Abstract
The diastereomeric spiroiminodihydantoin-2′-deoxyribonucleoside (dSp) lesions resulting from 2′-deoxyguanosine (dG) or 8-oxo-7,8-dihydro-2′-deoxyguanosine (dOG) oxidation have generated much attention due to their highly mutagenic nature. Their propeller-like shape leads these molecules to display mutational profiles in vivo that are stereochemically dependent. However, there exist conflicting absolute configuration assignments arising from electronic circular dichroism (ECD) and NOESY-NMR experiments; thus, providing definitive assignments of the 3D structure of these molecules is of great interest. In the present body of work, we present data inconsistent with the reported ECD assignments for the dSp diastereomers in the nucleoside context, in which the first eluting isomer from a Hypercarb HPLC column was assigned to be the S configuration and the second was assigned the R configuration. The following experiments were conducted: (1) Determination of the diastereomer ratio of dSp products upon one-electron oxidation of dG in chiral hybrid or propeller G-quadruplexes that expose the re or si face to solvent, respectively, (2) absolute configuration analysis using vibrational circular dichroism (VCD) spectroscopy, (3) reinterpretation of the ECD experimental spectra using time-dependent density functional theory (TDDFT) with the inclusion of 12 explicit H-bonding waters around the Sp free bases, and (4) reevaluation of calculated specific rotations for the Sp enantiomers using the hydration model in the TDDFT calculations. These new insights provide a fresh look at the absolute configuration assignments of the dSp diastereomers in which the first eluting from a Hypercarb-HPLC column is (-)-(R)-dSp and the second is (+)-(S)-dSp. These assignments now provide the basis for understanding the biological significance of the stereochemical dependence of enzymes that process this form of DNA damage.
INTRODUCTION
Electron-deficient species that result from oxidative and inflammatory stress have the potential to affect oxidation reactions on biomolecules, particularly in DNA.1,2 2′-Deoxyguanosine (dG) is the most electron-rich of the nucleosides and has the lowest one-electron redox potential3 resulting in dG being the major site for oxidation reactions.4 Studies concerning the products that arise from dG oxidation have identified myriad compounds, in which the two-electron oxidation product 8-oxo-7,8-dihydro-2′-deoxyguanosine (dOG) is consistently identified as a key product (Scheme 1).4–8 Additionally, in vivo levels of dOG are ~0.1–4 per 106 dG providing a marker to monitor a cell’s exposure to oxidative stress.9–11 If left unrepaired, dOG is moderately mutagenic causing dG → T (thymidine) transversion mutations.12 Furthermore, dOG is more susceptible toward one-electron oxidations than dG because its redox potential is ~600 mV lower.13 Accordingly, the two-electron oxidation of dOG furnishes two hydantoin compounds, 5-guanidinohydantoin-2′-deoxyribonucleoside (dGh) and spiroiminodihydantoin-2′-deoxyribonucleoside (dSp; Scheme 1).14–20 These hydantoins have been detected from both dG and dOG oxidations effected by a plethora of oxidants including HO•, CO3•−, 1O2, and high-valent transition metal species.14–18,20–23 The relative yields of dSp and dGh vary with context wherein dGh is greatest in duplex contexts or at low pH (<5.8), while dSp is greatest in nucleoside and G-quadruplex contexts or reactions in oligomers conducted at higher pH (>5.8).14,19,20,24,25 The dSp lesion has been observed in Escherichia coli,26 and both dSp and dGh have been quantified in a mouse model of infection-induced colitis leading to colon cancer.27 Moreover, dSp concentrations demonstrated a modest increase correlating with the progression of colon cancer in this model.27 In vivo studies demonstrated that both hydantoins show 100% mutagenesis causing dG→dT and dG→dC (2′-deoxycytidine) transversion mutations if unrepaired.28–30 These biological observations have implicated the hydantoins as potential culprits for causing mutations that result from oxidative and inflammatory stress leading to disease progression.
Scheme 1.
G-oxidation pathway to dOG, dSp and dGh.
The heterocycles dSp and dGh both possess a stereocenter giving rise to a pair of diastereomers (Scheme 1) that are readily separable by chromatography, even in long oligodeoxynucleotides (ODN) using ion-exchange HPLC. The dGh diastereomers were previously shown to be interconvertible making their individual study very challenging;21 in contrast, the dSp diastereomers are each chemically stable and do not interconvert. In this report, assignments of the R and S configurations for dSp are based on the conjugated exocyclic amine tautomers that were established computationally to be the lowest in energy (Scheme 1; see Supporting Information S1 for further details).31 In this discussion, the dSp diastereomers are numbered based on the elution order of ODNs using an ion-exchange HPLC column (Figure 1; Dionex DNApac 100).
Figure 1.

Ion-exchange and Hypercarb chromatograms demonstrating the switching of elution order for the dSp diastereomers. The dSp diastereomers were synthesized and purified separately in the sequence 5′-d(CGT CCA XGT CTA)-3′ where X = dSp1 or dSp2 following literature methods.32 Digestion of the individual diastereomerically pure strands by exhaustive nuclease and phosphatase treatment following literature methods33 gave nucleoside samples that were analyzed on a Hypercarb column to demonstrate the switching of elution order. Nomenclature for the two columns is defined as dSp1 and dSp2 on the ion-exchange column, and dSpA and dSpB on the Hypercarb column, i.e., dSp1 = dSpB and dSp2 = dSpA. See Supporting Information S2 for complete details.
Interestingly, studies from our laboratory and others have demonstrated a stereochemical dependence for dSp processing by enzymes in ODNs. The first instance of a dSp diastereomer difference was reported from our laboratory when Klenow Fragment (exo-) was allowed to insert dATP opposite the isomers for which dSp1-containing ODNs were found to be the more favorable substrate for insertion and bypass.32 This study was extended to in vivo experiments by Neeley, et al. to include pol II and pol IV, also showing more favorable bypass kinetics for dSp1.34 As further evidence for in vivo stereochemical dependency in dSp processing, Henderson, et al. utilized the restriction endonuclease and postlabeling assay (REAP) to determine the mutation profiles for dSp1 and dSp2 in Escherichia coli.28 In the sequence context 5′-d(GSpA)-3′, they found a strong stereochemical dependence in the relative amounts of dG→dT and dG→dC transversion mutations for dSp1 and dSp2. In subsequent experiments, Delaney, et al. showed a somewhat different dependence of mutation profiles on the dSp diastereomers in the sequence context 5′-(TSpG)-3′ using the REAP and competitive replication of adduct bypass assays (CRAB).29,30 An additional observation from our laboratory quantified the digestion kinetics of dSp isomers in the dinucleotides 5′-d(Np[Sp])-3′ (N = A, T, G or C) by nuclease P1 that showed dSp1 was the kinetically preferred substrate in all sequence combinations.33 Lastly, in collaboration with the David laboratory, we showed that the base excision repair glycosylase, hNEIL1 initiates repair of dSp1 more efficiently than dSp2 in all sequence and structural contexts studied.35–37 Molecular dynamics (MD) simulations conducted by Jia, et al. identified the S configuration for dSp as providing better contacts with the active site of hNEIL1, and therefore it should be the better substrate.38 As a result, discussions in our laboratory leaned toward dSp1 having the S configuration and dSp2 having the R configuration,33,36 leading to the question of how these absolute configurations could be assigned in the absence of a crystal structure.
The elution order for the dSp diastereomers on an ion-exchange (ODN) and on an aminosilica (nucleoside) HPLC column are the same; therefore, for these columns the nomenclature dSp1 and dSp2 will be used. However, on a Hypercarb HPLC column (Thermo Scientific) the elution order is switched, and to avoid confusion, these peaks will be referred to here as dSpA and dSpB. That is to say, dSpA = dSp2 and dSpB = dSp1 (Figure 1). This HPLC elution order observation has been described by many laboratories, and we reconfirmed these results independently (see Supporting Information S2 for details).19,33,39–42 Utilizing the Hypercarb HPLC column, Durandin, et al. prepared the free-base Sp enantiomers and measured their optical rotatory dispersion (ORD) values and then coupled their experimental observations with density functional theory (DFT) calculations.39 They continued their optical experiments with the Sp enantiomers in a second report by Ding, et al., in which the experimental electronic circular dichroism (ECD) spectra were compared against time-dependent DFT (TDDFT) calculated spectra.43,44 The same experimental results were also observed with the dSp nucleosides.43 In these two studies, they concluded that SpA (Sp2) has the S configuration, while SpB (Sp1) has the R configuration.39,43,44 In a separate study, Karwowski, et al. utilized an amino-silica column to purify the dSp nucleosides, and they conducted NOESY-NMR complemented with DFT studies on the dSp nucleosides and assigned dSp1 (dSpB) as the S isomer and dSp2 (dSpA) as the R isomer.45 These two laboratories, at NYU and Grenoble, came to opposite conclusions about the absolute configuration for the (d)Sp isomers that set the stage for the controversy surrounding their stereochemical assignments.
Our previous enzyme kinetic studies support the conclusion of the Grenoble group45 that dSp1 = (S)-dSp and dSp2 = (R)-dSp. Therefore, additional studies were pursued in an attempt to resolve the discrepancy that exists in the literature; the studies completed and presented in this paper are: (1) dSp product distributions resulting from oxidation of the chiral hybrid or propeller folded G-quadruplex topologies of the human telomere sequence that expose either the re or si face of dG to solvent, respectively, (2) absolute configuration assignments using vibrational circular dichroism (VCD) spectroscopy, (3) reevaluation of the ECD experiments and TDDFT calculations to include the effect of solvent interactions, (4) reevaluation of the ORD experimental and calculated signs to include the effect of solvent interactions, and (5) interpretation of the enzymatic studies based on these assignments. These experiments corroborate the NMR report,45 and provide an explanation why the previous ECD and ORD results cannot be reconciled.43
METHODS
dSp and Sp Synthesis
dSp nucleosides were prepared in a 100-mL solution containing dG (1 mM) and methylene blue (500 μM) in NaPi buffer (100 mM, pH 8) that was allowed to react while being irradiated with a 350-nm light (300-W light bulb suspended ~8 cm above the reaction) for 1 h at 22 °C. The methylene blue was removed by passing the mixture through a NAP-25 column (GE Healthcare) following the manufacturer’s protocol. Next, the unreacted dG was removed by passing the reaction mixture through a reversed-phase HPLC column while collecting the void volume where the dSp elutes. Upon lyophilization of the collected void volume, the mixture of dSp diastereomers were individually purified using a Hypercarb HPLC column (Thermo Scientific; 150 X 4.6 mm, 5 μm). The resolving method had line A = ddH2O with 0.1% acetic acid, and B = MeOH, while running an isocratic gradient of 0% B for 20 min followed by a linear increase in B to 75% over 20 min, with a flow rate of 1 mL/min while monitoring the absorbance at 240 nm. The retention times for dSp1 and dSp2 were 10 and 15 min, respectively. The yield after purification for dSp1 and dSp2 was 20 and 22%, respectively. The identity of the dSp isomers was confirmed by ESI+-MS/MS analysis of the free bases that gave the same daughter fragments that were previously reported: (M+H)+ = 184, 156, 141, 113, 86 (Supporting Information S3).15 The free-base Sp enantiomers were prepared from the diastereomerically pure dSp nucleosides. To an anhydrous pellet of dSp, 40% HF in pyridine was added to give a final concentration of 10 mg/mL Sp. The samples were incubated at 45 °C for 90 min. Next, the HF was neutralized with 80 mg of CaCO3 and the resulting mixture was filtered. The Sp enantiomers were purified using a Hybercarb HPLC column achieving a yield of ~75% for each enantiomer.
G-Quadruplex One-electron Oxidation and dSp Diastereomer Distributions
The natural human telomere sequence, 5′-d(TAGGG TTAGGG TTAGGG TTAGGG TT)-3′ was allowed to fold to the hybrid, basket or propeller topologies following our previously reported methods (Supporting Information S4).24 All oxidations were conducted in 20 mM MPi (pH 7.4) 120 mM MCl (M = Na or K) and 10 μM ODN at 37 °C. Oxidations were achieved as follows: Riboflavin (Type I photosensitizer) was added to a 50 μM concentration, and then the solution was exposed to 350-nm light (300-W light bulb placed ~8 cm above the reaction) for 10 min at 37 °C. For the CO3• − oxidations, the radical was produced when 3-morpholinosydnonimine (SIN-1; <3 mM) thermally decomposed (37 °C) to generate ONOO− that was allowed to react over 1 h with 25 mM MHCO3 (M = Na or K) to generate ONOOOCO2−, which is the precursor that undergoes homolysis to yield CO3• −.46 These oxidation conditions affect ~10% conversion to product.24 Product analysis was conducted by exhaustive nuclease and phosphatase digestion of the oxidized strands as previously described.33,47 Next, the dG-oxidation products were analyzed by Hypercarb HPLC analysis. The complete details for these methods are described in the Supporting Information.
VCD Measurements
Experimental VCD spectra for the SpA enantiomer or dSpA and dSpB diastereomers were recorded in DMSO-d6 at concentrations of 3.3 mg/0.15 mL, 6.0 mg/0.15 mL and 8.0 mg/0.2 mL, respectively. The samples were placed in a 100 μm path length cell with BaF2 windows. IR and VCD spectra were recorded on a ChiralIR2X™ VCD spectrometer (BioTools, Inc.) equipped with a dual PEM accessory that was optimized at 1400 cm−1 with a resolution of 4 cm−1. Data acquisition times were 20 h for SpA, 12 h for dSpA, 13 h for dSpB and 20 h for DMSO-d6. The IR and VCD spectra of the solvent were subtracted for baseline correction purposes.
ECD and UV-vis Measurements
Experimental ECD spectra for the dSp nucleosides and Sp-free bases were recorded at a concentration of 300 μM in ddH2O at 20 °C. The experimental UV-vis spectra were recorded at a concentration of 20 μM in ddH2O also at 20 °C. All recorded spectra had the solvent background subtracted.
Specific Rotation Measurements
The specific rotation at the sodium-D line was measured for the Sp-free bases in ddH2O at a concentration of 3.8 mg/mL at 20 °C.
Calculations
All calculations were performed using DFT methods as implemented in the Gaussian09 package.48 The 6-311++G(2d,2p) basis set was used in all calculations.49,50 During the course of this work several different functionals were used, specifically: B3LYP,51,52 M06-2X,53 CAM-B3LYP,54 and PBE1PBE.55 Calculations were performed on the compound of interest in the gas phase, with implicit solvation via the Gaussian09 implementation of the Polarizable Continuum Model (PCM) solvation,56,57 and with explicit solvation by water molecules added at the hydrogen bonding sites of the compound. Specific details of the different calculations are presented below.
For the VCD calculations, the geometry optimizations, vibrational energies and rotational strengths of the (d)Sp isomers were completed using the B3LYP functional on the isomers both in the gas phase and with implicit DMSO solvation.58 The data was visualized using Gaussview v5.59 For the UV-vis, ECD, and ORD calculations,60–62 the geometry optimizations of the Sp enantiomers were achieved using both the B3LYP and M06-2X functionals (Supporting Information Table S6). The B3LYP optimizations were completed on gas phase molecules and with implicit solvation by water, whereas the M06-2X functional was used for the optimization of a series of model systems in the gas phase and with both explicit waters added at hydrogen-bonding sites of the molecule along with the addition of implicit solvation to account for any solvation effects not treated by the implicit waters. The explicit water molecules were added incrementally as described in the discussion, with calculations performed with structures having seven, ten and twelve water molecules about the Sp enantiomers. Computed UV-vis and ECD spectra were completed using time-dependent DFT (TDDFT) theory to calculate 25 excitations.60,63–65 These calculations were done on the optimized structures using four different functionals: B3LYP, CAM-B3LYP, M06-2X, and PBE1PBE. The specific rotation at the sodium D-line ([α]D, deg dm−1 cm3 g−1) along with several other wavelengths between 300 and 589 nm were calculated for both the B3LYP and the M06-2X optimized geometries from the rotational lengths computed by TDDFT. The ECD spectra were plotted from the excitation energies and the rotational lengths by overlapping the Gaussian functions (eq. 1) for each transition.
| eq. 1 |
where Ei = excitation energy, Ri = rotatory strength, and σ = line-broadening factor. The value of σ was chosen to be 2500–3000 cm−1 to give a good fit to the broadening observed in the experimental spectra. The UV-vis data was visualized using Gaussview v5.59
RESULTS
G-Quadruplex Oxidations Yield dSp-Diastereomer Ratios that are Topologically Specific
Hybrid, basket and propeller folds of the human telomere sequence (5′-d(TA GGG TTA GGG TTA GGG TTA GGG TT)-3′) provide a chiral topology to conduct G-oxidation reactions with one-electron oxidants. In our previous studies, it was shown that G oxidation occurred at the 5′-G nucleotides with the one-electron oxidants riboflavin/hυ and CO3• − (generated in situ with SIN-1/HCO3−) in the hybrid, basket and propeller folding topologies (Figure 2).24 Oxidation of these chiral structures (~10% conversion) yields dSp as the major product (riboflavin/hν ~ 40–50%, and CO3• − ~60–70%)24 that was determined post oxidation by exhaustive nuclease treatment followed by product quantification utilizing a Hypercarb HPLC column. Interestingly, the dSp diastereomer ratio was highly influenced by the topological fold in which the oxidation occurred. Specifically, the hybrid structure gave a dSpA to dSpB ratio of nearly 3:1, the basket fold gave a 1:1 ratio, and the propeller fold gave a 1:3 ratio, respectively (Figure 3). These observations provide compelling evidence that blocking one side of dG by π-stacking against the middle G-quartet influenced the reaction profile. Thus, these observations set the stage for the mechanistic arguments that follow.
Figure 2.
Sites of dG oxidation in G-quadruplex topologies occurring at 5′-Gs with one-electron oxidants. The hybrid, basket and propeller folds are drawn based on previous structural studies.66–68 dG oxidation sites were previously reported by our laboratory.24 For the sake of brevity only the hybrid-1 fold is shown and not hybrid-2. The hybrid-2 fold gives the same sites of reactivity and products as observed in the hybrid-1 fold.24,67
Figure 3.

dSp diastereomer distributions observed from oxidation of the hybrid, basket and propeller G-quadruplex folds. The one-electron oxidants used were riboflavin/hν and CO3• − that was generated in situ by thermal decomposition (37 °C) of SIN-1 in the presence of HCO3−.24 dSp diastereomer ratios were determined from exhaustive digestion of oxidized G-quadruplexes followed by analysis on a Hypercarb HPLC column. The error on each measurement was ~8% of the value.
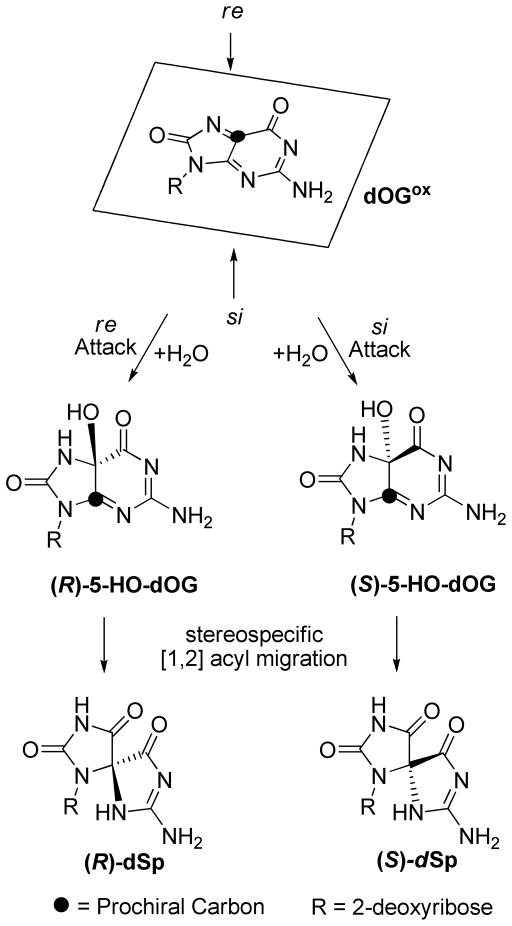
Interpretation of the topology-dependent dSp diastereomer yield is chemically reasoned through the oxidation mechanism of dG to dSp. Only the pathway to dSp will be described; other reaction channels exist and were discussed in the original studies, but they give products that were detected in lower absolute yield.24 All reactive 5′-G nucleotides, whether in the hybrid-1 fold (shown) or the hybrid-2 fold (see Supporting Information S5), expose the re face of C5, the prochiral center, to solvent (Figure 4).67 In contrast, all reactive 5′-Gs in the propeller fold expose the si face of C5,68 and by way of comparison, the reactive 5′-Gs in the basket fold expose the si and re faces equally (Figure 4).66 Four-electron oxidation of dG to dSp by one-electron oxidants is mechanistically reasoned to be initiated by a one-electron oxidation via Type I photosensitization (riboflavin/hν) or CO3• − to yield a radical cation (dG•+). In ODN contexts, dG•+ can add water at C8 and lose another electron and proton to furnish dOG, or it can deprotonate (pKa ~3.9) to give the neutral radical dG• (Scheme 2A). In CO3• − oxidations, radical coupling between dG• and CO3• − occurs followed by ester hydrolysis to yield dOG (Scheme 2A).69 dOG is stable but readily undergoes one-electron oxidation to giving a weak-acid radical intermediate dOG•+/dOG• (pKa ~6.6; Scheme 2B).13 The oxidation continues with one-electron oxidants to furnish two-electron oxidized dOG (dOGox; Scheme 2B) that adds water as a nucleophile at the prochiral C5 carbon to yield 5-hydroxy-8-oxo-7,8-dihydro-2′-deoxyguanosine (5-HO-dOG; Scheme 3).15 Alternatively, dOG• can couple with CO3• − at the prochiral C5 carbon and undergo ester hydrolysis to yield 5-HO-dOG (Scheme 2B) ;69 or water can add in a stereospecific fashion to dOG•+ followed by a second oxidation by the oxidant and proton loss to solvent to yield a specific diastereomer of 5-HO-dOG (Scheme 2B).70 Additions to the prochiral C5 position on the re face yield (R)-5-HO-dOG while on the si face (S)-5-HO-dOG is formed. The lifetime of 5-HO-dOG is short,71 and it undergoes acyl migration stereospecifically to yield dSp,70 such that (R)-5-HO-dOG undergoes acyl migration to give (R)-dSp, and (S)-5-HO-dOG gives (S)-dSp. Therefore, it is reasoned that in the hybrid G-quadruplex context (re face of C5 exposed, Figure 4), where dSpA was the major diastereomer observed (Figure 3), that dSpA (dSp2) = (R)-dSp, and in the propeller context (si face of C5 exposed, Figure 4), where dSpB was the major diastereomer observed (Figure 3) that dSpB (dSp1) = (S)-dSp. The basket context provides a convenient control because both faces are represented in equal amounts (Figure 4), and the dSp diastereomer ratio was found to be 1:1 (Figure 3). To further support these assignments an experiment that better predicts the absolute configurations of organic molecules, namely VCD, was then conducted.
Figure 4.
G-Quadruplex topology gives rise to different re vs. si faces of dG solvent exposure with respect to C5. The G-quadruplex structures were taken from pdb files 2JSQ and 1K8P.67,68
Scheme 2.
Proposed pathways for dG oxidation to intermediates that lead to dSp. (A) Oxidation pathway to dOG and (B) pathway to stereospecific formation 5-HO-dOG diastereomers and dOGox.
Scheme 3.
Stereospecific formation of dSp diastereomers based on the nucleophilic water attack trajectory to the electrophilic intermediate dOGox. For the sake of brevity, only the pathway leading to dSp from dOGox is shown.
VCD Analysis to Determine Absolute Configurations for the dSp Diastereomers
VCD spectroscopy has proven to be a robust method to determine absolute configurations by measuring the differential response of chiral molecules to left vs. right circularly polarized light during a vibrational transition.72 This method avoids the need for proton handles to make assignments that are necessary in NMR analysis, which is a problem for the Sp free base because it only possesses exchangeable protons. In addition, a defractable crystal does not need to be grown for X-ray crystallographic analysis. Furthermore, DFT-computed spectra match very well with those obtained experimentally. Carbonyl-,73,74 lactam-,75 and bicycle-containing73,76,77 molecules have all been correctly assigned by VCD making this method an excellent candidate for assigning the absolute configuration of the dSp (Sp) stereoisomers.
To conduct the VCD analysis the dSp diastereomers and Sp enantiomers were first purified individually on a Hypercarb HPLC column. Next, IR and VCD spectra were recorded for the SpA (Sp2) enantiomer in DMSO-d6 (Supporting Information S7–9). To achieve reasonable overlap between the experimental spectrum and those calculated using the B3LYP/6-311++G(2d,2p) method two critical steps had to be taken. (1) The inclusion of implicitly defined DMSO must be accounted for during the simulation, because calculations conducted in the gas phase did not match the experimental result (Supporting Information S8). (2) Calculating the conjugated exocyclic amine tautomer form of the Sp B-ring (Figure 5) was required to give a match with the experiment, while calculations conducted on the imino tautomer did not match the experiment (Supporting Information S9). From this initial data set it was concluded that the experimental spectrum for SpA matches the (R)-Sp calculated spectrum (Supporting Information S8 and 9). Armed with these pieces of information VCD analysis was conducted on both nucleoside diastereomers to further confirm this initial assignment.
Figure 5.

Numbering scheme for dSp.
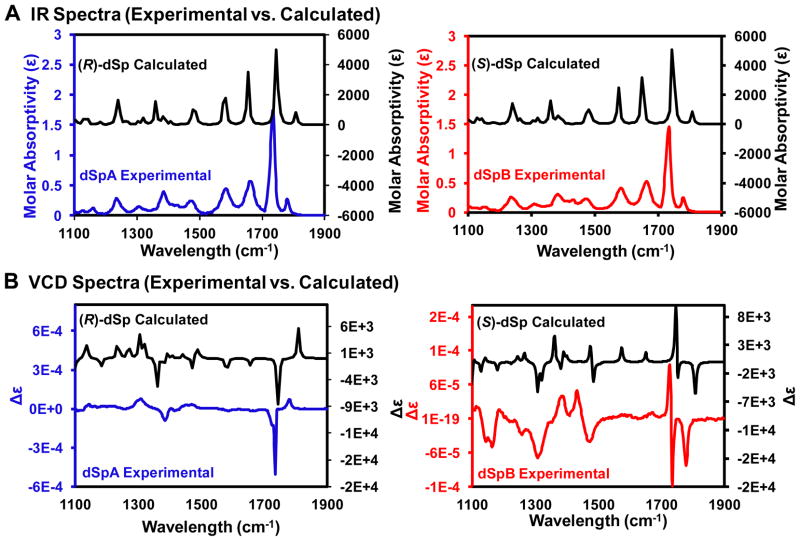
IR and VCD spectra were recorded for the two diastereomers in DMSO-d6, and the calculations were conducted with the S isomer placed in the anti glycoside bond orientation and the R isomer placed in the syn glycosidic bond orientation using the B3LYP functional while implicitly defining the solvent as DMSO and maintaining the conjugated tautomeric form. These configurations have previously been assigned as the lowest in energy via DFT energy minimization calculations.31,45 In the IR spectra for both nucleosides, the peak alignment between the calculated and experimental spectra matched closely with respect to peak spacing and relative intensities (Figure 6A). Next, the experimental VCD spectrum for dSpA was aligned with the calculated spectrum for (R)-dSp, and the experimental spectrum for dSpB was aligned with the calculated spectrum for (S)-dSp. This alignment gave the best overlap, with the opposite alignment between experimental and calculated VCD spectra not showing any similarity (Supporting Information S10 and S11). The most definitive observation used to assign the dSpA experimental spectrum and the (R)-dSp calculated spectrum was the two carbonyl stretching peaks around 1775 cm−1 (expt = 1780 and 1736 cm−1; calcd = 1808 and 1748 cm−1) that gave a positive sign of rotation for the higher frequency signal and a negative sign of rotation for the lower frequency signal (Figure 6B). Two additional peaks were observed in the experimental VCD spectrum (Figure 5 for dSp numbering) that gave similar frequency and sign with the calculated spectrum that include the N7 C-N bending vibration with a negative rotation (exp = 1386 cm−1; calcd = 1359 cm−1) and the N9 N-C bending frequency giving a positive rotation (expt = 1305 cm−1; calcd = 1305 cm−1; Figure 6B).
Figure 6.
Comparison of experimental vs. calculated IR and VCD spectra for the dSp diastereomers. Experimental spectra were recorded in DMSO-d6. Calculated spectra were obtained using a B3LYP/6-311++G(2d,2p) method with implicit DMSO solvation. (A) Comparison of IR spectra for dSpA (blue) with the calculated (R)-dSp, and dSpB (red) with the calculated (S)-dSp. (B) VCD spectrum comparison for dSpA (blue) to that calculated for (R)-dSp, and dSpB (red) to that calculated for (S)-dSp.
Next, the experimental IR spectrum for dSpB and the one calculated for (S)-dSp gave similar patterns in peak spacing and relative intensities (Figure 6A). In the experimental VCD spectrum for dSpB and the one calculated for (S)-dSp three carbonyl stretching peaks with the same sign and similar frequency were observed around 1775 cm−1 (Figure 6B; expt = 1778, 1735 and 1726 cm−1; calcd = 1808, 1753 and 1744 cm−1). Additional signals (see Figure 5 for dSp numbering) were observed in the experimental and calculated spectra with the same sign and similar frequency include the C-N bend at the C2 carbon (expt = 1470 cm−1; calcd = 1480 cm−1), and two C-N bending signals from the N7 and N9 nitrogens (expt = 1381 and 1305 cm−1; calcd = 1359 and 1305 cm−1; Figure 6B). A table of all signals calculated and experimentally observed is provided in the Supporting Information (S11). To reiterate, the most prominent signals confirming the configuration assignment for these two diastereomers were the carbonyl stretches in which the two isomers were experimentally observed and computationally predicted to give different numbers and signs of rotation (S isomer = 3 peaks and R isomer = 2 peaks, Figure 6B). From these comparison plots, we again conclude that dSpA (dSp2) = (R)-dSp and dSpB (dSp1) = (S)-dSp.
ECD Analysis to Determine the Absolute Stereochemistry for the Sp Enantiomers
Because all of our previous conclusions (enzyme kinetics,33,35,36 product distributions and VCD) support the NMR-based stereochemical assignments45 for the dSp diastereomers and not the previous ORD and ECD assignments,39,43,44 we elected to reevaluate the ECD spectroscopic studies along with the TDDFT calculations conducted by the NYU group to determine why the ECD data led to an opposite assignment. In the first step, the ECD experiments and calculations were repeated as previously reported.43 Utilizing a Hypercarb HPLC column, the dSp diastereomers and Sp enantiomers were individually prepared and analyzed by ECD in water, and as expected, we obtained the same experimental results as previously reported by Ding, et al. (Supporting Information S12). Next, to determine the absolute configuration for the Sp enantiomers the experimental spectra were interpreted based on first optimizing the Sp structure in the gas phase using a B3LYP/6-311++G(2d,2p) method, followed by TDDFT calculations to calculate the ECD spectrum. These calculations provided ECD spectra similar to the method reported by the NYU group.43 As expected, the experimental and calculated ECD spectra we obtained led to the same conclusions as the NYU group (Supporting Information S12–14). However, the discrepancy with all of our other results was so compelling that we elected to delve deeper into the calculations of the ECD and ORD spectra.
A number of literature sources have addressed several aspects of UV-vis, ECD and ORD calculations that can be explored, including the increasing the number of excitations calculated, the choice of functional used to account for any conformational flexibility, and the need to account for solvent effects.57,61–64,78 Specifically, studies on other systems have clearly determined that the solvent effect on both UV-vis and ECD spectra can be very pronounced for polar molecules studied in aqueous environments.79 These studies generally employ both implicit solvation methodology, the addition of explicit solvent molecules to the system, or a combination of both to account for the effects of solvation. It has also been shown that the choice of the functional used to model excited state interactions during the TDDFT calculations (CAM-B3LYP) can impact the calculated results for both UV-vis and ECD.54 The use of explicit solvation models also lends to the choice of a functional that reproduces intermolecular interactions, such as M06-2X or PBE1PBE.53,55 Therefore, the same calculations (both geometry optimization and then TDDFT calculations) were repeated under several other conditions. First, the number of excitations calculated was increased from 25 to 50 for (S)-Sp using the B3LYP method, and the same UV-vis and ECD spectra were observed, ruling out this as a possible error (Supporting Information S15). Second, a different functional was used for the TDDFT calculations (M06-2X), while still using the B3LYP optimized geometry in the gas phase. The resulting ECD spectra obtained in this manner were very similar to those obtained in the gas phase calculations and led to the same stereochemical assignments for Sp as reported by the NYU group (Supporting Information S16).43 Third, the geometry optimization was repeated with the M06-2X functional in the gas phase followed by TDDFT calculation using the same functional. These calculations provided the same ECD spectrum that was observed with the B3LYP under the same conditions, though the UV-vis spectrum predicted using the M06-2X geometry matched the experimental much better than the B3LYP (Supporting Information S17). Finally, the geometry optimization was repeated with the B3LYP functional while incorporating implicit solvation by water utilizing the PCM solvation model.56,57 Again, the resulting ECD spectra obtained in this manner showed little difference from those obtained in the gas phase calculations and therefore led to the same stereochemical assignments for Sp (Supporting Information S18 and S19).43
As neither functional choice nor implicit solvation changed the assignment, additional calculations were performed with explicit solvation. A set of seven, ten and twelve explicit H-bonding water molecules were added to the (S)-Sp isomer with twelve waters representing the complete first hydration shell around Sp (Figure 7). The seven-water model has one hydrogen-bonded water for each carbonyl oxygen or ring nitrogen atom. The increase to ten hydrogen bonded water molecules added a second water molecule to each of the carbonyl oxygens. Finally, two additional water molecules were added to the exocyclic amine group, resulting in the twelve-water model. These structures were optimized with the 6-311++G(2d,2p) basis set using the M06-2X functional, while still including implicit solvation by water (Supporting Information S20). The M06-2X functional was chosen as it has previously been shown to perform well when treating explicit solvent interactions.53,80 To ensure the explicitly solvated (R)-Sp enantiomer was indeed a mirror image of the S enantiomer, the mirror invert function in GaussView was used to obtain the structure.59 TDDFT calculations were then performed on these structures with the above mentioned four functionals to obtain the simulated ECD and UV-vis spectra.
Figure 7.
Representative (S)-Sp structures with explicit H-bonded waters. (S)-Sp was geometrically optimized with a B3LYP/6-311++G(2d,2p) method with implicit definition of water. All (S)-Sp structures with explicit waters were geometrically optimized with a M06- 2X/6-311++G(2d,2p) method with implicit definition of water.
The calculated UV-vis and ECD spectra for both (R)- and (S)-Sp with an increasing number of explicit waters modeled with the M06-2X functional are shown in Figure 8. The M06-2X (Figure 8) and CAM-B3LYP methods (Supporting Information S21 and 22) gave the best energies and intensities in comparison to the experimental spectra, and B3LYP and PBE1PBE gave similar results that can be found in Supporting Information (S23–S26). Taking the (S)-Sp calculations as the first example, once all four UV-vis spectra are lined up on the arrow, chosen based on the experimental spectrum (Figure 9), the shoulder centered at 233 nm becomes less pronounced as more waters were added and gives a shape similar to the experimental spectrum (Figure 8A). Next, in the ECD spectra it was observed that the trough at 260 nm without any explicit waters evolves in a water-dependent fashion to a peak when 12 waters were added (Figure 8B). This same evolution occurs with (R)-Sp in the UV-vis spectra giving a less pronounced shoulder at 233 nm with added water (Figure 8A). In the ECD spectrum for (R)-Sp, the peak evolution also occurs with the opposite sign, in which the peak at 260 nm with 0 waters evolves in a water-dependent fashion into a trough when twelve explicit waters were added (Figure 8B).
Figure 8.
Calculated UV-vis and ECD spectra for (R)- and (S)-Sp via the M06-2X method with 0, 7, 10 and 12 explicit water interactions. The optimized structures were submitted to TDDFT calculations with a M06-2X/6-311++G(2d,2p) method with PCM solvation by water. (A) The calculated UV-vis spectra, and (B) calculated ECD spectra. The spectra were red-shifted by 0 waters = 5 nm, 7 waters = 5 nm, 10 waters = 15 nm, and 12 waters = 18 nm to line up on the minima or maxima labeled with the number 1 at 233 nm. The peak at 233 nm corresponds to the shoulder in the experimental UV-vis spectrum and a peak in the ECD spectrum (Figure 9). The arrow placed in the ECD spectra points out the peak evolution that occurs as increasing explicit waters were added to Sp.
Figure 9.
UV-vis and ECD spectra comparison plots with the experimental and calculated spectra. The gas phase structures were optimized with a B3LYP/6-311++G(2d,2p) method in the gas phase and TDDFT calculations were conducted with the same method and basis set. The 12 explicit waters structures were geometrically optimized with a M06-2X/6-311++G(2d,2p) method with PCM solvation by water and the TDDFT calculations were achieved with the same method, basis set and solvation model. Plots are shown for the UV-vis spectra (A) and the ECD spectra (B). The calculated spectra were red-shifted by 15 nm for the gas phase and 18 nm for the solvated calculations, respectively, to match with the experimental spectra at 233 nm.
These calculations for Sp, once lined up with the experimental spectra (in water), immediately point to the inconsistency of the previous ECD reports43,44 with the NMR,45 enzymology,33,35,36 dSp product distributions from the oxidation of G-quadruplex folds, and the VCD data reported here. Overlaying the experimental spectra with the gas phase or calculated UV-vis spectra with twelve explicit waters for both isomers shows that the inclusion of water gives a much better overlap with the experimental UV-vis spectrum than the gas phase calculation (Figure 9). Next, taking the (S)-Sp example, once solvent interactions are accounted for, the overlap between the calculated and experimental ECD spectra supports the conclusion that SpB (Sp1) = (S)-Sp while the gas phase calculations led to the opposite conclusion (Figure 9). Assigning SpB (Sp1) as the S isomer is fully consistent with all previous studies that led to this conclusion.33,36,45 This same phenomenon also occurs in the UV-vis and ECD spectra with the (R)-Sp enantiomer when gas phase and explicit water calculations are compared to the SpA (Sp2) experimental spectra. The conclusion with the addition of water in the calculation is that SpA (Sp2) = (R)-Sp, which is the opposite conclusion obtained from gas phase calculations by the NYU group. Furthermore, the calculated ECD spectra utilizing the other three functionals (B3LYP, CAM-B3LYP and PBE1PBE) with twelve explicit waters led to the same conclusions with respect to the absolute stereochemistry for the Sp enantiomers (Supporting Information S23–S26).
Specific Rotation to Determine the Absolute Stereochemistry for the Sp Enantiomers
A final experiment to establish the absolute configuration for the Sp enantiomers was achieved by measuring the specific rotation at the sodium D-line ([α]D, 589 nm). The specific rotation was measured for SpA and SpB and compared to TDDFT calculations using either the B3LYP or M06-2X methods (Table 1). The experimental [α]D value for SpA (Sp2) was −29 deg SpB (Sp1) was +30 deg (Table 1). The calculated [α]D value for (S)-Sp in the gas phase with B3LYP was −18 deg; however, using the M06-2X functional in the gas phase gave a value of +38 deg that is opposite in sign relative to the B3LYP functional and agrees with the experimental value for SpB. Next, the same methods were used but water was included by implicit definition and the calculated signs were both positive, supporting SpB as the S isomer (Table 1). The most interesting observation was the sign change on the B3LYP method. The inclusion of solvent in this calculation causing a sign change for the specific rotation immediately points to an explanation as to why the ORD values leading to the assignment of Sp absolute configurations by the NYU group are opposite to all other methods.39,45 The last comparison was to a TDDFT calculation using (S)-Sp with twelve explicit waters running the M06-2X method to obtain an [α]D value of +39 deg (Table 1) that gave a reasonable match to the experimental value of SpB (+30 deg, Table 1). From these data, using B3LYP in the gas phase vs. B3LYP with implicit water or the M06-2X functional lead to opposite conclusions about the absolute configuration for SpB. It should also be noted that the specific rotations calculated at other wavelengths between 300 and 600 nm also show the trend of being negative for the B3LYP gas phase calculations, as shown by the NYU group,39 but are positive, as expected for the S configuration, with all other computational methods (Supporting Information S27). These TDDFT calculations further support our conclusion that SpA(Sp2) = (−)-(S)-Sp and SpB (Sp1) = (+)-(S)-Sp.
Table 1.
Specific rotations SpB (Sp1) compared to TDDFT values.
|
|
||||
|---|---|---|---|---|
| [α]D, deg dm−1 cm3 g−1 | ||||
|
| ||||
| Conditions | Computedfor (S)-Sp | Experimental | ||
|
| ||||
| B3LYP | M06-2X | SpA | SpB | |
|
| ||||
| ddH2O | --- | --- | − 29a | + 30b |
| Gas Phase | − 18 | + 38 | --- | --- |
| Implicit Water | + 55 | + 68 | --- | --- |
| 12 Waters | N.D. | + 39 | --- | --- |
This value was obtained at 20 °C from a solution with a concentration of 3.6 mg/mL.
This value was obtained at 20 °C from a solution with a concentration of 3.8 mg/mL.
N.D. = Not Determined
DISCUSSION
dSp Product Distributions in G-Quadruplex Contexts
Oxidation of dG to dSp in the chiral topology of the G-quadruplex gave preferential yields of dSp diastereomers. The context-dependent yields for dSp diastereomers were found to be 3:1 in the hybrid fold, and 1:3 in the propeller fold for dSpA and dSpB, respectively (Figure 3). Mechanistic arguments presented above based on the solvent exposure of the re vs. si face of dG (Scheme 3) led to the conclusion that dSpA (dSp2) = (R)-dSp, and dSpB (dSp1) = (S)-dSp. These arguments assume that the orientation of the purine heterocycles remain more or less the same during the multi-step oxidation process. Indeed, the fact that the ratios are only 3:1 and not higher suggest that a considerable amount of disorder does exist at the stereochemically defining moment of water attack at C5 (Scheme 3). The basket-folded context was also studied, in which the re and si faces of reactive 5′ dGs are solvent exposed in equal amounts, and the dSp diastereomer ratio was found to be 1:1 (Figure 3), supporting our mechanistic claims. Thus, the unequal bifurcation of dG oxidation to the dSp diastereomers demonstrates that the chiral G-quadruplex topology drives the stereochemical outcome of chiral products. Although this observation provides a rational argument for assignments of configurations for dSp, we sought a more definitive answer using VCD spectroscopy.
VCD Assignment of the Absolute Configuration for the dSp Diastereomers
In the second test, VCD analysis provided spectroscopic data to further support our previous claims regarding the dSp configurations. From studies on the dSp nucleosides in DMSO-d6, the recorded spectrum for dSpA (dSp2) was found to line up with the calculated spectrum for the R isomer of dSp, while the spectrum for dSpB (dSp1) lined up with the calculated spectrum for the S isomer of dSp (Figure 6). The most telling feature in the experimental and calculated spectra lies in the number and sign of carbonyl stretching bands that were observed around 1775 cm−1. In the dSpA (dSp2) experimental spectrum and the calculated spectrum for the R isomer, two carbonyl stretching bands were observed, while in the experimental spectrum for dSpB (dSp1) and the calculated spectrum for the S isomer of dSp, three carbonyl stretching bands were observed (Figure 6B). While the carbonyl signals were unique in number and sign for the diastereomers and the absolute configuration could be called from these signals alone, we extended our conclusions to include additional C-N bending signals (Figure 6B and Supporting Information S9). Some signals were predicted but not observed in the experimental spectra for both nucleosides and these unidentified signals fall into two types. (1) Bending signals lost due to solvent-mediated diffusional broadening in DMSO. Examples of these losses include the N-C bends for the C2 carbon with N3 (calcd ~ 1580 cm−1) and N2 (calcd ~1650 cm−1, See Figure 6B for dSp numbering) in both nucleoside samples (Figure 6B and Supporting Information S11). Observation of this effect has previously been experimentally observed and calculated via inclusion of explicit solvent interactions in DMSO.81,82 (2) Signal loss of C-C and C–H bending frequencies in the 1100–1400 cm−1 region were lost due to the conformational flexibility of the sugar, and literature sources support this hypothesis.81 Furthermore, there were a few vibrational transitions with larger (~20 cm−1) deviation in frequency between experiment and theory, however, all had the correct sign of rotation (Supporting Information S11). Signals with the best agreement between experiment and theory came from atoms closest to the asymmetric center (e.g., N9 N-C bends) while those with the greatest deviation were not directly bonded with the asymmetric carbon (e.g., C2 N-C bend). The likely cause of this difference lies in the glycosidic bond orientation that was selected for calculation because as previously demonstrated changes in this bond angle can cause changes in the placement of the base relative to the sugar and alter the vibrational frequencies.31 In contrast, the NOESY-NMR assignments relied on weak NOEs between the NH3 and H2′/H2″ protons that differed between the dSp diastereomers based on DFT calculations.45 However, in this case any minor structural differences do not change our conclusion, because the carbonyl stretching signals were so unique for the two diastereomers (Figure 6B). In conclusion, dSpA (dSp2) is the R isomer and dSpB (dSp1) is the S isomer.
In an additional experiment, VCD analysis of the SpA (Sp2) free base further supported the R absolute configuration for this isomer (Supporting Information S7–9). In this experiment and the corresponding DFT calculations the importance of taking solvent into account during the simulation was identified, as the gas phase calculations did not show a pattern of peaks that were at all similar in sign and frequency to what was observed in the experiment (Supporting Information S8). Additionally, this experiment further supports the result that the preferred tautomer is the one with the exocyclic amine conjugated with the carbonyl in the B-ring (Scheme 1 and Supporting Information S1) as the most representative structure for Sp. These observations again demonstrate the power of VCD analysis to provide absolute configuration assignments that are achievable by interpretation with routine DFT calculations.72
ECD Assignments of the Absolute Configuration for the dSp Diastereomers
ECD spectra for aqueous solutions of the Sp enantiomers were interpreted with TDDFT calculations to make the final determination of the absolute configurations and to understand the inconsistency in the literature reports.43,44 Based on explicit solvent-dependent TDDFT simulations using the M06-2X (Figures 8), CAM-B3LYP, B3LYP or PBE1PBE (Supporting Information S23–S26) functionals in comparison with experimental spectra determined that the addition of explicit waters in the calculation gave rise to the opposite absolute assignments compared to gas phase calculations (Figure 9).43 As a consequence, the calculations including twelve H-bonding waters were used to further support SpA (Sp2) = (R)-dSp and SpB (Sp1) = (S)-Sp. Previous studies have shown that TDDFT calculations to obtain ECD spectra have led to incorrect determination of rotatory lengths and their sign and taking solvent effects into account can rectify the problem.83,84,85 Also, accounting for solvent effects on the electronic excitation properties has previously been demonstrated to influence the rotatory strengths and their signs, which are the calculated values used to make the simulated ECD spectra.74,83,86–89 From these data, the M06- 2X (Figure 9) and CAM-B3LYP (Supporting Information S21 and S22) provide the best overlap with the UV-vis and ECD experimental values. Because the present ECD-based assignments are further supported by dSp diastereomer distributions found upon dG oxidation in the chiral G-quadruplex fold, as well as the absolute configuration assignment by VCD, we conclude that consideration of the solvent influence on the computed ECD spectra is essential when conducting TDDFT calculations on the Sp system. Lastly, the inclusion of solvent interactions in TDDFT studies caused a reversal in the sign of the specific rotation for SpB compared to gas phase calculations (Table 1). This observation further supports all of our other claims regarding the Sp stereochemistry.
The solvent effect that resulted in the TDDFT calculations giving different calculated chiroptical properties was then examined more closely. The inclusion of solvent can alter the ground-state optimization geometry that in turn can impact the TDDFT calculation results. To test this hypothesis the optimized structure with twelve waters was stripped of the water molecules and then submitted to TDDFT using the B3LYP functional in the gas phase. This calculation provided the same ECD spectrum as observed with the B3LYP gas phase optimized structure, thus ruling out structure as the source of the new interpretation (Supporting Information S28).
Literature sources point to several potential roles of the solvent can play in altering rotatory strengths (both sign and intensity), the achiral solvent can form a chiral solvent cage around the chiral solute that must be accounted for, solvent effects on molecular polarization, or solvent effects on the vibrational contribution to the electron excitation response (both in formation and decay). All of these can be challenging to predict a priori and to model by TDDFT calculations.90–94 Furthermore, the locked structure for the Sp heterocycle places the carbonyls in a configuration that places the O6 carbonyl over the hydantoin ring. This structural effect has previously been shown to dramatically alter the calculated pKa for Sp,95 and it is anticipated that this same through-space electronic effect is at play during the chirooptical response in these studies. Thus, to tease out how all of these parameters are at play in determining the chiroptic response in ECD and optical rotation experiments is beyond the scope of the current in which we used complementary methods to reconcile the absolute configuration for the (d)Sp isomers.
Comparison of All Reports Concerning the Absolute Configuration for the dSp Diastereomers
Prior to this report, two laboratories set out to determine the absolute configuration for the dSp diastereomers and came to opposite conclusions: the Grenoble group based their conclusion on NOESY-NMR results45 and the NYU group on ORD and ECD coupled with TDDFT calculations.43,44 ORD and ECD experiments interpreted through TDDFT calculations in the gas phase led to the opposite assignments for the dSp diastereomers (Table 2, using the dSp1 vs. dSp2 nomenclature). However, the NMR-based report45 and the data reported herein came to the same conclusion that dSp1 = (S)-dSp and dSp2 = (R)-dSp (Table 2). These updated assignments are in accord with our previous enzyme kinetic studies concerning repair of the dSp diastereomers in an ODN by hNEIL135,36 as well as the nuclease P1 digestion of dinucleotides bearing the dSp isomers (Table 2).33 In conclusion, the NMR-based analysis,45 enzyme studies,33,36 and the studies reported herein all complement one another in defining the S isomer as the first eluting and the R as the second eluting isomer from either an ion-exchange (ODN, Figure 1) or amino-silica (nucleoside) HPLC column (Supporting Information S2). Moreover, accounting for the elution order switch between columns, the R isomer is the first and the S is the second-eluting isomer when nucleosides are separated on a Hypercarb HPLC column (Figure 1).
Table 2.
Comparison of dSp Absolute Configuration Assignments Determined by Each Method of Study
| Method | R Isomera | S Isomera | Reference |
|---|---|---|---|
| NOESY-NMR | dSp2 | dSp1 | 45 |
| G-Quad Product Distributions | dSp2 | dSp1 | This Work |
| VCD | dSp2 | dSp1 | This Work |
| ECD & TDDFT with 12 watersb | dSp2 | dSp1 | This Work |
| [α]D value & TDDFT with implicit water | dSp2 | dSp1 | This Work |
| hNEIL I Glycosylase Kinetics | dSp2 | dSp1 | 36 |
| Nuclease P1 Digestion Kinetics | dSp2 | dSp1 | 34 |
| ECD & TDDFT Gas Phase | dSp1c,d | dSp2c,d | 43 and 44 |
| ORD & TDDFT Gas Phase | dSp1c,e | dSp2c,e | 39 |
dSp nomenclature is based on the elution order from an ion-exchange or amino-silica HPLC column.
This interpretation was achieved using either a B3LYP, CAM-B3LYP, M06-2X or PBE1PBE method, and all methods were conducted with the 6-311++G(2d,2p) basis set.
In the referenced works the dSp nomenclature was determined on a Hypercarb HPLC column that has the opposite order of elution when compared to the ion-exchange or amino-silica HPLC column. Therefore, in the referenced work, the dSp1 and dSp2 labels are the opposite of those written in the table.
The data was reproduced in the current report (Figure 9 and Supporting Information S12–14).
The data was reproduced in the current report (Table 1 and Supporting Information S27).
Implications for DNA-processing enzymes
In vivo mutagenesis experiments conducted in E. coli utilizing the REAP assay found a stereochemical dependence in the mutation profile observed for the dSp diastereomers. More specifically, dSp1 caused 72% dG→dC (dSp•dG base pair) and 27% dG→dT (dSp•dA base pair) transversion mutations, while dSp2 caused 57% dG→dC and 41% dG→dT transversion mutations in the sequence context 5′-d(GSpA)-3′.28 However, when the dSp isomers were studied by the REAP assay in MutY deficient E. coli in the sequence context 5′-d(TSpG)-3′ the mutation profile changed to dSp1 having 19% dG→dC and 80% dG→dT transversion mutations, and dSp2 gave nearly equal amounts of dG→dC and dG→dT transversions.27 A similar mutation profile was observed for the dSp isomers in the same sequence context studied by the CRAB assay in E. coli.30 From these data it is concluded that (R)-dSp (dSp2) gives similar mutation profiles in both sequence contexts and (S)-dSp (dSp1) gives sequence-context dependent mutation profiles. The updated absolute configurations for the dSp diastereomers will allow future studies to determine if there is a stereospecific explanation for these mutations.
In an additional study, Jia, et al. conducted MD simulations on an 11-mer duplex that had the R or S isomers of dSp embedded in the sequence context 5′-d(CSpC)-3′. In their studies, the R isomer was found to be less distorting to the duplex structure than the S isomer, regardless of the base pairing partner.96 In a follow up study, Khutsishvili, et al. used the same 11-mer sequence to determine experimentally the differences the dSp diastereomers have on the duplex. Although the two dSp isomers paired with dC were indistinguishable by thermal melting and calorimetric analysis, NMR studies pointed to differences in perturbation of adjacent base pairs between the two isomers.41 The “S” isomer (assigned by ECD calculations in the gas phase) was found to be less perturbing to the local structure than the “R” isomer. This NMR observation41 runs counter to the MD studies of the same laboratory.96 Reinterpretation of the absolute configuration for the dSp diastereomers to those presented in this report would now allow this data inconsistency to be reconciled.
dSp-containing DNA strands were found to be substrates for a host of DNA repair enzymes in both the nucleotide excision repair (NER)37 and base excision repair (BER) pathways. In the BER pathway, dSp is repaired by Fpg,97 Nth,98 and Nei98 of E. coli, yOGG1 and yOGG2 of yeast,99 and the NEIL1 class of repair enzymes from viral100 and mammalian sources,35,36 NEIL2101 and NEIL3.102–105 Studies with hNEIL1 have demonstrated stereochemically dependant repair of the two dSp diastereomers in single-stranded, bubble and bulged ODN contexts, as well as double-stranded ODN contexts in all four possible base pairing schemes.35,36 In all of these studies, dSp1 was the better substrate for repair, and it was hypothesized that dSp1 was the S isomer based on structural arguments, but this was never confirmed. Prior to these repair experiments, Jia, et al. conducted MD simulations showing that the S diastereomer would form better contacts with the hNeil1 active site, and therefore, it would be the better substrate for hNEIL1.38 With the current assignment of the S configuration to dSp1, the experimental and calculated data now agree.
In a final experimental example, the dSp diastereomers were digested from the dinucleotide 5′-d(Np[Sp])-3′ (N = A, T, G or C) by nuclease P1.33 In this study, it was found that dSp1 was hydrolyzed to the 5′-dSp-monophosphate 2–6 fold faster than the dSp2-bearing dinucleotide. The kinetic difference was hypothesized to result from differences in glycosidic bond angle preferences between the dSp isomers. Previous DFT calculations showed that the R isomer favors the anti conformation by ~3 kcal/mol more than the syn conformation,31,45 and the anti conformation places the B ring of dSp (Scheme 1) in a path that sterically inhibits hydrolysis of the phosphodiester bond. Therefore, the substrate with the slower reaction rate, dSp2, was hypothesized to be the R isomer. In light of the current results, the dSp absolute configurations based on nuclease P1 reaction kinetics are now further supported.
CONCLUSION
A fresh look at the absolute configurations for the dSp diastereomers was achieved by analysis of oxidation product distributions in the human telomere G-quadruplexes, absolute configuration assignments using VCD, and reinterpretation of the ECD and ORD spectra using explicit waters in the TDDFT calculations. The latter highlights the significant effect of H-bonding to solvent on the chiroptical properties of this heterocycle. Furthermore, VCD analysis provides a wealth of structural data that can readily be interpreted through DFT calculations. These studies in combination with the NOESY-NMR results45 allow the conclusion that dSp1 = (S)-dSp and dSp2 = (R)-dSp based on the diastereomer elution order from an ion-exchange or amino-silica HPLC column, and on the Hypercarb HPLC column, the first eluting isomer is (R)-dSp and the second is (S)-dSp. This new insight further complements the stereochemically dependent reaction kinetics that hNEIL135,36 and nuclease P133 display toward the dSp diastereomers. Furthermore, the biological consequence of oxidations in the human telomere G-quadruplex fold giving the R isomer of dSp, and the observation that it is the least efficient substrate for hNEIL1 repair, suggest that this isomer might be the major source of cellular mutations from dSp leading to nearly equal amounts of dG→dT and dG→dC transversion mutations.28–30 Additionally, the R isomer of dSp might preferentially be detected from oxidative and inflammatory stressed cell samples. As the data becomes available, the in vivo dSp isomer ratios will be instructive in understanding mutation profiles from these diastereomers.
Supplementary Material
Acknowledgments
A.M.F. and C.J.B would like to thank Profs. H. Bernhard Schlegel, Joel Harris and James Muller (University of Utah) for helpful discussions. Computational resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged.
Funding
This work was supported by an NIH grant (CA090689).
ABBREVIATIONS
- CRAB
competitive replication of adduct bypass assay
- dA
2′-deoxyadenosine
- dC
2′-deoxycytidine
- DFT
density functional theory
- dG
2′-deoxyguanosine
- dGh
5- guanidinohydantoin-2′-deoxyribonucleoside
- dOG
8-oxo-7,8-dihydro-2′-deoxyguanosine
- dOGox
two-electron oxidized 8-oxo-7,8-dihydro-2′-deoxyguanosine
- dSp
spiroiminodihydantoin-2′-deoxyribonucleoside
- ECD
electronic circular dichroism
- 5-HO-dOG
5-hydroxy-8-oxo-7,8-dihydro-2′-deoxyguanosine
- MD
molecular dynamics
- ORD
optical rotatory dispersion
- REAP
restriction endonuclease and postlabeling assay
- SIN-1
3- morpholinosydnonimine
- Sp
spiroiminodihydantoin
- TDDFT
time-dependent density functional theory
- T
thymidine
- VCD
vibrational circular dichroism
Footnotes
A.M.F., A.M.O., J.Z., C.J.B. declare no competing financial interests. Y.H. and R.K.D are employed by the VCD manufacturer, BioTools Inc.
Detailed methods, HPLC, mass spectra, G-quadruplex structures, free base VCD experiment and calculated spectra, TDDFT UV-vis, ECD spectra and complete reference 48. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Cadet J, Douki T, Ravanat JL. Free Radical Biol Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Halliwell B, Whiteman M. Br J Pharmacol. 2004;142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steenken S, Jovanovic SV. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 4.Cadet J, Douki T, Gasparutto D, Ravanat JL. Mutat Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Delaney S, Jarem DA, Volle CB, Yennie CJ. Free Radical Res. 2012;46:420–441. doi: 10.3109/10715762.2011.653968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gimisis T, Cismas C. Eur J Org Chem. 2006;2006:1351–1378. [Google Scholar]
- 7.Pratviel G, Meunier B. Chem Eur J. 2006;12:6018–6030. doi: 10.1002/chem.200600539. [DOI] [PubMed] [Google Scholar]
- 8.Burrows CJ, Muller JG. Chem Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 9.Mangal D, Vudathala D, Park JH, Lee SH, Penning TM, Blair IA. Chem Res Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matter B, Malejka-Giganti D, Csallany AS, Tretyakova N. Nucleic Acids Res. 2006;34:5449–5460. doi: 10.1093/nar/gkl596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins AR, Cadet J, Moller L, Poulsen HE, Vina J. Arch Biochem Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 12.David SS, O’Shea VL, Kundu S. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steenken S, Jovanovic SV, Bietti M, Bernhard K. J Am Chem Soc. 2000;122:2373–2374. [Google Scholar]
- 14.Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. 2012 doi: 10.1021/ja306077b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo W, Muller JG, Rachlin EM, Burrows CJ. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 16.Luo W, Muller JG, Rachlin EM, Burrows CJ. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 17.Rokhlenko Y, Geacintov NE, Shafirovich V. J Am Chem Soc. 2012;134:4955–4962. doi: 10.1021/ja212186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shafirovich V, Cadet J, Gasparutto D, Dourandin A, Geacintov NE. Chem Res Toxicol. 2001;14:233–241. doi: 10.1021/tx000204t. [DOI] [PubMed] [Google Scholar]
- 19.Gremaud JN, Martin BD, Sugden KD. Chem Res Toxicol. 2010;23:379–385. doi: 10.1021/tx900362r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niles JC, Wishnok JS, Tannenbaum SR. Chem Res Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 21.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. J Am Chem Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 22.Angelov D, Spassky A, Berger M, Cadet J. J Am Chem Soc. 1997;119:11373–11380. [Google Scholar]
- 23.Cui L, Ye W, Prestwich E, Wishnok JS, Taghizadeh K, Dedon PC, Tannenbaum SR. Chem Res Toxicol. 2013;26:195–202. doi: 10.1021/tx300294d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming AM, Burrows CJ. Chem Res Toxicol. 2013;26:593–607. doi: 10.1021/tx400028y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller JG, Duarte V, Hickerson RP, Burrows CJ. Nucleic Acids Res. 1998;26:2247–2249. doi: 10.1093/nar/26.9.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hailer MK, Slade PG, Martin BD, Sugden KD. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Proc Natl Acad Sci USA. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 29.Delaney S, Neeley WL, Delaney JC, Essigmann JM. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 30.Delaney S, Delaney JC, Essigmann JM. Chem Res Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Biochemistry. 2005;44:6043–6051. doi: 10.1021/bi0473657. [DOI] [PubMed] [Google Scholar]
- 32.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Fleming AM, Muller JG, Burrows CJ. New J Chem. 2013;37:3440–3449. doi: 10.1039/C3NJ00418J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. J Biol Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 35.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. J Am Chem Soc. 2013;135:13851–13861. doi: 10.1021/ja4059469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia L, Shafirovich V, Geacintov NE, Broyde S. Biochemistry. 2007;46:5305–5314. doi: 10.1021/bi062269m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durandin A, Jia L, Crean C, Kolbanovskiy A, Ding S, Shafirovich V, Broyde S, Geacintov NE. Chem Res Toxicol. 2006;19:908–913. doi: 10.1021/tx060078e. [DOI] [PubMed] [Google Scholar]
- 40.Joffe A, Geacintov NE, Shafirovich V. Chem Res Toxicol. 2003;16:1528–1538. doi: 10.1021/tx034142t. [DOI] [PubMed] [Google Scholar]
- 41.Khutsishvili I, Zhang N, Marky LA, Crean C, Patel DJ, Geacintov NE, Shafirovich V. Biochemistry. 2013;52:1354–1363. doi: 10.1021/bi301566v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravanat JL, Cadet J. Chem Res Toxicol. 1995;8:379–388. doi: 10.1021/tx00045a009. [DOI] [PubMed] [Google Scholar]
- 43.Ding S, Jia L, Durandin A, Crean C, Kolbanovskiy A, Shafirovich V, Broyde S, Geacintov NE. Chem Res Toxicol. 2009;22:1189–1193. doi: 10.1021/tx900107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding S, Kolbanovskiy A, Durandin A, Crean C, Shafirovich V, Broyde S, Geacintov NE. Chirality. 2009;21:E231–E241. doi: 10.1002/chir.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karwowski B, Dupeyrat F, Bardet M, Ravanat JL, Krajewski P, Cadet J. Chem Res Toxicol. 2006;19:1357–1365. doi: 10.1021/tx060088f. [DOI] [PubMed] [Google Scholar]
- 46.Lonkar P, Dedon PC. Int J Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleming AM, Muller JG, Ji I, Burrows CJ. Org Biomol Chem. 2011;9:3338–3348. doi: 10.1039/c1ob05112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frisch MJ, et al. Gaussian 09, Revision C.01. Wallingford, CT: 2010. [Google Scholar]
- 49.Krishnan R, Binkley JS, Seeger R, Pople JA. J Chem Phys. 1980;72:650–654. [Google Scholar]
- 50.Frisch MJ, Pople JA, Binkley JS. J Chem Phys. 1984;80:3265–3269. [Google Scholar]
- 51.Becke AD. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 52.Lee C, Yang W, Parr RG. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Y, Truhlar DG. Theor Chem Acc. 2008;120:215–241. [Google Scholar]
- 54.Yanai T, Tew DP, Handy NC. Chem Phys Lett. 2004;393:51–57. [Google Scholar]
- 55.Adamo C, Barone V. J Chem Phys. 1999:110. [Google Scholar]
- 56.Scalmani G, Frisch MJ. J Chem Phys. 2010:132. doi: 10.1063/1.3359469. [DOI] [PubMed] [Google Scholar]
- 57.Tomasi J, Mennucci B, Cammi R. Chem Rev. 2005;105:2999–3094. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 58.Cheeseman JR, Frisch MJ, Devlin FJ, Stephens PJ. Chem Phys Lett. 1996;252:211–220. [Google Scholar]
- 59.Dennington R, Keith T, Millam J. GaussView, Version 5. Semichem Inc; Shawnee Mission, KS: 2009. [Google Scholar]
- 60.Autschbach J, Ziegler T, van Gisbergen SJA, Baerends EJ. J Chem Phys. 2002;116:6930–6940. [Google Scholar]
- 61.Stephens PJ, Devlin FJ, Cheeseman JR, Frisch MJ. J Phys Chem A. 2001;105:5356–5371. [Google Scholar]
- 62.Mennucci B, Tomasi J, Cammi R, Cheeseman JR, Frisch MJ, Devlin FJ, Gabriel S, Stephens PJ. J Phys Chem A. 2002;106:6102–6113. [Google Scholar]
- 63.Bauernschmitt R, Ahlrichs R. Chem Phys Lett. 1996;256:454–464. [Google Scholar]
- 64.Stratmann RE, Scuseria GE, Frisch MJ. J Chem Phys. 1998;109:8218–8224. [Google Scholar]
- 65.Scalmani G, Frisch MJ, Mennucci B, Tomasi J, Cammi R, Barone V. J Chem Phys. 2006;124:1–15. doi: 10.1063/1.2173258. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Patel DJ. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 67.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parkinson GN, Lee MPH, Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 69.Crean C, Geacintov NE, Shafirovich V. Angew Chem, Int Ed. 2005;44:5057–5060. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- 70.Munk BH, Burrows CJ, Schlegel HB. J Am Chem Soc. 2008;130:5245–5256. doi: 10.1021/ja7104448. [DOI] [PubMed] [Google Scholar]
- 71.McCallum JEB, Kuniyoshi CY, Foote CS. J Am Chem Soc. 2004;126:16777–16782. doi: 10.1021/ja030678p. [DOI] [PubMed] [Google Scholar]
- 72.He Y, Bo W, Dukor RK, Nafie LA. Appl Spectrosc. 2011;65:699–723. doi: 10.1366/11-06321. [DOI] [PubMed] [Google Scholar]
- 73.Stephens PJ, McCann DM, Devlin FJ, Flood TC, Butkus E, Stoncius S, Cheeseman JR. J Org Chem. 2005;70:3903–3913. doi: 10.1021/jo047906y. [DOI] [PubMed] [Google Scholar]
- 74.Polavarapu PL, Scalmani G, Hawkins EK, Rizzo C, Jeirath N, Ibnusaud I, Habel D, Nair DS, Haleema S. J Nat Prod. 2011;74:321–328. doi: 10.1021/np100512w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vass E, Hollósi M, Forró E, Fülöp F. Chirality. 2006;18:733–740. doi: 10.1002/chir.20312. [DOI] [PubMed] [Google Scholar]
- 76.Monde K, Taniguchi T, Miura N, Kutschy P, Curillova Z, Pilatova M, Mojzis J. Bioorg Med Chem. 2005;13:5206–5212. doi: 10.1016/j.bmc.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 77.Devlin FJ, Stephens PJ, Osterle C, Wiberg KB, Cheeseman JR, Frisch MJ. J Chem Soc. 2002;67:8090–8096. doi: 10.1021/jo020225n. [DOI] [PubMed] [Google Scholar]
- 78.Polavarapu PL. Chirality. 2012;24:909–920. doi: 10.1002/chir.22015. [DOI] [PubMed] [Google Scholar]
- 79.Sebek J, Gyurcsik B, Sebestik J, Kejik A, Bednarova L, Bour P. J Phys Chem A. 2007;111:2750–2760. doi: 10.1021/jp068811y. [DOI] [PubMed] [Google Scholar]
- 80.Jacquemin D, Perpete EA, Ciofini I, Adamo C, Valero R, Zhao Y, Truhlar DG. J Chem Theory Comput. 2010;6:2071–2085. doi: 10.1021/ct100119e. [DOI] [PubMed] [Google Scholar]
- 81.Lassen PR, Skytte DM, Hemmingsen L, Nielsen SFk, Freedman TB, Nafie LA, Christensen SBg. J Nat Prod. 2005;68:1603–1609. doi: 10.1021/np0502995. [DOI] [PubMed] [Google Scholar]
- 82.Poopari MR, Dezhahang Z, Xu Y. Phys Chem Chem Phys. 2013;15:1655–1665. doi: 10.1039/c2cp42722b. [DOI] [PubMed] [Google Scholar]
- 83.Mennucci B, Cappelli C, Cammi R, Tomasi J. Chirality. 2011;23:717–729. doi: 10.1002/chir.20984. [DOI] [PubMed] [Google Scholar]
- 84.Stephens PJ, McCann DM, Devlin FJ, Flood TC, Butkus E, Stoncius S, Cheeseman JR. J Org Chem. 2005;70:3903–3913. doi: 10.1021/jo047906y. [DOI] [PubMed] [Google Scholar]
- 85.Mukhopadhyay P, Zuber G, Goldsmith MR, Wipf P, Beratan DN. ChemPhysChem. 2006;7:2483–2486. doi: 10.1002/cphc.200600477. [DOI] [PubMed] [Google Scholar]
- 86.Pikulska A, Hopmann KH, Bloino J, Pecul M. J Phys Chem B. 2013;117:5136–5147. doi: 10.1021/jp400562n. [DOI] [PubMed] [Google Scholar]
- 87.Mennucci B, Martinex JM. J Phys Chem B. 2005;109:9818–9829. doi: 10.1021/jp050034z. [DOI] [PubMed] [Google Scholar]
- 88.Mennucci B, Martinez JM. J Phys Chem B. 2005;109:9830–9838. doi: 10.1021/jp050035r. [DOI] [PubMed] [Google Scholar]
- 89.Si Y, Yang G, Hu M, Wang M. Chem Phys Lett. 2011;502:266–270. [Google Scholar]
- 90.Kongsted J, Pedersen TB, Jensen L, Hansen AE, Mikkelsen KV. J Am Chem Soc. 2005;128:976–982. doi: 10.1021/ja056611e. [DOI] [PubMed] [Google Scholar]
- 91.Isborn CM, Mar BD, Curchod BFE, Tavernelli I, Martinez TJ. J Phys Chem B. 2013;117:12189–12201. doi: 10.1021/jp4058274. [DOI] [PubMed] [Google Scholar]
- 92.Pecul M, Marchesan D, Ruud K, Coriani S. J Chem Phys. 2005;122:024106. doi: 10.1063/1.1829046. [DOI] [PubMed] [Google Scholar]
- 93.Kongsted J, Pedersen TB, Osted A, Hansen AE, Mikkelsen KV, Christiansen O. J Phys Chem A. 2004;108:3632–3641. [Google Scholar]
- 94.Fidler J, Rodger PM, Rodger A. J Am Chem Soc. 1994;116:7266–7273. [Google Scholar]
- 95.Verdolino V, Cammi R, Munk BH, Schlegel HB. J Phys Chem B. 2008;112:16860–16873. doi: 10.1021/jp8068877. [DOI] [PubMed] [Google Scholar]
- 96.Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 97.Krishnamurthy N, Muller JG, Burrows CJ, David SS. Biochemistry. 2007;46:9355–9365. doi: 10.1021/bi602459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Nucleic Acids Res. 2001;29:1967–1974. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Biochemistry. 2003;42:11373–11381. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 100.Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. DNA Repair. 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. DNA Repair. 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 102.Sejersted Y, Hildrestrand GA, Kunke D, Rolseth Vy, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, Burrows CJ, Luna L, Bjoras M. Proc Natl Acad Sci USA. 2011;108:18802–18807. doi: 10.1073/pnas.1106880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. Proc Natl Acad Sci USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjoras M. DNA Repair. 2013 doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. J Biol Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.