TO THE EDITOR
Epidermal Langerhans cells (LCs) are central in various aspects of skin biology, but the fact that the cells are the sole cell type constitutively expressing high levels of CD1a molecules has been appreciated only when researchers use CD1a as a specific marker of human LCs. Although we (Pena-Cruz et al., 2003; Sugita et al., 1999) and others (de Jong et al., 2010; Hunger et al., 2004) recently reported studies focusing on a functional role for CD1a in lipid antigen presentation and T-cell activation in the skin, a fundamental question remains to be addressed how such high levels of CD1a expression is maintained in LCs. Certain factors and stimuli have been implicated in CD1a expression in myelomonocytic cells, among which granulocyte-macrophage colony-stimulating factor (GM-CSF) is most prestigious because of its ability to induce CD1a transcription and translation as well as differentiation into LC-like cells in combination with other bioactive factors (Caux et al., 1992; Geissmann et al., 1998; Mohamadzadeh et al., 2001). Given that keratinocytes, the major symbionts of LCs, are the potential source of GM-CSF, one can reasonably hypothesize that the constitutive CD1a expression in LCs may be induced and sustained by GM-CSF present in the epidermal microenvironment. To test this, however, is not an easy task as mice and rats, for which gene manipulation protocols are readily available, have deleted the CD1A gene and thus, LCs in these animals lack the expression of CD1a molecules. Alternatively, we took advantage of this genetic defect and established a murine transgenic (Tg) line carrying a cloned fragment of the human genomic CD1A gene (Fig. 1a). We then mated the human CD1a Tg mice with GM-CSF knockout (KO) mice (Dranoff et al., 1994), hoping to directly address the hypothesis raised above.
Fig. 1.
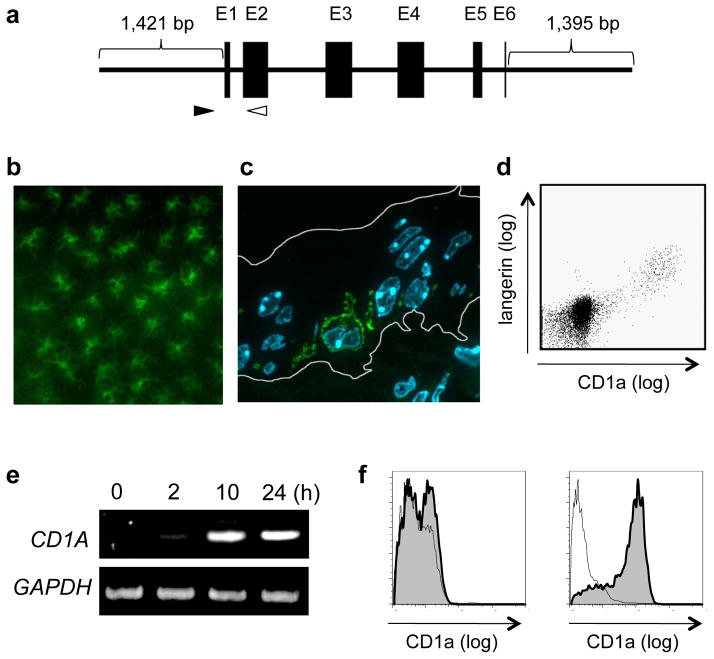
CD1a expression in LCs of the CD1a Tg mice. a. The schematic view of the cloned human CD1A genome (5,833 bp) used for the generation of the Tg mice is shown. Black boxes indicate exons (E). The filled and unfilled arrowheads indicate the position of the sense and antisense primers, respectively, designed for the PCR-based genotyping. b. The epidermal sheet obtained from the ear auricle of the CD1a Tg mouse was fixed, permeabilized and labeled with FITC-conjugated antibodies to human CD1a molecules. Note that the reticular network of CD1a-positive dendritic cells is visualized. c. An ultrathin cryosection of the ear skin obtained from the CD1a Tg mouse was generated, followed by labeling with FITC-conjugated antibodies to CD1a. Nuclei were stained with DAPI. The border of the epidermis is indicated with while lines. Note the presence of a CD1a-positive dendritic cell with a convoluted nucleus in the suprabasal layer of the epidermis. d. The epidermal cell suspension of the CD1a Tg mouse was double-labeled with FITC-conjugated antibodies to CD1a and PE-conjugated antibodies to langerin, and analyzed by flow cytometry. Note that the langerin-positive LCs express CD1a molecules. e. Bone marrow-derived macrophages isolated from the CD1a Tg mouse were stimulated in vitro with recombinant GM-CSF. The cells were then harvested at indicated time points, and RNA was extracted, followed by RT-PCR for detection of CD1A and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts. f. The GM-CSF-stimulated (left panel) and the mock-stimulated (right panel) bone marrow macrophages from the CD1a Tg mouse were labeled with FITC-conjugated antibodies to CD1a and analyzed by flow cytometry.
It is an essential requirement for this study that the cellular distribution of CD1a molecules in humans should be reconstituted faithfully in the CD1a Tg mice. We confirmed that LCs (Fig. 1, b, c, and d) and immature thymocytes (data not shown) in these mice specifically and almost exclusively expressed the transgene. Furthermore, macrophages derived from the bone marrow of the CD1a Tg mice were induced to transcribe the human CD1A transgene upon in vitro stimulation with recombinant GM-CSF (Fig. 1e) and expressed the gene product on the cell surface (Fig. 1f). These observations revealed a trans-species gene activation pathway in which murine transcription factors that would be required for transcription of the human CD1A genome functioned properly, resulting in the constitutive and GM-CSF-induced expression of CD1a molecules.
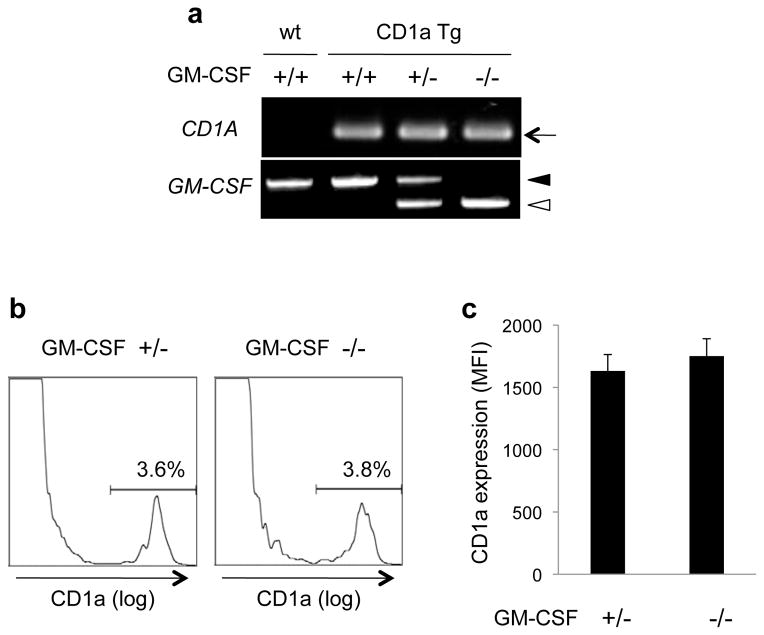
We then analyzed the CD1a expression on LCs either in the presence or in the absence of endogenous GM-CSF. The CD1a Tg mice carrying heterozygous (+/−) or homozygous (−/−) mutations in the GM-CSF genes were identified by the genomic polymerase-chain reaction (PCR) (Fig. 2a), and LCs were isolated from the epidermis, followed by flow cytometric analysis for CD1a expression. Representative histograms are shown in Fig. 2b, indicating that the profiles are comparable between the two groups. Analysis of six animals in each group revealed that the level of CD1a expression on LCs, determined by the mean fluorescence intensity (MFI), was unaffected in the absence of endogenous GM-CSF (Fig. 2c). The present study demonstates that the 1,421 bp upstream sequence of the CD1A genome is sufficient for its highly cell type-specific expression. The identification of LC-specific, but GM-CSF-independent, cellular signals and pathways directed toward the short stretch of the genome would eventually provide important implications in LC and CD1a biology.
Fig. 2.
GM-CSF-independent expression of CD1a molecules by LCs. a. The PCR-based genotyping patterns are shown for the wild-type mouse (wt, +/+), the CD1a Tg mouse carrying the wild-type GM-CSF genes (CD1a Tg, +/+), and the CD1a Tg mice carrying either heterozygous (CD1a Tg, +/−) or homozygous (CD1a Tg, −/−) mutations in the GM-CSF genes. b and c. The epidermal cell suspensions obtained from the CD1a Tg mice carrying heterozygous (left panel in b) or homozygous (right panel in b) mutations in the GM-CSF genes were labeled with FITC-conjugated antibodies to CD1a and analyzed by flow cytometry. The mean MFI values as well as the standard deviations obtained after the analysis of six animals in each group are shown in c.
All animal experiments were performed according to institutional guidelines on animal welfare and the humane treatment of laboratory animals.
Acknowledgments
Funding support was provided by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research(B)).
References
- Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat Immunol. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranoff G, Crawford AD, Sadelain M, Ream B, Rashid A, Bronson RT, Dickersin GR, Bachurski CJ, Mark EL, Whitsett JA, et al. Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science. 1994;264:713–716. doi: 10.1126/science.8171324. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. Transforming growth factor beta1, in the presence of granulocyte/macrophage colony-stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J Exp Med. 1998;187:961–966. doi: 10.1084/jem.187.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger RE, Sieling PA, Ochoa MT, Sugaya M, Burdick AE, Rea TH, Brennan PJ, Belisle JT, Blauvelt A, Porcelli SA, et al. Langerhans cells utilize CD1a and langerin to efficiently present nonpeptide antigens to T cells. J Clin Invest. 2004;113:701–708. doi: 10.1172/JCI19655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadzadeh M, Berard F, Essert G, Chalouni C, Pulendran B, Davoust J, Bridges G, Palucka AK, Banchereau J. Interleukin 15 skews monocyte differentiation into dendritic cells with features of Langerhans cells. J Exp Med. 2001;194:1013–1020. doi: 10.1084/jem.194.7.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Cruz V, Ito S, Dascher CC, Brenner MB, Sugita M. Epidermal Langerhans cells efficiently mediate CD1a-dependent presentation of microbial lipid antigens to T cells. J Invest Dermatol. 2003;121:517–521. doi: 10.1046/j.1523-1747.2003.12429.x. [DOI] [PubMed] [Google Scholar]
- Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]