atrial tachycardia arising from the tricuspid annulus: electrophysiologic and electrocardiographic characteristics. J Cardiovasc Electrophysiol. 2001; 12:653-659 [DOI] [PubMed] [Google Scholar]
- 48.Coppen SR, Severs NJ, Gourdie RG. Connexin45 (alpha 6) expression delineates an extended conduction system in the embryonic and mature rodent heart. Dev Genet. 1999; 24:82-90 [DOI] [PubMed] [Google Scholar]