SUMMARY
Balancing the acquisition, allocation and storage of energy during periods of food deprivation is critical for survival. We show that natural variation in the foraging (for) gene, which encodes a cGMP-dependent protein kinase (PKG) in the fruit fly Drosophila melanogaster, affects behavioral and physiological responses to short-term food deprivation. Rover and sitter, natural allelic variants of for, differ in their stored carbohydrate reserves as well as their response to short-term deprivation. Fewer carbohydrates are stored in the fat body of rovers compared with sitters, and more labeled glucose is allocated to lipid stores compared with carbohydrate stores during a short feeding bout. Short-term food deprivation decreases hemolymph glucose levels in rovers but not in sitters. After food deprivation, rovers increase their food intake more slowly than sitters, and rover hemolymph levels take longer to respond to re-feeding. Finally, rovers have lower adipokinetic hormone (akh) mRNA levels than sitters. Our data suggest that for mediates larval responses to short-term food deprivation by altering food intake and blood glucose levels.
Keywords: adipokinetic hormone, behavior genetics, cGMP-dependent protein kinase, food intake, foraging, glucose homeostasis
INTRODUCTION
The ability to balance the acquisition and storage of energy is essential for survival in nature because the abundance and quality of food can change drastically over time and this can result in periods of food deprivation. When food is limited, the homeostatic control system regulates energy balance and storage (Stubbs and Tolkamp, 2006). Despite tight regulation of energy homeostasis, there are individual differences in energy balance (Speakman, 2004). This is due, in large part, to interactions between the nutritive environment and the genes that contribute to variation in energy balance (Speakman, 2004). Energy acquisition, use and storage all contribute to energy balance.
The foraging (for) gene in Drosophila melanogaster (L.) provides a rare example of a naturally occurring genetic polymorphism that contributes to individual differences in energy acquisition (Kaun et al., 2007). for encodes a D. melanogaster cGMP-dependent protein kinase (PKG); two naturally occurring alleles of for are rover (forR) and sitter (fors) (de Belle et al., 1989). In Drosophila, the adult heads and larval nervous systems of rovers have higher PKG activity and for-transcript levels than those of sitters (Osborne et al., 1997). for affects larval food acquisition in an environmentally dependent fashion (Kaun et al., 2007). When food is plentiful, rover larvae have lower food intake and a higher proportion of glucose absorbed than sitters. When food is scarce, rover and sitter larvae increase food intake to a common maximal level with rovers retaining their increased glucose absorption. These phenotypes affect larval survival and development in nutritionally depleted environments (Kaun et al., 2007).
Interestingly, PKG has also been associated with human disorders in energy balance, including obesity and diabetes. High levels of cGK1, the mammalian ortholog of for, have been associated with obesity in mammals (Engeli et al., 2004; Su et al., 2003) whereas low levels have been associated with high glucose concentrations and diabetes (Wang et al., 2002; Zanetti et al., 2005).
During short periods of food deprivation, organisms use their stored carbohydrate reserves to supply energy for survival. In insects, the mobilization of these stored carbohydrates is mediated by peptide hormones called adipokinetic hormones (AKHs) (Gade and Auerswald, 2003). In Drosophila, one of these peptides is called AKH or dAKH. It is encoded by the akh gene and it mobilizes glucose after starvation (Kim and Rulifson, 2004; Lee and Park, 2004). Targeted cell ablation of akh-producing cells decreases hemolymph sugar levels whereas over-expression of akh increases them (Isabel et al., 2005; Kim and Rulifson, 2004; Lee and Park, 2004). A decrease in akh expression is also associated with a lack of starvation-induced hyperactivity and resistance to starvation-induced death (Isabel et al., 2005; Lee and Park, 2004). Thus, akh appears to mediate hemolymph sugar levels and starvation-induced changes in behavior.
In the present study, we explore how natural variation in for affects glucose homeostasis by investigating the physiological and behavioral responses to short-term food deprivation. Firstly, we investigate whether for affects the absorption and storage of carbohydrates. Secondly, we examine whether for affects glucose expenditure following short periods of food deprivation. Finally, we investigate whether different levels of akh mRNA are found in the rover and sitter variants.
MATERIALS AND METHODS
Strains
The wild-type rover and sitter strains used are isogenic for chromosomes 2 and 3 and homozygous for the forR or fors alleles, respectively (deBelle and Sokolowski, 1987). To ensure that any differences found between the two natural variants was specific to for, the same measurements were made using fors2, a sitter mutant generated on a rover genetic background (deBelle et al., 1989; Pereira and Sokolowski, 1993). Flies were maintained in 170ml plastic culture bottles with 40ml of standard culture medium at 25±1°C and a 12L:12D photocycle. Standard culture medium contained: 50g Baker's yeast; 100g sucrose; 16g agar; 0.1gKPO4; 8gKNaC4H4O6·4H2O; 0.5gNaCl; 0.5gMgCl2; and 0.5gFe2(SO4)3 per liter of tap water. Larvae were reared from egg-hatch to mid-third-instar (96±2h post-hatch) in 25°C at densities of 100 larvae per 35ml of medium in 100mm×15mm Petri dishes.
Glucose absorption and allocation
The glucose absorption protocol was modified from Riha and Luckinbill (Riha and Luckinbill, 1996) and is described in Kaun et al. (Kaun et al., 2007). Glucose allocation experiments were modified from Bligh and Dyer (Bligh and Dyer, 1959) and Westphal and Jann (Westphal and Jann, 1965) as described below. Groups of 30 larvae were homogenized and lipids, carbohydrates and proteins were extracted in layers from the same homogenate. Sample sizes were 30 larvae per vial with six vials per strain. Larvae were removed from −80°C and homogenized in 200μl of 2:1 chloroform:methanol in a 1.5ml tube using a hand-held motorized mortar with plastic pestle. An additional 800μl of 2:1 chloroform:methanol was added, samples were then vortexed and centrifuged at 10,000g for 5min at 4°C. The chloroform phase, composed of soluble material containing the lipid fraction, was removed and transferred to scintillation vials. 500μl of phosphate-buffered saline solution (PBS: 2.5mmoll−1 NaH2PO4; 8.5mmoll−1 Na2HPO4; and 175mmoll−1 NaCl; pH7.4) was added to the remaining precipitate and methanol phase. Samples were vortexed, 500μl of phenol was added and samples were heated at 65°C in a water bath for 1h with occasional vortexing. Samples were cooled on ice for 1h and then centrifuged at 3000g for 30min at 4°C for biphasic separation. The lower phenol layer, composed of soluble material containing the protein fraction, was removed and transferred to scintillation vials. The upper layer, containing the aqueous carbohydrate fraction, was transferred to separate scintillation vials. The remaining interphase was also transferred to scintillation vials. We then added 10ml scintillation fluid to each scintillation vial, and samples were vortexed for 30s, shaken for two hours then left at room temperature for 24h. The amount of 14C in each vial was calculated using counts observed over 60s per sample in a scintillation counter (Wallac 1409 Liquid Scintillation Counter, Perkin Elmer Life Sciences, Woodbridge, ON, Canada).
Prior to calculating the specific activity (fmoles) of intake and absorption of [14C]-6-glucose per larva, a conversion factor taking into account the specific activity of the radiolabeled substance was calculated. Preferential storage of glucose consumed in the form of one macronutrient over the other, was calculated as percentage of 14C absorbed per strain compared with the total 14C absorption per strain, where total 14C was defined as the sum of 14C in lipids, carbohydrates, proteins and interphase.
Protein assays
Protein levels were determined using the bicinchoninic acid (BCA) method [modified from Marron et al. (Marron et al., 2003)]. Individual larvae were homogenized in 300μl double-distilled water (ddH2O) and centrifuged at 10,000g for 2min. After centrifugation, 10μl of the supernatant was removed from each sample and placed in a SPECTRAplate™ Quartz UV Transparent 96-well Microplate (Molecular Devices, Sunnyvale, CA, USA) with 100μl of Sigma BCA Protein Assay Reagent (Sigma Chemical Co., St Louis, MO, USA). Samples were incubated overnight at room temperature and protein concentrations were determined by comparing the absorbance at 562nm with standard curves. Standard curves were constructed using bovine serum albumin (BSA; Sigma Chemical Co.) with concentrations ranging from 0.20mgml−1 to 0.45mgml−1.
Lipid assays
Lipid levels were determined by hydrolyzing triglycerides and then measuring the resulting glycerol levels [modified from Lee and Park (Lee and Park, 2004)]. Individual larvae were homogenized on ice in 150μl 0.1% Tween-20 in PBS in a 1.5ml tube using a hand-held motorized mortar with plastic pestle. Tween-20 is a non-ionic surfactant that is used to disperse and emulsify, and acts to disperse lipids into globules, which are suspended in the water solution. Samples were heated at 70°C for 5min to inactivate endogenous enzymes, then vortexed briefly. 30μl of the homogenate was removed and incubated on a rocking platform at 37°C overnight in a 1.5ml tube with 30μl of 2mgml−1 Candida rugosa lipase (Benjamin and Pandey, 1998) (Sigma Chemical Co.) suspended in 1×PBS. The lipase acts to hydrolyze triglycerides resulting in the production of free fatty acids and glycerol. Samples were then vortexed briefly and centrifuged at 13,000g for 10min. 10μl of the supernatant was removed and incubated with 100μl of Sigma Free Glycerol Reagent (Sigma Chemical Co.) at 37°C for 2h in a SPECTRAplate™ Quartz UV Transparent 96-well Microplate (Molecular Devices). Glycerol concentrations were determined by comparing absorbance at 540nm with standard curves. Standard curves were constructed using Sigma glycerol standards (Sigma Chemical Co.) with concentrations ranging from 0.20mgml−1 to 0.45mgml−1. Ten larvae per food deprivation condition per strain were assayed.
Carbohydrate assays
Whole larval assays
Whole animal carbohydrate assays were as in Marron et al. (Marron et al., 2003). Individual larvae were homogenized in 300μl ddH2O in a 1.5 ml tube using a hand-held motorized mortar with plastic pestle. 10μl of Rhizopus mold amyloglucosidase (8mgml1 suspended in ddH2O; Sigma Chemical Co.) was added to 10μl of homogenate in a SPECTRAplate™ Quartz UV Transparent 96-well Microplate (Molecular Devices). Rhizopus amyloglucosidase catalyzes the conversion of glycogen and trehalose into glucose (Parrou and Francois, 1997). Samples were left overnight at room temperature. 100μl of Sigma Glucose Assay Reagent (Sigma Chemical Co.) was added and samples were left for 1 h. Glucose was assayed using the hexokinase and G6PDH reactions and measuring the increase in absorbance at 340nm due to NADP+ reduction to NADH. Glucose concentrations were determined by comparing absorbance by NADH at 340 nm with standard curves. Standard curves were constructed using Sigma glucose standards (Sigma Chemical Co.) with concentrations ranging from 0.05mgml−1 to 0.30mgml−1. Ten larvae per food deprivation condition per strain were assayed.
Larval hemolymph carbohydrate assays
Hemolymph carbohydrate assays were performed as above with the following changes [modified from Kim and Rulifson, and Lee and Park (Kim and Rulifson, 2004; Lee and Park, 2004)]. Ten groups of five mid-third-instar larvae were washed with distilled water (dH2O) and blot dried. Hemolymph was extracted by tearing the cuticle and allowing the hemolymph to bleed out onto a glass slide. 2μl of hemolymph was rapidly withdrawn and mixed with 38μl 1×PBS. The sample was vortexed and centrifuged for 10 min to precipitate blood cells and tissue debris. 10μl of supernatant from each sample plated with 10μl of 8.0mgml−1 Rhizopus amyloglucosidase (Sigma Chemical Co.) was left overnight at room temperature. 100μl of Sigma Glucose Assay Reagent (Sigma Chemical Co.) was added and left for 1 h, after which, glucose concentrations were determined as above.
Larval fat body carbohydrate assays
Fat body carbohydrate assays were performed as for the whole larval carbohydrate assays with the following modifications. Fat bodies were dissected from individual animals in 1×PBS and transferred to 1.5 ml vials with 50μl 0.1% Tween-20, which were kept on ice. Samples were homogenized using a hand-held motorized mortar with plastic pestle. 20μl of 8.0mgml−1 Rhizopus amyloglucosidase was added, and samples were vortexed briefly. Samples were left overnight in 37°C, vortexed again, then centrifuged for 5 min at 13,000g to precipitate tissue debris. 10μl of supernatant was plated with 90μl of Sigma Glucose Assay Reagent (Sigma Chemical Co.) and left for 1 h. Glucose concentrations were determined as above using standard curves with concentrations ranging from 0.01mgml−1 to 0.25mgml−1. Glucose concentrations were standardized by protein concentration from the same samples, determined using the BCA method described above. Eight larvae per food deprivation condition per strain were assayed.
Acute food deprivation and re-feeding
Larvae were removed from food plates, washed in dH2O, and placed in groups of 30–40 in 45mm×10mm Petri plates with three 20mm×3mm plugs of 1.4% agar on the bottom. Lids were held firmly on top by a 0.2 kg weight. Larvae were left for 1, 2 or 3h on agar before testing for nutrient storage or food intake. For re-feeding assays, larvae were removed from agar plates to 45mm×10mm Petri plates with three 20mm×3mm plugs of standard laboratory fly culture medium for 30 min, 1 or 2 h.
Food intake
Food intake protocols were performed as in Kaun et al. (Kaun et al., 2007). Briefly, larvae were removed from food plates, washed in dH2O, and groups of ten were placed into circular wells (86mm in diameter and 0.5 mm deep) previously filled with yeast paste (2:1 water:yeast) mixed with 0.08% Brilliant Blue R dye (Sigma Chemical Co.). The wells were then covered with 9 cm Petri plate lids. Larvae remained on this dyed yeast paste for varying amounts of time depending on the experiment. They were then boiled for 10s, aligned on a microscope slide, placed under a dissecting microscope (Zeiss, Toronto, ON, Canada) and imaged using Northern Eclipse software (Empix Imaging, Mississauga, ON, Canada). Food intake was measured as the number of pixels (square pixels were used for quantification) in the image colored by the dye relative to the total number of pixels in the whole larval body taken as a percentage. Image J software was used (ImageJ v. 1.28j, 2002 and ImageJ v. 1.32j, 2004; http://rsweb.nih.gov.ij) for the digital quantification. Thirty larvae per food deprivation condition per strain were assayed.
Quantitative real-time PCR (qRT-PCR)
RNA extraction
RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). Larvae were collected and frozen in liquid nitrogen in groups of ten, then stored at −80°C until RNA extraction was performed. Ten larvae per strain per condition were homogenized on ice in 500μl TRIzol Reagent in a 1.5 ml tube using a hand-held motorized mortar with RNAse-free plastic pestle. They were left for 5 min at room temperature. 100μl of chloroform was then added and samples were shaken vigorously for 15 s and then left for 3 min at room temperature. Samples were centrifuged at 12,000 g for 15 min at 4°C. The supernatant was then extracted and placed in a 1.5 ml RNase-free microtube. 250μl of isopropanol was added, samples were shaken gently, and left at room temperature for 10 min. Samples were then centrifuged at 12,000g for 10 min at 4°C and the supernatant was disposed of. 500μl of 75% ethanol was added, samples were vortexed briefly, then centrifuged at 7500g for 5 min at 4°C. Supernatant was disposed of and samples were left for 15 min in the fumehood to dry. 50μl of RNase-free water was added and samples were heated to 55°C for 10 min. Concentration of RNA was quantified using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
Reverse transcript synthesis
Transcripts were synthesized using SuperScript II Reverse Transcriptase (Invitrogen). 2μg of RNA was added to 10μl of RNase-free water, samples were then treated with DNase by adding 0.5μl of DNase (200Uμl−1, Invitrogen) and 1μl of 10X DNase Buffer (Invitrogen) and left at room temperature for 15 min. 1μl of 25mmoll−1 EDTA was added and samples were incubated at 75°C for 5 min to inactivate the enzyme. Samples were centrifuged briefly (13,000g), then 0.4μl random heximers (Qiagen, Mississauga, ON, Canada) and 1μl of Biolase dNTP (Bioline, Randolph, MA, USA) were added. Samples were incubated at 65°C for 5 min, quick chilled on ice and centrifuged briefly (13,000g). 2μl of 0.1mol DTT (Invitrogen) and 4μl of 5×Superscript II Buffer (Invitrogen) was mixed in with gentle pipetting. Samples were left at room temperature for 2 min after which 1μl of Superscript II Reverse Transcriptase (200Uμl−1, Invitrogen) was gently pipetted into each sample. Samples were left at room temperature for 10 min and then incubated at 42°C for 50 min. The reaction was stopped by incubating samples at 70°C for 15 min.
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed with the SYBR green method (Eurogentec, San Diego, CA, USA) using the ABI 7700 Sequence Detection (Applied Biosystems, Foster City, CA, USA). cDNA was diluted 1:10 and plated as 5μl samples with 2.5μl each of 3μm forward (5′-GGCAACTGCAAGACCTCCAA-3′) and backward (5′-TGTGCCTGAGATTGCACGAA-3′) akh primers, 2.5μl ddH2O and 12.5μl SYBR-green Master Mix (Eurogentec). A standard curve using mixed cDNA of the samples was composed using dilutions 1:2, 1:5, 1:10, 1:20, 1:50 and 1:100. Samples were standardized against Rp49 using 50nmoll−1 final concentration for forward (5′-ATCGGTTACGGATCGAACAA-3′) and backward (5′-GACAATCTCCTTGCGCTTCT-3′) primers. Three wells per sample were plated on a 96well-plate (Applied Biosystems) and a mean of these wells was taken as a sample size of one. Relative expression was calculated using Relative Expression Software Tool and Multiple Condition Solver REST-MCS (v. 2, http://bioinformatics.gene-quantification.info). Sample size was six independent groups of larvae per strain.
Statistical analysis
JMP/IN 5.1 was used for all statistical analyses (SAS Institute, Cary, NC, USA). Means ± s.e.m. are presented on all graphs. One-way and two-way analyses of variance (ANOVA) were performed when sample sizes were larger than six per condition per strain and when tests for unequal variances showed non-significance. Equality of variance was analysed using two tests, i.e. Levene's test and Bartlett's test. Pairwise parametric comparisons were performed with Student Neuman–Keuls (SNK) post hoc tests. Non-parametric Kruskal–Wallis tests were followed with non-parametric Wilcoxon two-group tests. P<0.05 was considered significant. All experiments were replicated at least once.
RESULTS
Rovers absorb more total [14C]-6-glucose compared with sitters and the sitter mutants; this was true for each macronutrient (protein, F2,15=15.09, P=0.0003; lipid, F2,15=196.79, P<0.0001; carbohydrate, F2,15=18.18, P<0.0001) (Fig.1A). Compared with sitters, rover larvae store a larger proportion of ingested 14C in total lipid stores and a smaller proportion in total carbohydrate stores. There were no significant differences in the proportion of ingested 14C in total protein stores (lipids, Kruskal–Wallis , P=0.007, and Wilcoxon forR vs fors P=0.03, forR vs fors2 P=0.004, fors vs fors2 P=0.3; carbohydrates, , P=0.002, forR vs fors P=0.004, forR vs fors2 P=0.004, fors vs fors2 P=0.1; proteins, −, P=0.9).
Fig. 1.
Natural variation in nutrient storage due to for. (A) Rover (forR) larvae store a larger proportion of ingested 14C in their lipid stores whereas sitter larvae (fors and fors2) store a larger proportion in their carbohydrate stores. (B) Well-fed mid-third-instar forR, fors and fors2 larvae differ in total body carbohydrate but not lipid or protein stores. (C) Well-fed forR larvae have significantly lower carbohydrate levels in fat body but not in hemolymph (D) compared with fors and fors2 larvae. Error bars indicate means ± s.e.m.
Well-fed, mid-third-instar rover (forR) and sitter (fors and fors2) larvae differ in their total body carbohydrates but not in their lipid or protein stores (Fig.1B) (carbohydrate, F2,57=18.11, P=0.0001; SNK, forRvs forsP=0.02, forRvs fors2P=0.0003, forsvs fors2P=0.1; lipid, F2,57=0.27, P=0.8; protein, F2,57=0.33, P=0.7). Rover larvae have significantly lower carbohydrate levels in fat body than sitters (Fig.1C) (F2,21=4.34, P=0.03; SNK, forRvs forsP=0.03, forRvs fors2P=0.01, forsvs fors2P=0.8). No differences in carbohydrate levels are found in larval hemolymph of the variants (Fig.1D) (F2,57=0.80, P=0.5).
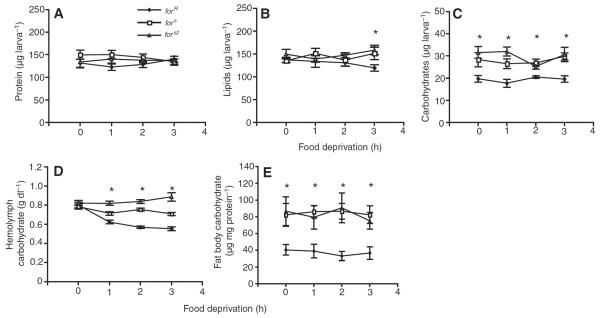
Acute food deprivation does not affect total protein levels (Fig.2A) (strain, F2,104=0.92, P=0.40; food deprivation, F3,104=0.48, P=0.70; strain by food deprivation interaction, F6,104=1.16, P=0.33) or total carbohydrate levels (Fig.2C) (strain, F2,105=11.51, P<0.0001; food deprivation, F3,105=0.86, P=0.47; strain by food deprivation, F6,105=1.16, P=0.33) in rovers or sitters. Lipid levels are decreased in rovers but not in sitters after three hours of food deprivation (Fig.2B) (F2,26=4.12, P=0.03, forRvs forsP=0.03, forRvs fors2P=0.01, forsvs fors2P=0.6).
Fig. 2.
Natural variation in nutrient store expenditure following acute food deprivation. (A) Acute food deprivation does not affect total protein levels in rovers (forR) or sitters (fors and fors2). (B) Three hours of acute food deprivation significantly decreases total body lipid levels in rover but not in sitter larvae. (C) Acute food deprivation does not affect whole body carbohydrate levels despite rover larvae showing significantly lower total body carbohydrate levels compared with sitter larvae. (D) Acute food deprivation decreases hemolymph carbohydrate levels in forR larvae but not fors or fors2 larvae. (E) Acute food deprivation does not affect fat body carbohydrate levels; forR larvae have significantly less carbohydrate in their fat bodies than fors or fors2 larvae. Error bars indicate means ± s.e.m.
Acute food deprivation decreases hemolymph carbohydrate levels in rover but not in sitter larvae (Fig.2D) (strain, F2,107=0.70, P=0.50; food deprivation, F3,107=25.78, P<0.0001; strain by food deprivation, F6,107=9.67, P<0.0001). By contrast, food deprivation does not affect carbohydrate levels in the fat body of either variant (Fig.2E) (strain, F2,84=5.84, P=0.004; food deprivation, F3,84=0.09, P=0.97; strain by food deprivation, F6,84=0.20, P=0.97).
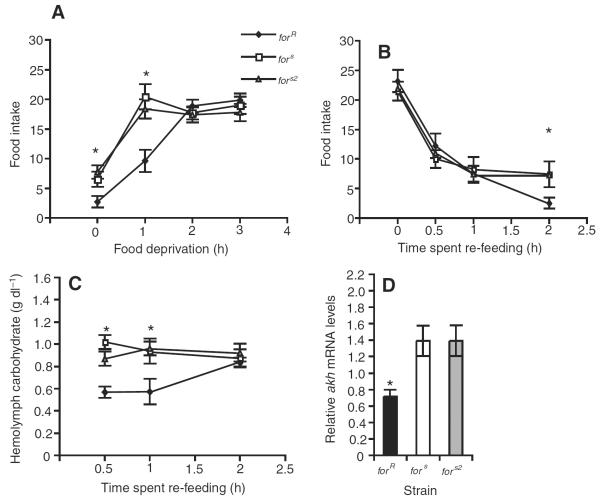
After one hour of food deprivation, sitter larvae increase their food intake more than rover (Fig.3A) (strain, F2,348=5.74, P=0.004; food deprivation, F3,348=57.36, P<0.0001; strain by food deprivation, F6,348=5.14, P<0.0001). There are no differences in food intake between the strains after two or three hours of food deprivation (Fig.3A) (2h, F2,87=0.44, P=0.65; 3h, F2,87=0.60, P=0.55).
Fig. 3.
for affects recovery from acute food deprivation. (A) After one hour of food deprivation, sitter (fors and fors2) larvae increase their food intake to a greater extent than rover (forR) larvae. After two and three hours of food deprivation, there were no significant differences in food intake between rovers and sitters. (B) Re-feeding experiments show that after three hours of acute food deprivation, followed by 0, 30 min, 1 h or 2 h of re-feeding, food intake levels in forR larvae return to their initial well-fed food intake levels more slowly than in fors and fors2 larvae. (C) Hemolymph carbohydrate levels in forR larvae returned to well-fed levels after 2 h of re-feeding, whereas hemolymph carbohydrate levels in fors and fors2 were not changed by acute food deprivation (see Fig. 2D) or re-feeding. (D) Well-fed forR larvae have significantly lower akh mRNA levels than fors and fors2. Relative akh mRNA levels were measured using quantitative real-time PCR and standardized against forR larvae. Error bars indicate means ± s.e.m.
We examined recovery from acute food deprivation by re-feeding larvae following a period of acute food deprivation. Upon re-feeding, sitter food intake returns to initial well-fed levels sooner than in rover (Fig.3B) (strain, F2,339=4.01, P=0.02; time spent re-feeding, F3,339=22.23, P<0.0001; strain by time spent re-feeding, F6,339=2.57, P=0.02). For example, after three hours of food deprivation and 30min of re-feeding, food intake levels of sitters return to initial well-fed levels (fors, F1,58=2.91, P=0.09; fors2, F1,56=2.86, P=0.10); however, after two hours of re-feeding, rover food intake levels are still higher than rover well-fed food intake levels (F1,58=14.89, P<0.0003). Similar recovery profiles are found after one and two hours of food deprivation (data not shown). Hemolymph carbohydrate levels in rover larvae return to well-fed levels more slowly than in sitters (Fig.3C) (strain, F2,80=11.90, P<0.0001; time spent re-feeding, F2,80=7.39, P=0.001; strain by time spent re-feeding, F4,80=2.89, P=0.03).
Relative abundance of akh RNA using qRT-PCR shows that akh RNA is more abundant in well-fed sitters compared with rovers (Fig.3D) (F2,17=6.17, P=0.01; SNK, forR vs forsP=0.007, forRvs fors2P=0.007, forsvs fors2P=1.0).
DISCUSSION
The ability to balance the acquisition, storage and use of energy is essential for all organisms. We previously showed that well-fed rovers ingest less food but absorb more glucose than well-fed sitters (Kaun et al., 2007). In the present study, we show that for affects the allocation and storage of food in well-fed animals and hemolymph carbohydrate reserves in food-deprived animals. Together, these data suggests that for plays an integral role in energy homeostasis in Drosophila.
Our measures of energy allocation and storage in fed animals suggest that rovers and sitters exhibit differences in their metabolic strategies. Rovers tend towards fat metabolism and sitters towards carbohydrate metabolism. Sitters allocated more of their ingested 14C-glucose to carbohydrate reserves and had higher total fat body carbohydrate stores. We did not find higher total lipid stores in rovers than in sitters. Further studies of lipid storage and metabolism in the larval fat body are needed to understand for's affect on lipid levels. These differences in nutrient metabolism may be linked to the foraging behavior of rover and sitter larvae. Rovers move more on and between food substrates compared with sitters and, thus, may need stored energy resources that can sustain longer bouts of locomotion. In other organisms, such as locusts, sustained flight results in mainly lipid metabolism (Candy et al., 1997). There is some evidence suggesting a role for PKG in lipid metabolism (Lafontan et al., 2005; Langdin, 2006). Atrial natriuetic peptides (ANP) increase intracellular cGMP, which, in turn, activates PKG leading to lipase phosphorylation and lipolysis (Lafontan et al., 2005).
Homeostatic regulation of blood sugar levels is a fundamental physiological process in both invertebrates and vertebrates. Disruptions in glucose homeostasis are associated with health problems such as diabetes (Rosen and Speigelman, 2006). In humans, diabetes-related disorders are associated with low PKG levels. Our results with D. melanogaster suggest a conserved role for PKG in glucose homeostasis.
Our results also hint at possible mechanisms through which PKG may mediate glucose homeostasis. In insects such as locusts, AKHs mobilize carbohydrates from the fat bodies via binding to a Gq-dependent receptor thus stimulating a phospholipase C (PLC). The resulting inositol triphosphate (IP3) releases Ca2+ from internal stores (Gade and Auerswald, 2003). This cascade activates glycogen phosphorylase leading to release of stored carbohydrates into the hemolymph.
How might PKG affect akh? It could act directly or indirectly on akh by changing its transcription (Pilz and Broderick, 2005). Alternatively, PKG could indirectly affect akh by acting on molecules that disrupt glucose homeostasis, such as insulin or neuropeptide F (npf). In Drosophila, the insulin signaling pathway is integral for proper storage of carbohydrates and lipids, especially during growth and development (Mirth and Riddiford, 2007). npf signaling has been linked to insulin signaling and starvation-induced differences in behavior (Wu et al., 2005a; Wu et al., 2005b). PKG may be linked to npf-dependent food acquisition (Kaun, 2007). Thus, disruption of insulin signaling could potentially disrupt glucose homeostasis and indirectly affect akh.
PKG is also known to directly phosphorylate PLC-β, leading to inhibition of PLC-β3 activity (Xia et al., 2001). Inhibition of PLC could decrease glycogen phosphorylase activity, potentially slowing the release of stored carbohydrates into the hemolymph. This is consistent with rovers showing decreased hemolymph sugar levels after food deprivation compared with sitters. It would be interesting to determine if PKG and AKH proteins interact and co-localize in Drosophila tissue.
The difference in both foraging behavior and response to food deprivation between rover and sitter Drosophila bears an intriguing resemblance to the polyphenism seen in Locusta migratoria migratoriodes gregarious and solitary locusts, which also differ in adipokinetic strategies (Pener et al., 1997). Parallels can be drawn between rovers and gregarious locusts, and sitters and solitary locusts. Both rovers and gregarious locusts move greater distances to forage. Gregarious locusts and potentially rovers preferentially metabolize lipids over carbohydrates (Pener et al., 1997). Solitary locusts have higher resting hemolymph sugar levels similar to sitters after a short period of food deprivation (Pener et al., 1997). Solitary locusts also have higher AKH levels similar to the higher akh mRNA levels found in sitters (Pener et al., 1997). Whether for plays a role in foraging behavior or adipokinetic balance in solitary and gregarious locusts remains to be determined.
for has been implicated in food-related behaviors in a variety of organisms, including Caenorhabditis elegans (Fujuwara et al., 2002; You et al., 2008), honey bees (Ben-Shahar et al., 2002), ants (Ingram et al., 2005) and Drosophila (Osborne et al., 1997). Whether and how PKG plays a role in energy balance in these organisms remains to be investigated.
Acknowledgments
We thank Allen Gibbs for advice on protein, lipid and carbohydrates assays and helpful comments on this manuscript, Evan Ardiel and Reza Azanchi for help with sample collection, Yehuda Ben Shahar helped develop the fed, food-deprived and re-fed protocol, Joel Levine gave support and advice on these experiments, Josh Krupp helped with qRT-PCR, Scott Douglas edited an earlier version of this manuscript and all the Sokolowski lab members for support advice and input on this project. Thanks also to two anonymous reviewers who provided valuable comments on an earlier version of this manuscript. Funding was provided by NIDDK grants 5R01DK070141-03 to M.B.S. A Natural Sciences and Engineering Research Council CGS-D and Ontario Graduate Scholarship in Science and Technology supported K.R.K.
Footnotes
The authors declare that they have no competing financial interests.
REFERENCES
- Benjamin S, Pandey A. Candida rugosa lipases: molecular biology and versatility in biotechnology. Yeast. 1998;14:1069–1087. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1069::AID-YEA303>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Influence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Candy DJ, Becker A, Wegener G. Coordination and integration of metabolism in insect flight. Comp. Biochem. Physiol. 1997;117B:497–512. [Google Scholar]
- de Belle JS, Sokolowski MB. Heredity of rover-sitter alternative foraging strategies of Drosophila melanogaster larvae. Heredity. 1987;59:73–84. [Google Scholar]
- de Belle JS, Hilliker AJ, Sokolowski MB. Genetic localization of foraging (for): a major gene for larval behavior in Drosophila melanogaster. Genetics. 1989;123:157–163. doi: 10.1093/genetics/123.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeli S, Janke J, Gorzelniak K, Bohnke J, Ghose N, Lindschau C, Luft FC, Sharma AM. Regulation of the nitric oxide system in human adipose tissue. J. Lipid Res. 2004;45:1640–1648. doi: 10.1194/jlr.M300322-JLR200. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Gade G, Auerswald L. Mode of action of neuropeptides from the adipokinetic hormone family. Gen. Comp. Endocrinol. 2003;132:10–20. doi: 10.1016/s0016-6480(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Ingram KK, Oefner P, Gordon DM. Task-specific expression of the foraging gene in harvester ants. Mol. Ecol. 2005;14:813–818. doi: 10.1111/j.1365-294X.2005.02450.x. [DOI] [PubMed] [Google Scholar]
- Isabel G, Martin JR, Chidami S, Veenstra JA, Rosay P. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R531–R538. doi: 10.1152/ajpregu.00158.2004. [DOI] [PubMed] [Google Scholar]
- Kaun KR. PhD Thesis. University of Toronto; Canada: 2007. Neurogenetic and plastic components of food-related behaviours due to the foraging gene in Drosophila melanogaster. [Google Scholar]
- Kaun KR, Riedl CAL, Chakaborty-Chatterjee M, Belay AT, Douglas S, Gibbs A, Sokolowski MB. Natural variation in food acquisition due to a Drosophila cGMP-dependent protein kinase. J. Exp. Biol. 2007;210:3547–3558. doi: 10.1242/jeb.006924. [DOI] [PubMed] [Google Scholar]
- Kim SK, Rulifson EJ. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Lafontan M, Moro C, Sengenes C, Galitzky J, Crampes F, Berlan M. An unsuspected metabolic role for atrial natriuretic peptides: the control of lipoysis, lipid mobilization, and system nonesterfied fatty acids levels in humans. Arterioscler. Thromb. Vasc. Biol. 2005;25:2032–2042. doi: 10.1161/01.ATV.0000183728.14712.d8. [DOI] [PubMed] [Google Scholar]
- Langdin D. Adipose tissue lipolysis as a metabolic pathway to define pharmacological strategies against obesity and the metabolic syndrome. Pharm. Res. 2006;53:482–491. doi: 10.1016/j.phrs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marron MT, Markow TA, Kain KJ, Gibbs AG. Effects of starvation and dessication on energy metabolism in desert and mesic Drosophila. J. Insect Physiol. 2003;49:261–270. doi: 10.1016/s0022-1910(02)00287-1. [DOI] [PubMed] [Google Scholar]
- Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. BioEssays. 2007;29:344–355. doi: 10.1002/bies.20552. [DOI] [PubMed] [Google Scholar]
- Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, Coulthard A, Pereira HS, Greenspan RJ, Sokolowski MB. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- Parrou JL, Francois J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 1997;248:186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- Pener MP, Ayali A, Golsenser E. Adipokinetic hormone and flight fuel related characteristics of density-dependent locust phase polymorphism: a review. Comp. Biochem. Physiol. B. 1997;117:513–524. [Google Scholar]
- Pereira HS, Sokolowski MB. Mutations in the larval foraging gene affect adult locomotory behavior after feeding in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1993;90:5044–5046. doi: 10.1073/pnas.90.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz RB, Broderick KE. Role of cyclic cGMP in gene regulation. Front. Biosci. 2005;10:1239–1268. doi: 10.2741/1616. [DOI] [PubMed] [Google Scholar]
- Riha VF, Luckinbill LS. Selection for longevity favors stringent metabolic control in Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 1996;51:B284–B294. doi: 10.1093/gerona/51a.4.b284. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR. Obesity: the integrated roles of environment and genetics. J. Nutr. 2004;(Suppl):2090S–2105S. doi: 10.1093/jn/134.8.2090S. [DOI] [PubMed] [Google Scholar]
- Stubbs RJ, Tolkamp BJ. Control of energy balance in relation to energy expenditure in animals and man: an ecological perspective. Br. J. Nutr. 2006;95:657–676. doi: 10.1079/bjn20041361. [DOI] [PubMed] [Google Scholar]
- Su J, Zhang S, Tse J, Scholz PM, Wiess HR. Alternations in nitric oxide-cGMP pathway in ventricular myocytes from obese leptin-deficient mice. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H2111–H2117. doi: 10.1152/ajpheart.00316.2003. [DOI] [PubMed] [Google Scholar]
- Wang S, Shiva S, Poczatek M, Schultz-Cherry S, Villain M, Murphy-Ullrich JE. Nitric-oxide and cGMP-dependent protein kinase regulation of glucose-mediated thrombospondin 1-dependent transforming growth factor-beta activation in mesangial cells. J. Biol. Chem. 2002;277:9880–9888. doi: 10.1074/jbc.M108360200. [DOI] [PubMed] [Google Scholar]
- Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Meth. Carbohydr. Chem. 1965;5:83–91. [Google Scholar]
- Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. USA. 2005a;102:13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat. Neurosci. 2005b;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- Xia C, Bao Z, Yue C, Sanborn BM, Liu M. Phosphorylation and regulation of G-protein activated phospholipase C-β3 by cGMP protein kinases. J. Biol. Chem. 2001;276:19770–19777. doi: 10.1074/jbc.M006266200. [DOI] [PubMed] [Google Scholar]
- You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescente in C. elegans: a model for satiety. Cell. Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M, Barazzoni R, Stebel M, Roder M, Roder E, Biolo G, Baralle FE, Cattin L, Guarnieri G. Dysregulation of the endothelial nitric oxide synthase-soluble guanylate cyclase pathway is normalized by insulin in the aorta of the diabetic rat. Atherosclerosis. 2005;181:69–73. doi: 10.1016/j.atherosclerosis.2005.01.011. [DOI] [PubMed] [Google Scholar]