Summary
Balancing selection refers to a variety of selective regimes that maintain advantageous genetic diversity within populations. We review the history of the ideas regarding the types of selection that maintain such polymorphism in flowering plants, notably heterozygote advantage, negative frequency-dependent selection, and spatial heterogeneity. One shared feature of these mechanisms is that whether an allele is beneficial or detrimental is conditional on its frequency in the population. We highlight examples of balancing selection on a variety of discrete traits. These include the well-referenced case of self-incompatibility and recent evidence from species with nuclear-cytoplasmic gynodioecy, both of which exhibit trans-specific polymorphism, a hallmark of balancing selection. We also discuss and give examples of how spatial heterogeneity in particular, which is often thought unlikely to allow protected polymorphism, can maintain genetic variation in plants (which are rooted in place) as a result of microhabitat selection. Lastly, we discuss limitations of the protected polymorphism concept for quantitative traits, where selection can inflate the genetic variance without maintaining specific alleles indefinitely. We conclude that while discrete-morph variation provides the most unambiguous cases of protected polymorphism, they represent only a fraction of the balancing selection at work in plants.
Keywords: negative frequency-dependent selection, nuclear-cytoplasmic gynodioecy, overdominance, spatial heterogeneity, trans-specific polymorphism
I. Introduction
The factors that maintain genetic and phenotypic variation within natural populations have long interested evolutionary biologists. Is this variation neutral and governed by random factors? Is it transient because of selection for or against particular alleles? Or, alternatively, does selection maintain variation (Ennos, 1983)? Biologists refer to this last case as balancing selection (e.g. Hedrick, 2006; Mitchell-Olds et al., 2007; Hurst, 2009; Andrés, 2011).
Balancing selection is a concept with a complicated history in evolutionary biology. It revolves around the idea that the persistence of adaptive polymorphism within populations over the long term demands special explanation, because as stated by Dobzhansky (1951, p. 109), the ‘absolute equality of adaptive values of two biological forms is…highly unlikely.’ Hence, one form replaces the other eventually and polymorphism is lost. How then do we explain high and sometimes extreme levels of polymorphism, such as the 45 different self-sterility alleles in a plant with fewer than 1000 individuals representing the whole species (Wright, 1969, p. 402)? Balancing selection models grew out of ideas meant to account for this polymorphism. The mechanisms include heterozygote advantage, negative frequency-dependent selection, spatial or temporal habitat heterogeneity, antagonistic pleiotropy, and sexual antagonism, among others. While differing in details, these mechanisms share the feature that whether an allele is beneficial or detrimental is conditional in some way. An allele cannot be described as advantageous or deleterious, except in a particular context. A second common attribute of most balancing selection mechanisms is that selection favors an allele when it is rare (see e.g. Clarke, 1979; Maynard Smith, 1989 (p. 65); Mokkonen et al., 2011, plus more on this later). We provide a historical perspective of the development of these ideas, give examples of balancing selection in plants, and discuss the gradient between the maintenance of genetic variants within and among populations. Finally, we consider balancing selection in the context of quantitative-trait variation, where allelic fitnesses are highly context dependent. Here, selection can maintain genetic variation without preserving specific alleles for long intervals.
II. History
Take a look in Dobzhansky’s (1951) book ‘Genetics and the origin of species’ and you will find nothing in the index on ‘balancing selection’. However, in his chapter on adaptive polymorphism, he refers to ‘balanced polymorphism’ in reference to heterozygote advantage. This simplest model of balancing selection, often called ‘overdominance’, was first proposed by plant geneticists (East, 1908; Shull, 1908) as a mechanism for the very general observations of hybrid vigor and inbreeding depression (Darwin, 1876; Crow, 1987). Dobzhansky championed overdominance as a major explanation for polymorphism in natural populations (Dobzhansky, 1955; Lewontin, 1974), arguing that polymorphism achieved via means other than heterosis could lead to an ‘adaptive accident’ and loss of one of the variants.
Dobzhansky alluded to a second form of balancing selection in the 1951 book. On page 132, he writes, ‘Balanced polymorphism, based on adaptive superiorities of heterozygotes, is not the only possible kind of adaptive polymorphism. A species will be polymorphic if it contains a variety of genotypes each of which is superior in adaptive value to the others in some habitats which occur regularly in the territory occupied by this species.’ Moreover, he goes on to state that ‘…populations which occupy many habitats in a given territory should be genetically more diversified than populations restricted or specialized for occupation of only few habitats.’ (p. 133). This idea – that spatial variation in selection on particular alleles can lead to balanced polymorphism – is the basis for the multi-niche selection models that were developed extensively during the 1960s and 1970s. While the first widely cited mathematical model is that of Levene (1953), the essential idea is clearly stated in Dobzhansky’s chapter.
What about other balancing-selection mechanisms such as negative frequency-dependent selection? Each of the founders of theoretical population genetics – Fisher, Wright and Haldane – evoked this model of selection to address major questions in evolutionary biology. Perhaps the most famous negative frequency-dependent selection model is ‘Fisher’s principle’ (Fisher, 1930; chapter 6; Edwards, 1998). Fisher argued that males and females are equally frequent in most dioecious species because if one sex were to become more frequent, the alternate sex would enjoy a per capita reproductive advantage. Rare advantage is ensured by the fact that the total reproductive success of each sex is equal, at least in species where every zygote has one mother and one father.
In 1939, Sewall Wright developed a model to understand the large number of self-sterility alleles that exist in the hermaphrodite Oenothera organensis. This plant is remarkable in many ways, including its extremely narrow endemism (it grows only in canyons of the Organ Mountains of New Mexico) and its rather exceptionally long styles, ranging from 150 to 180 mm (Emerson, 1938). These long styles make looking at pollen-tube growth, and hence self-incompatibility of pollen grains, relatively easy. In his model, Wright (1939) recognizes that self-incompatibility alleles will be under negative frequency-dependent selection, mentioning the ‘strong selection pressure tending to increase the frequency of rare alleles’. Haldane also applied the idea of rare advantage maintaining polymorphism in plants. In 1949, he discussed how a rare genotype of a plant host species would be resistant to diseases that can attack hosts on the basis of their genotypes (Haldane, 1949; see also Haldane, 1954).
Wright (1969) also discusses how local differences in the direction of selection within a randomly breeding population, combined with density-dependent mortality, would lead to a balanced polymorphism. Here, rare genotypes benefit from the favorable effect of low density. In other words, Wright realized that the inclusion of rare advantage into the mechanism of opposing selection caused by spatial heterogeneity was how polymorphism is maintained; or as stated by Clarke (1979), ‘Selection in multiple niches is not an alternative to frequency-dependent selection…but a way of generating it.’ Wright specifically mentions plants in this context, as a way of pointing out that habitat selection is not an essential component of spatial heterogeneity maintaining polymorphism (p. 124). By the 1970s, articles including the phrase ‘frequency-dependent selection’ were not hard to come across in evolution journals. In 1979, Clarke referred to the evidence for it as ‘pervasive’ while Trotter & Spencer (2007) called frequency dependence the ‘most intuitively obvious explanation’ for polymorphism in nature.
While interest in frequency-dependent selection waxed over the latter part of the 20th century, enthusiasm for overdominance as a general explanation for both polymorphism and inbreeding depression waned. There were several reasons for this, but studies of inbreeding depression provide some of the most relevant data. If overdominant loci are common, they should cause substantial reductions in fitness when organisms are inbred, and indeed, inbreeding depression is very common (Charlesworth & Charlesworth, 1987). However, data returning from long-term genetic studies of corn began to suggest that heterosis is caused more by pseudo-overdominance than genuine heterozygote advantage (Gardner, 1963; Moll et al., 1964; Crow, 1987; but also see Birchler et al., 2006). Here, pseudo-overdominance refers to elevated fitness in hybrids owing to complementation of recessive deleterious alleles at two closely linked genes (see Table 1 for definitions of some relevant terms).
Table 1.
Definitions are given for relevant terms
| Antagonistic pleiotropy – when a polymorphism affects multiple fitness components (e.g. survival and mating success) and a genotype that is beneficial with respect to one component (survival) is detrimental with regard to another (mating success) |
| Balancing selection – a collection of different selection regimes that maintain genetic variation |
| Emergent overdominance – when the heterozygote advantage becomes evident only after averaging genotypic fitnesses over different ‘populations’ (plants occupying different locations, experiencing different environments, or living in different generations) |
| Migration-selection balance – when gene flow (migration into a local population) and local selection have conflicting effects on allele frequency |
| Negative frequency-dependent selection – when the relative fitness of a genotype or phenotype changes with its frequency such that it is favored when rare but not when common |
| Overdominance (heterozygote advantage) – when the heterozygote of a gene confers higher fitness than either homozygote |
| Pseudo-overdominance – when recessive mutations at two closely linked loci, in repulsion, are masked in hybrids; the phenotypic effect mimics heterozygote advantage |
| Purifying selection – a collection of different selection regimes (e.g. directional, stabilizing) that reduce genetic variation |
| Sexual antagonism – when an allele favored in one sex, is disfavored in the other sex |
| Simple overdominance – when the heterozygote advantage is evident within a specific population in a single environment |
| Trans-specific polymorphism – polymorphism that is shared among related species as a consequence of balancing selection maintaining alleles over a very long time |
A second key piece of evidence, particularly relevant to natural populations, relates to the fact that inbreeding depression is caused by the combined effects of many genetic loci. When considering a trait affected by many loci, geneticists often partition the overall genetic variation into components such as the additive variance and the dominance variance (Falconer & Mackay, 1996). These variance components can be estimated even if one does not know anything about the number of genes affecting the trait or how alternative alleles act at these genes. This is useful because the relative magnitude of additive vs dominance variance is informative about the genetic basis of inbreeding depression. If inbreeding depression is caused by overdominant loci, with each allele at a locus reasonably frequent in the population, there should be substantial dominance variance relative to the additive variance in fitness. By contrast, if inbreeding depression is a result of many loci that harbor rare, partially recessive, deleterious alleles, then the dominance variance should be minimal relative to the additive variance. Experimental estimates of variance components (additive ≫ dominance) favor rare deleterious alleles as the major cause of inbreeding depression (Charlesworth & Hughes, 2000; Charlesworth & Willis, 2009).
III. Concepts and terminology
The term balancing selection has been used inconsistently in the literature. We here advocate a relatively inclusive definition of balancing selection as the general alternative to purifying selection. Selection is purifying if it reduces genetic variation, balancing if it maintains genetic variation. By this classification, purifying and balancing selection each encompass a range of distinct scenarios and models. Spatial scale is critically important in describing the variance effects of selection – the same selection regime may be purifying at the local scale (within populations) but balancing at the global scale (maintaing alleles within the species as a whole). In this section, we discuss balancing selection with respect to the related but distinct concepts of overdominance, frequency-dependent selection and protected polymorphism. The scale dependence of balancing/purifying selection is discussed in the section on Spatial and temporal variation in selection.
While overdominance is only one form of balancing selection, there are multiple forms of overdominance. We suggest that it is useful to distinguish ‘simple overdominance’ from ‘emergent overdominance’ (Table 1) when considering experiments on the genetic basis of heterosis and inbreeding depression. The bulk of the experimental evidence noted above evaluates simple overdominance, the circumstance in which the heterozygote at a single gene has higher fitness than either homozygote within a specific population living in a specific environment. By contrast, numerous selection regimes generate a sort of heterozygote advantage, but it is not simple overdominance. This happens when heterozygote advantage emerges by averaging over different populations or different environments or different generations. An example is the model of Gillespie & Turelli (1989) who consider genotype–environment interactions for the many loci affecting a quantitative trait. They assume the trait is subject to constant stabilizing selection, but that the effect of alternative genotypes on the phenotype changes with the environment. Within an environment, however, alleles act additively. In other words, there is no dominance, never mind any overdominance. Yet despite additive gene action and constant stabilizing selection, these authors find that environmental heterogeneity across the entire populaton causes the fitness of a multi-locus genotypes to increase with the amount of heterozygosity.
Why is it important to distinguish simple overdominance from emergent overdominance? Experimental evidence bearing strongly against simple overdominance may not exclude emergent overdominance. If overdominance in fitness is generated by environmental heterogeneity in space or time, it will not be evident in an experiment based on individuals considered in one environment. Environmental heterogeneity was purposefully excluded in most of the experiments considering the genetic basis of heterosis. The distinction between simple and emergent overdominance was alluded to over 50 yr ago by Dempster (1955) when he wrote, ‘Overdominance could result in a large amount of fitness variation which might be largely additive within regions or generations if selection pressures were variable’ (p. 31, italics ours).
Another terminological distinction worth noting is the difference between protected polymorphism and frequency-dependent selection (Table 1). A polymorphism is ‘protected’ if the selection regime acts to increase each of the alternative alleles when they are rare. This seems like frequency dependence, but the statement refers to alleles and not genotypes. Protection requires that the average fitness of an allele, an average over the genotypes in which it occurs, depends on frequency. This can occur with constant genotypic fitnesses, like simple overdominance. By contrast, frequency-dependent selection occurs when the fitness of genotypes, or the distinct morphs they determine, are directly influenced by population frequency. In the next section, we review a number of different ways in which negative frequency-dependent selection acts in plants.
IV. Balanced polymorphisms within plant species
1. Self-incompatibility
‘It has been known for a long time that a very large number of alleles is the rule in the loci that determine self-incompatibility in many species of plants’
(Wright, 1969, p. 402)
Self-incompatibility is a genetic mechanism in hermaphroditic plants that prevents inbreeding, and leads to a high rate of diversification within the lineages that exhibit it (Goldberg et al., 2010). It is an oft-cited example of balancing selection for two reasons. First, as mentioned above, negative frequency-dependent selection on alleles of the S-locus (or S-genes complex), which controls both pollen and pistil specificity (de Nettancourt, 2001), results in the maintenance of dozens of alleles within populations and species (see summary of studies in table 1 of Castric & Vekemans, 2004). This occurs because a pollen grain with a common allele will be limited in terms of mates, while a pollen grain with a rare allele will not. Hence, plants with rare alleles have a selective advantage in terms of siring success that is strong enough to overcome their loss by genetic drift (see model by Wright, 1939).
S-locus polymorphisms also illustrate how balancing selection can increase the longevity of alleles. Neutral polymorphisms, which are typically used as the null model in molecular population genetics, have a finite lifespan contingent on N, the population size (Kimura, 1983). If we randomly sample two alleles from a population, the expected number of generations back in time until they share a common ancestor is 2N. However, some S-locus alleles are sufficiently divergent in sequence that the common ancestor was estimated to exist 40 million yr ago (Goldberg et al., 2010). If balanced alleles are sufficiently ancient, they will have been segregating in the ancestral population for entire clades. This produces ‘trans-specific polymorphism’ (Table 1), where the same alleles are identified in multiple, related species. The negative frequency dependence of selection on S-loci is sufficiently strong that some groups may even have trans-generic polymorphisms (Ioerger et al., 1990; reviewed by Castric & Vekemans, 2004). Trans-specific polymorphism is a striking signature of persistent balancing selection, but it may not be very common (Hedrick, 2006). Oftentimes, it requires the same selection regime to be maintained continuously for millions of years. As we discuss later, however, the absence of ancient alleles within a population does not indicate an absence of balancing selection. Full-genome surveys of variation may now allow researchers to identify more subtle signatures of balancing selection, such as clusters of polymorphisms with unusually intermediate allele frequencies (e.g. Andrés et al., 2009; Amambua-Ngwa et al., 2012).
2. Nuclear-cytoplasmic gynodioecy
Approximately 10% of all flowering plant species are gynodioecious. In populations of such species there are two morphs: hermaphrodites, which produce pollen and seeds, and females, which produce only seeds (Darwin, 1877). Females are ‘male sterile.’ In order to understand how balancing selection might be involved in the maintenance of this dimorphic breeding system, one must understand how selection operates on the genes conferring and countering male sterility.
While hermaphrodites acquire fitness through both male and female function, because females are present in the population, hermaphrodites gain more of their fitness via pollen than via seeds: every seed has one mother and one father and hermaphrodites father all of the seeds produced. This interesting polymorphism is therefore difficult to maintain if the gene(s) controlling male sterility are in the nucleus, because females need to compensate for not gaining fitness via pollen. To do so, they must have twice the seed fitness of hermaphrodites (Lewis, 1941). This can be achieved by producing higher quality seeds (no pollen leads to no selfing, hence no uniparental inbreeding depression (Lloyd, 1975)), higher numbers of seeds (by not making pollen, they save resources that can be spent on seed production (Darwin, 1877)), or some combination of the two (reviewed in Shykoff et al., 2003).
By contrast, if the gene(s) controlling male sterility are located within the mitochondrial genome of the cytoplasm (and hence referred to as cytoplasmic male sterility (CMS) genes), then the fitness comparison is between cytoplasms with functioning CMS genes and cytoplasms without functioning CMS genes, because the cytoplasm is normally inherited maternally (but see McCauley, 2013). Hence, the loss of fitness via nuclear genes will not impact the relative fitness of cytoplasmic genes. As a consequence, the fitness of a cytoplasm with CMS genes need only be marginally higher than that of one without CMS genes – no two-fold difference is needed (Lewis, 1941). It is therefore not surprising that CMS genes confer male sterility in most cases of gynodioecy in natural plant species (Bailey et al., 2003).
That said, the nuclear genome contained within an individual with a CMS gene, but without a two-fold seed-fitness advantage, will have lower fitness than one without CMS genes, because they are biparentally inherited. This sets up selection for a nuclear gene that can counteract the action of the CMS genes and restore male fertility – such genes are called nuclear-restorer genes (Delannay et al., 1981). Hence, this breeding system commonly involves two sets of genes (CMS genes and their nuclear restorers), and there can be multiple CMS genes, each with its own restorer, within a species. In fact, the maintenance of nuclear-cytoplasmic gynodioecy, as it is called, requires polymorphism of the loci in both genomes. To state it more concretely, phenotypic polymorphism (hermaphrodites and females) has underlying polymorphism at a locus (or loci) in both the mitochondrial genome (either multiple CMS genes or a combination of male sterile and male fertile cytoplasms) and the nuclear genome (again, multiple restorers, or a combination of restoring and nonrestoring nuclear genomes).
Two different evolutionary dynamics can lead to this polymorphism, and they start out the same way: as a CMS gene rises in frequency, the restorer able to counteract its effect increases in frequency (Charlesworth, 1981; Frank, 1989; Gouyon et al., 1991; Bailey et al., 2003; Bailey & Delph, 2007). At this point one of two things is thought to occur. Under one scenario the restorer sweeps to fixation, and gynodioecy is maintained only by the invasion of a new CMS gene (Frank, 1989). Under this episodic scenario, the cytoplasm with the highest seed fitness fixes and balancing selection does not come into play. Alternatively, at some point a restorer is sufficiently common that the transmission advantage of the cytotype carrying the CMS gene it restorers is negated, and the frequency of the CMS gene decreases. Moreover, because a cost of restoration exists, this results in a time-lagged decrease in the frequency of the restorer (e.g. Gouyon et al., 1991; Bailey et al., 2003). At this point, there is once again selection causing the CMS gene to increase and the cycle continues, often with new CMS genes and their restorers coming into play. In other words, the CMS genes are under negative frequency-dependent selection, and they cycle in frequency without going to fixation or being lost. If the latter scenario is maintained over a sufficient length of time, signatures of balancing selection on the CMS genes should exist.
Ways to distinguish between these two evolutionary scenarios have been conceived in spite of the fact that CMS genes and their restorers are, for the most part, unidentified in natural plant species exhibiting gynodioecy. The logic is as follows. The mitochondrial genome of flowering plants is haploid and all genes within it are predominantly maternally co-inherited. Hence, if balancing selection acts on CMS genes, it will effectively act on the entire set of cytoplasmic genes. Moreover, any mutations that occur in mitochondrial genes will accumulate among and help differentiate different CMS cytotypes.
This means that if multiple mitochondrial haplotypes exist within populations of a gynodioecious species, and the divergence time of these haplotypes is older than the species themselves (i.e. trans-specific polymorphism exists), then balancing selection is likely at play. Städler & Delph (2002) found support for this prediction in their study of the nucleotide diversity of a mitochondrial gene of Silene acaulis: a large number of divergent haplotypes existed and these haplotypes were ancient, with divergence time estimated to be at least 15 million yr (see also Houliston & Olson (2006) for a similar study of Silene vulgaris). Furthermore, if balancing selection is operating on CMS genes, then gynodioecious species should exhibit greater mitochondrial haplotype diversity than nongynodioecious relatives. Exactly this result was documented in a study of two mitochondrial genes from ten Silene species, three of which were gynodioecious (Touzet & Delph, 2009). Remarkably, two closely related gynodioecious species in this study showed evidence of trans-specific polymorphism and contained very large numbers of haplotypes, for example, 13 cob haplotypes from 19 individuals of S. acaulis (Fig. 1) and 12 from 23 individuals of S. nutans.
Fig. 1.
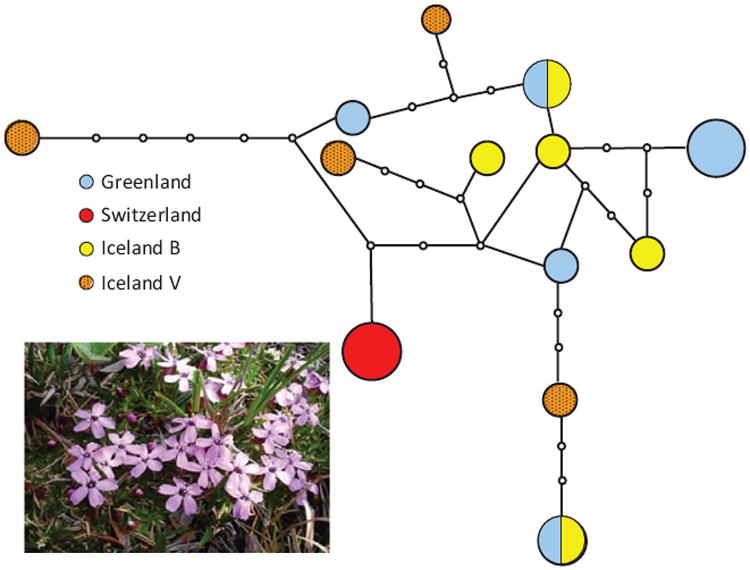
Haplotype network of cob in gynodioecious Silene acaulis from four different populations using statistical parsimony (from data used in Touzet & Delph, 2009). The small circles represent hypothetical haplotypes not found in the sampled individuals. Each branch between two haplotypes (sampled or hypothetical) indicates a single mutational step. The area of the circles is proportional to the haplotype frequency. Three of the four populations contain multiple, highly diverged haplotypes.
Moreover, while cytoplasmic haplotype diversity (both mitochondrial and chloroplastic) should be higher in gynodioecious species, nuclear gene diversity should not. A recent comparison of sequence variability between these three genomes in S. nutans and a closely related dioecious species, S. otites, found more haplotypes and more nucleotide diversity in the two cytoplasmic genomes of the gynodioecious species, but no difference for nuclear gene diversity between the species (Lahiani et al., 2013). This result was found in spite of a likely higher rate of mitochondrial mutations in the dioecious species, discounting mutation rate differences as being responsible, and supporting the conclusion that balancing selection was operating.
Lastly, female frequency in gynodioecious populations should be correlated with mitochondrial haplotype diversity if the latter is a good proxy for the diversity of CMS genes. Comparisons of closely related gynodioecious and nongynodioecious species of Lobelia revealed this relationship, as well as higher mitochondrial haplotype diversity within the gynodioecious Lobelia siphilitica than in the hermaphroditic species L. cardinalis (L. Delph and B. Montgomery, unpublished data). Taken together, these studies strongly implicate long-term persistence of balancing selection in the maintenance of nuclear-cytoplasmic gynodioecy.
V. Other discrete-trait polymorphisms
In addition to self-incompatibility alleles and CMS variants, balancing selection has been shown to be responsible for polymorphism in a variety of other discontinuous traits in plants. We review a few examples here, acknowledging this list is far from exhaustive (see also Ford, 1971; Mitchell-Olds et al., 2007, and references therein).
1. Heterostyly and heterodichogamy
Just as dioecy is protected via negative frequency-dependent selection as noted by Fisher (1930), other polymorphisms associated with sex are similarly protected. One clear example is distyly/heterostyly, in which two or three floral morphs are maintained within populations via dissortative mating (Fig. 2) caused by intra-morph pollen–pistil incompatibilities and reciprocal positioning of anthers and stigmas (Fisher, 1941). This interesting polymorphism exists in at least 28 different angiosperm families (Barrett & Shore, 2008).
Fig. 2.
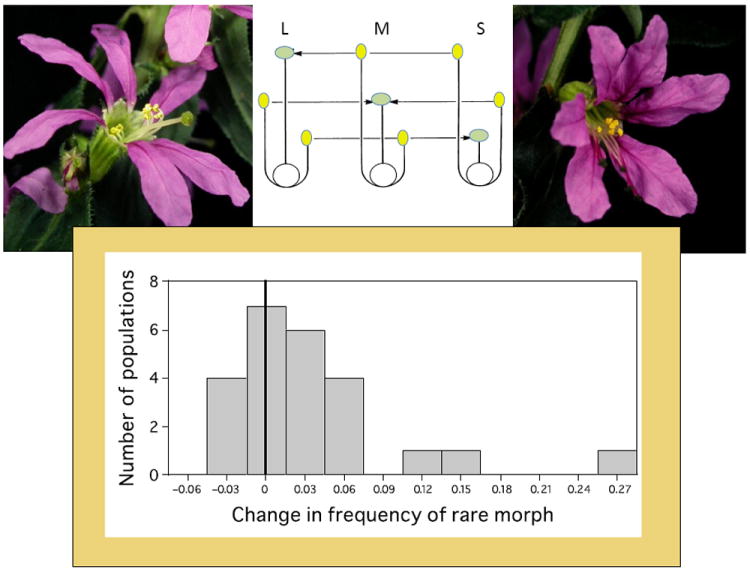
Two of the three morphs of the tristylous species, Lythrum salicaria – (left) the long-styled morph, (right) the short-styled morph (photos courtesy of C. Eckert). The middle panel depicts the positions of the styles (light green ovals) and stamens (yellow ovals) of the three morphs of this species, with the arrows indicating legitimate pollinations (e.g. pollen from the long-styled morph (L) can pollinate the mid- (M) or short-styled (S) morphs). The lower graph depicts evidence for negative frequency-dependent selection in the wild: the frequency of the rarest morph of a set of rapidly expanding natural populations of L. salicaria tended to increase in frequency over a 5-yr observation period (the solid vertical line marks the zero-change category; data from Eckert et al., 1996).
Evidence of negative frequency-dependent selection maintaining heterostyly comes from a study of tristylous Lythrum salicaria, purple loosestrife (Eckert et al., 1996). The authors were able to make use of the invasive nature of this wetland species by studying 24 natural, rapidly expanding populations, each of which had one rare morph (< 11%), over a 5-yr period. This study is noteworthy as an example of detecting whether negative frequency-dependent selection operates in nature by quantifying how rare morphs fared in natural rather than experimental settings. They found that morph evenness and the frequency of the rare morph (Fig. 2) both increased significantly over time.
Another example is heterodichogamy, a sexual system that has evolved independently many times; it occurs in at least 14 angiosperm families, although new cases continue to be found (Wang et al., 2012). It refers to a genetic dimorphism in which one morph presents its pollen for removal at the same time that the other morph presents its stigma for pollen deposition (Lloyd & Webb, 1986). Hence, one morph has flowers that are protandrous (pollen is shed before stigma receptivity) and the other has flowers that are protogynous (the reverse). Modifications on this theme exist, however, with the commonality being that outcrossing is enhanced. For example, a novel type of heterodichogamy occurs in Hernandia, wherein the unisexual flowers of this monoecious species open at different times of the day (pistillate flowers in the morning and staminate flowers in the afternoon or the reverse; Endress & Lorence, 2004). While we know of no studies that have attempted to perturb morph ratios to see whether the rare morph would increase in frequency, the vast majority of species exhibit 1 : 1 morph ratios indicative of balancing selection as for sex ratios (Renner, 2001; Wang et al., 2012).
2. Flower color
Flower-color polymorphism exists in a wide range of species. While other mechanisms have been identified as maintaining this polymorphism (e.g. opposing pleiotropic effects, see Frey, 2004; Carlson & Holsinger, 2010), a number of cases involve negative frequency-dependent selection, either directly or indirectly exerted by pollinators. Some of the best-studied cases are with ‘deceptive’ orchids. Orchids are characterized as being deceptive to pollinators when they make showy flowers that attract pollinators, but then offer no nectar reward (Sprengel, 1793). The behavior of bees is such that they will switch away from a flower type after visiting it and finding it unrewarding, and this behavior is thought to lead to several instances of flower-color and odor polymorphism via negative frequency-dependent selection (reviewed in Schiestl, 2005). For example, manipulation of the relative frequency of the two color morphs (yellow and purple) of the rewardless orchid Dactylorhiza sambucina revealed that rare color morphs did indeed have greater reproductive success than common morphs (Gigord et al., 2001).
Balancing selection on flower-color polymorphism has also been shown in a nonorchid, rewarding species, Ipomoea purpurea, the common morning glory (Subramaniam & Rausher, 2000). Although the mechanism is different than that found for deceptive orchids, the result is the same: rare alleles increase in frequency. The w allele leads to white, rather than pigmented, flowers when homozygous. However, rather than white genotypes being over-visited when rare, they are under-visited, which leads to higher rates of selfing (Brown & Clegg, 1984; Epperson & Clegg, 1986). Nevertheless, when experimental populations are set up with low frequencies of the w allele, this allele increases in frequency in the next generation, as a consequence of inbreeding depression and pollen discounting not being strong enough to counteract the transmission advantage afforded by selfing. Hence, although it is usually less common than the W allele in natural populations, selection for the w allele when rare prevents its elimination by drift (Subramaniam & Rausher, 2000).
3. Meiotic drive in Mimulus
Self-incompatibility alleles, CMS, and flower-color morphs illustrate balanced polymorphism resulting from negative frequency-dependent selection. Another form of balancing selection, conflict between gametic and zygotic selection, is illustrated by the female meiotic drive locus in Mimulus guttatus (Fishman & Saunders, 2008). Here, distinct centromeric elements on one chromosome are the alternative alleles. The driving allele (D) exhibits a 58 : 42 transmission relative to the nondriving alternative (d) in female meiosis. However, in adult diploids, the D/D homozygote suffers 20% reduced pollen viability and may also have reduced seed set (L. Fishman, pers. comm.). Thus, selection at the gametic stage favors D while zygotic selection favors d. A simple one-locus population-genetic model predicts a balanced polymorphism given the estimated fitness effects (see supplementary materials of Fishman & Saunders, 2008). The D allele is predicted to increase when rare even if D/D homozygotes have very low fertility. In an outbred population, rare alleles occur almost entirely in heterozygotes, and in these D/d individuals, D enjoys the transmission advantage without any zygotic fitness cost. At the other boundary (when d is rare), d will increase if the zygotic fitness of D/d sufficiently exceeds the zygotic fitness of D/D.
The meiotic-drive polymorphism in Mimulus does not exhibit an ‘ancient allele’ sequence-diversity pattern. By contrast, it appears that the D allele has recently increased to intermediate population frequency. The D allele is associated with a specific constellation of alleles at surrounding polymorphic loci over a genomic region spanning at least 10 megabases (fig. 4 of Fishman & Saunders, 2008). By contrast, the d allele is associated with a diversity of haplotypes. This is the sequence-diversity pattern expected with a ‘partial sweep’ if the d allele is ancestral: a favorable mutation (d to D) occurs within a single genetic background and selection rapidly increases this haplotype to intermediate population frequency. The important point is that a simple genome wide survey of sequence diversity would not have identified D/d as a balanced polymorphism because the alternative alleles are not highly divergent in sequence. Ancient balanced polymorphisms, which yield the diagnostic sequence pattern evident for S-locus and CMS genes, are easy to spot, but they may be only a small fraction of balanced polymorphisms.
4. Host–pathogen systems
Although Haldane had hypothesized that rare disease-resistant genotypes should be at an advantage relative to common genotypes back in 1949 (see Section I. Introduction), and although boom-and-bust cycles had been documented for agricultural crops, data from natural plant populations documenting negative frequency-dependent selection was lacking until very recently (Barrett, 1988; Chaboudez & Burdon, 1995; Barrett et al., 2009). What data there were regarding variation among hosts for resistance to pathogens and for pathogens for infectivity of hosts, consisted mostly as catalogs of variation across varying spatial scales (Barrett, 1988; Laine et al., 2011). As a first step in 1995, it was shown that the most common clone of the aster Chondrilla juncea was infected by a rust pathogen in 13 out of 16 study populations; this result is consistent with negative frequency-dependent selection, but could also be caused by other processes (Chaboudez & Burdon, 1995). Part of the difficulty stemmed from the complexity of factors influencing host–pathogen interactions in the wild.
What was needed was evidence of fluctuating selection over time, wherein changes in the frequencies of common resistant host variants corresponded with changes in pathogen variants. Such selection has recently been shown in host–pathogen systems using time-shift inoculation/infection experiments (Decaestecker et al., 2007; Gomez & Buckling, 2011; Koskella, 2013). With such experiments, hosts from any given year are exposed to sympatric pathogens from the past, contemporary, and future time points, and vice versa. If negative frequency-dependent selection is at work, then various populations are likely to be out of sync with regard to which genotypes can be infected (hosts) and infect (pathogen). Furthermore, because the antagonists are expected to be responding and counter-responding (cycling), whether pathogens from a given time point are better or worse at infecting hosts will depend on what part of the cycle is encompassed by the experiment – overall one should see variation across time rather than consistent increases or decreases in susceptibility or infectivity.
This approach, together with population-level phenotype and genotype surveys, was taken to address whether negative frequency-dependent selection was acting to maintain variation in resistance of Australian native flax (Linum marginale) to its obligate rust fungus (Thrall et al., 2012). This fungus is sufficiently virulent that it can reduce the population size of the flax during severe epidemics, indicating that the host should be under strong selection to evolve resistance. The results of this 6-yr study revealed asynchrony across populations, and complex shifts in patterns of resistance and infectivity over time. Moreover, the population-level surveys revealed fluctuations in existing pathogen genotypes, rather than the introduction of new variants. Taken together, these results show a pattern of change consistent with negative frequency-dependent selection. That said the complex patterns observed could only be understood by taking a multi-population and multi-year approach, as much of the variation was attributable to the effect of population (space) and the interaction of space and time (Thrall et al., 2012). This leads us to the next section, on space and time.
VI. Spatial and temporal variation in selection
Environmental heterogeneity in space and time can maintain genetic variation. For spatially varying selection, the archetypal model of Levene (1953) considered a population occurring in two environments with random mating within generations and random assignment of genotypes to environments across generations. This model does allow protected polymorphism, albeit within a modest parameter space where there is an appropriate balance of positive and negative effects across environments. Conditions for protected polymorphism become much less restrictive if genotypic mixing across environments is incomplete owing to limited migration or habitat choice (Deakin, 1966; Christiansen, 1974). Exact results depend on genetic and demographic details (Karlin, 1982), but Wright’s continent-island model (Wright, 1931) provides a conservative ‘rule of thumb’: a locally advantageous allele with selection coefficient s, will be maintained against opposing gene flow occurring at rate m, if s > m.
Scale is a fundamental issue when considering spatial variation in selection. At the scale of the entire range of a species, s > m is not a very stringent condition. Plants are rooted in place and normally have limited seed and pollen dispersal. Spatially varying selection generates morphological differentiation across landscapes that is routinely strong enough such that botanists classify fully interfertile populations as ecotypes (Clausen et al., 1940, 1948; Bennington & McGraw, 1995; Kruckeberg et al., 1995). This can establish the interesting situation in which selection is locally purifying but globally balancing. Directional selection within sub-populations will reduce the local genic variance if the favored allele is locally common. However, at the scale of the entire species, the differing direction of selection within different sub-populations insures that no single allele is predominant. As noted by Tack et al. (2012), the variation that exists among populations may play ‘a crucial role in safeguarding the maintenance of variation.’
While geographically varying selection is often discussed as being distinct from balancing selection, and is usually referred to as local adaptation (e.g. Mitchell-Olds et al., 2007), it is not always easy to draw this distinction when dealing with natural populations. Plant species exhibit the full range of population structures, from well mixed to highly structured. This absence of discontinuity makes it very difficult to identify the natural analogs of the theoretical ‘sub-population.’ In the models, sub-populations are internally uniform with regard to selection and internally well mixed with regard to mating and gene flow. Operationally, sub-populations are identified as collections of plants that occupy an area, say a meadow or forest tract. Sub-populations are distinguished because they are spatially segregated from each other. However, sub-populations defined this way are not necessarily homogenous with regard to selection pressures, nor necessarily well mixed with regard to gene flow.
There is abundant evidence that the direction of selection can vary among microsites within contiguous plant populations (e.g. Silander, 1979; Stewart & Schoen, 1987; Tonsor et al., 1993). This kind of heterogeneity can maintain alternative alleles, although the conditions for this to happen depend on the relative scales of fitness heterogeneity vs seed and pollen migration. If there is sufficient gene flow, a Levene (1953) type population structure is obtained with the associated conditions for protected polymorphism (limited but not impossible). However, if seed/pollen dispersal is sufficiently restricted and localized selection regimes are sufficiently consistent across generations, microhabitat adaptation can occur within populations. In order to eliminate historical or random effects as being responsible for the pattern of variation, it is important to conduct reciprocal-transplant experiments (Ennos, 1983). Indeed, the empirical hallmark of local adaptation – resident genotype superiority in a reciprocal-transplant experiment – has been demonstrated in a number of short-lived plant species at scales of 3–50 m. Examples include Delphinium nelsonii (Waser & Price, 1985), Impatiens capensis (Schmitt & Gamble, 1990), Impatiens pallida (Schemske, 1984) and Trifolium repens (Turkington & Harper, 1979).
In some cases, investigators have demonstrated genetic differentiation associated with environmental gradients that exist within contiguous plant populations. For example, Audigeos et al. (2013) investigated highly localized geographic structure within stands of the tropical tree species Eperua falcate. Individuals are distributed over an edaphic gradient that ranges from flooded bottomlands to seasonally dry soils over a distance of a few hundred meters. Gene flow is probably substantial across this gradient given minimal differentiation at putatively neutral molecular markers. Lack of differentiation can be due to lack of time since a population is partitioned. However, Audigeos et al. (2013) note significant differentiation at genes related to stress response, consistent with divergent selection despite gene flow. A common-garden experiment further demonstrated habitat-specific divergence in ecologically relevant traits such as growth rate, leaf chemistry and physiological measurements (Brousseau et al., 2013). The Eperua studies nicely illustrate how a combination of methods can be used to identify genetic microhabitat selection and differentiation within populations. However, the general features of this example are not usual (see, e.g., Ager et al., 1993).
Counter to these positive examples, there are certainly situations where localized gene flow is sufficient to prevent local adaptation within populations (e.g. Stratton & Bennington, 1998). In fact, there are a number of reasons why the s > m rule is less likely to hold at small than large spatial scales (Galloway & Fenster, 2000). Small-scale environmental gradients may be lesser in magnitude or less temporally stable. If a microsite that is relatively wet in the current generation is not likely to be so in future generations, local adaptation will not occur even with very restricted gene flow. Also, the number of plants within a localized patch of a population is much smaller than the number of plants in large geographic regions. Selection is less effective at opposing genetic drift in small populations. A more subtle effect is that a single immigrant to a small population translates to a higher migration rate (m) than a single immigrant to a larger population. A correspondingly larger s is thus required to maintain a locally favorable allele in a small than a large population. Indeed, there is some indication that local adaptation is less frequent in smaller sub-populations (Leimu & Fischer, 2008).
Temporal fluctuations in the direction of selection, mentioned above as a factor that can prevent local adaptation, can also maintain variation. Haldane & Jayakar (1963) demonstrated the conditions for polymorphism when selection fluctuates between two different regimes; both alleles persisting when the geometric mean fitness of heterozygotes (through time) exceeds that of homozygotes (see also Dempster, 1955). This rather stringent condition, which may be further limited by genetic drift in finite populations (Hedrick, 1976), caused many researchers to dismiss temporally varying selection as a factor maintaining variation. However, more recent theoretical studies (Kondrashov & Yampolsky, 1996; Burger & Gimelfarb, 2002) suggest that fluctuating selection can greatly elevate the genetic variance relative to constant selection if there is positive autocorrelation of conditions between generations and mutation occasionally introduces novel mutations into the population. Unfortunately, despite considerable evidence for temporal fluctuations in selection (e.g. Kelly, 1992; Grant & Grant, 1995), we currently have limited evidence on whether these fluctuations typically increase or decrease variation, excepting the evidence from host–pathogen dynamics discussed previously.
VII. Selection and the maintenance of trait variation
The theory of protected polymorphisms is extensive and it provides a compelling conceptual framework for evaluation of discrete-trait polymorphisms. However, it is in key ways too limited to describe how selection maintains genetic variation for quantitative traits. Here, how one frames the question of variation becomes critical. The population genetic perspective is based on the maintenance of specific, alternative alleles. Oftentimes, however, it is more natural to think in terms of the total genetic variance in a trait (or set of traits) than in the persistence of alleles at particular loci. In terms of the trait variance, we can ask whether the net effect of selection is positive or negative relative to a null expectation. A natural null is the variance of a selectively neutral trait, all else equal (mutation rates, allelic effects, population structure, etc.). Considering whether selection is balancing (net positive effect on variance) or purifying (net negative effect on variance) in this context yields quite different conclusions than those that emerge from the single-locus equilibrium models of population genetics. In fact, it may be that the most important variance-positive effects of selection on quantitative traits do not involve protected polymorphisms.
In order to illustrate this, consider a mutation with additive effect a in a randomly mating population of size N. Ignoring complexities such as dominance and epistasis, the genetic variance contributed by this allele in any one generation is 2pqa2 (Falconer & Mackay, 1996). If the mutation is neutral and first appears as a single copy (p = 1/2N), the expected total variance that it contributes over its lifespan (until loss or fixation) is
(Crow & Kimura, 1970, p. 329). By contrast, consider the fate of a mutation responsive to a temporal fluctuation in the environment. Imagine that this mutation is initially favored by selection and increases to 50% frequency over its first thousand generations, but is then disfavored and declines to loss in the next thousand generations. The total variance contributed by this mutation will likely exceed 500 a2. Selection has greatly increased the variance contribution of this polymorphism despite that the sojourn of this allele is certainly not that of a protected polymorphism. The point of this example is simple: If selection routinely drives alleles from rarity to intermediate frequencies, it will generate substantial trait variation even if the typical fate of alleles is extinction (or fixation) on short evolutionary time scales – hundreds to thousands, but not millions of generations.
In the preceding example, we assumed that selection prevented immediate loss of the mutation and allowed a subsequent increase in frequency. Most mutations, even those that confer substantial fitness advantage, are lost as a consequence of sampling in the short term (Haldane, 1927). However, countering this stochastic loss is the continual input of new mutations. On a per locus rate, mutation is highly infrequent. However, the total mutation rate to new alleles affecting a trait (summed over the entire genome) may be quite high, perhaps a few percent per individual per generation (Turelli, 1984). In a large population or meta-population, hundreds of such alleles are introduced each generation.
Do realistic selection regimes generate dynamics where selection has a net-positive effect on the genetic variance but alleles are not preserved indefinitely? A concrete example is a simulation study motivated by the general features of geographically varying selection in plants (Kelly, 2006). The model considered Quantitative Trait Loci (QTLs) for a trait with different fitness optima in different populations. Migration was allowed at varying rates among populations. The mutational model and mutational parameters were based on standards from quantitative genetics (Latter, 1960; Bulmer, 1972; Turelli, 1984; Lynch & Walsh, 1998). Adjusting the relative strengths of selection and migration, this model reiterated realistic patterns of trait variation. Unsurprisingly, geographically varying selection routinely inflated the variance 10–100 fold relative to the neutral case. The variance positive effect of selection was largely caused by populations diverging in mean phenotype corresponding to different fitness optima (fig. 6 of Kelly, 2006; see also Slatkin, 1978).
This simulation is relevant here because the polymorphisms that emerge in this model do not qualify as protected in the population-genetic sense. In fact, alternative alleles typically failed to persist long enough to yield molecular signatures of balanced polymorphism. There are several reasons for this, but the most important is that the model invoked selection on the phenotype. This differs from the usual population genetic convention of assigning fitness values directly to genotypes. The usual convention is problematic for quantitative traits where mutations at different genes can have similar effects. As a consequence of this ‘genetic redundancy’ (Goldstein & Holsinger, 1992; Brookfield, 1997), a mutation at one locus can effectively substitute for an allele with comparable effects at another locus. Such substitutions might occur because of selection or drift, but regardless, they will tend to reduce the lifespan of individual alleles.
VIII. Conclusions
Plants provide clear examples of balanced polymorphism. We have reviewed classic cases such as self-incompatibility loci, as well as more recently described systems such as nuclear-cytoplasmic gynodioecy. In many of these examples, variation is concentrated into a limited number of discrete morphs. The selection regime on these morphs can be directly studied and quantified, and such studies have routinely revealed a frequency dependence to selection. It is noteworthy that these examples often involve the basic reproductive mode of the plant (self-incompatibility, dioecy, gynodioecy, heterostyly, etc.). By their nature, these systems may generate a frequency-dependent selection regime that is persistent in the long term. In other words, the rare advantage of alternative morphs is sustained through time within populations despite the inevitable fluctuations in environmental conditions and selection pressures experienced by an evolutionary lineage. In this situation, alternative alleles may persist for long intervals, in some cases long enough to accumulate the molecular signature of balanced polymorphism.
Negative frequency-dependent selection maintains polymorphism within populations. By contrast, spatially varying selection maintains adaptive polymorphism at the among-population level. It can also contribute to within-population variation, either through microhabitat selection and resulting intra-population spatial structuring or via gene flow between divergent populations. Recent work on plant host–pathogen systems illustrates how interactions among different mechanisms (e.g. spatiotemporal variation and underlying negative frequency-dependent selection) contribute to variation in both host resistance and disease pathogenicity. Molecular-genetic evidence for ancient resistance polymorphisms has been based largely on alleles sampled from different populations (e.g. Stahl et al., 1999; Bergelson et al., 2001; Tian et al., 2002; Bakker et al., 2006) and thus indicative of balancing selection at the whole-species level. However, given that among-population processes can heighten within-population variation, a more complete understanding of adaptive polymorphism will require study of both pattern and process across a variety of spatial scales (Laine et al., 2011; Tack et al., 2012; Thrall et al., 2012).
The conspicuous discrete morph examples we have discussed are likely a small subset of balanced polymorphisms within plant populations. Most are difficult to detect because either the balanced alleles are not ancient (and thus do not exhibit the hallmark molecular signature) and/or the fitness effects and hence fitness tradeoffs of genotypes are too small for direct measurement. Both difficulties are likely to be acute when considering genes affecting quantitative traits. Here, alternative genotypes at a locus will typically be associated with a full distribution of phenotypes; a challenging proposition if one intends to characterize selection at the scale of genes. Also, because different alleles (either at the same gene or different genes) can produce essentially the same effect on a quantitative trait, selection can maintain variation without preserving particular alleles indefinitely. As a consequence, bona fide protected polymorphism might represent only a minor component of balancing selection when considered from the perspective of a trait’s variance.
Acknowledgments
The authors thank C. Lively, M. Rausher, M. Wade, V. Koelling and H. Spencer for stimulating discussions and/or comments on drafts of this manuscript. C. Eckert provided data and shared photos. L.F.D. acknowledges support from grant NSF DEB-0813766 and J.K.K. from grants NIH GM073990-02 and NSF IOS-0951254.
References
- Ager AA, Heilman PE, Stettler RF. Genetic variation in red alder (Alnus rubra) in relation to native climate and geography. Canadian Journal of Forest Research. 1993;23:1930–1939. [Google Scholar]
- Amambua-Ngwa A, Tetteh KKA, Manske M, Gomez-Escobar N, Stewart LB, Deerhake E, Cheeseman IH, Newbold CI, Holder AA, Knuepfer E, et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genetics. 2012;8:e1002992. doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés AM. Encyclopedia of life sciences. Chichester, UK: John Wiley & Sons Ltd; 2011. Balancing selection in the human genome. [Google Scholar]
- Andrés AM, Hubisz MJ, Indap A, Torgerson DG, Degenhardt JD, Boyko AR, Gutenkunst RN, White TJ, Green ED, Bustamante CD, et al. Targets of balancing selection in the human genome. Molecular Biology and Evolution. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audigeos D, Brousseau L, Traissac S, Scotti-Saintagne C, Scotti I. Molecular divergence in tropical tree populations occupying environmental mosaics. Journal of Evolutionary Biology. 2013;26:529–544. doi: 10.1111/jeb.12069. [DOI] [PubMed] [Google Scholar]
- Bailey MF, Delph LF. Sex-ratio evolution in nuclear-cytoplasmic gynodioecy when restoration is a threshold trait. Genetics. 2007;176:2465–2476. doi: 10.1534/genetics.107.076554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MF, Delph LF, Lively CM. Modeling gynodioecy: novel scenarios for maintaining polymorphism. American Naturalist. 2003;161:762–776. doi: 10.1086/374803. [DOI] [PubMed] [Google Scholar]
- Bakker EG, Toomajian C, Kreitman M, Bergelson JA. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JA. Frequency-dependent selection in plant-fungal interactions. Philosophical Transactions of the Royal Society B: Biological Sciences. 1988;319:473–483. [Google Scholar]
- Barrett LG, Thrall PH, Dodds PN, van der Merwe M, Linde CC, Lawrence GJ, Burdon JJ. Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Molecular Biology and Evolution. 2009;26:2499–2513. doi: 10.1093/molbev/msp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH, Shore JS. New insights on heterostyly: comparative biology, ecology and genetics. In: Franklin-Tong VE, editor. Self-incompatibility in flowering plants: evolution, diversity and mechanisms. Berlin, Germany: Springer; 2008. pp. 3–32. [Google Scholar]
- Bennington CC, McGraw JB. Natural selection and ecotypic differentiation in Impatiens pallida. Ecological Monographs. 1995;65:303–323. [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian DC. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Unraveling the genetic basis of hybrid vigor. Proceedings of the National Academy of Sciences, USA. 2006;103:12957–12958. doi: 10.1073/pnas.0605627103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield JFY. Genetic redundancy. Advances In Genetics Incorporating Molecular Genetic Medicine. 1997;36:137–155. doi: 10.1016/s0065-2660(08)60308-9. [DOI] [PubMed] [Google Scholar]
- Brousseau L, Bonal D, Cigna J, Scotti I. Highly local environmental variability promotes intra-population divergence of quantitative traits: an example from tropical rainforest trees. Annals of Botany. 2013 doi: 10.1093/aob/mct176. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Clegg MT. Influence of flower color polymorphism on genetic transmission in a natural population of the common morning glory, Ipomoea purpurea. Evolution. 1984;38:796–803. doi: 10.1111/j.1558-5646.1984.tb00352.x. [DOI] [PubMed] [Google Scholar]
- Bulmer MG. The genetic variability of polygenic characters under optimizing selection, mutation and drift. Genetical Research. 1972;19:17–25. doi: 10.1017/s0016672300014221. [DOI] [PubMed] [Google Scholar]
- Burger R, Gimelfarb A. Fluctutating environments and the role of mutation in maintaining quantitative genetic variation. Genetical Research. 2002;80:31–46. doi: 10.1017/s0016672302005682. [DOI] [PubMed] [Google Scholar]
- Carlson JE, Holsinger KE. Natural selection on inflorescence color polymorphisms in wild Protea populations: the role of pollinators, seed predators, and intertrait correlations. American Journal of Botany. 2010;97:934–944. doi: 10.3732/ajb.0900348. [DOI] [PubMed] [Google Scholar]
- Castric V, Vekemans X. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Molecular Ecology. 2004;13:2873–2889. doi: 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- Chaboudez P, Burdon JJ. Frequency-dependent selection in a wild plant-pathogen system. Oecologia. 1995;102:490–493. doi: 10.1007/BF00341361. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Hughes KA. The maintenance of genetic variation in life history traits. In: Singh RS, Krimbas CB, editors. Evolutionary genetics from molecules to morphology. Cambridge, UK: Cambridge University Press; 2000. pp. 369–392. [Google Scholar]
- Charlesworth D. A further study of the problem of the maintenance of females in gynodioecious species. Heredity. 1981;46:27–39. [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annual Review of Ecology and Systematics. 1987;18:237–268. [Google Scholar]
- Charlesworth D, Willis JH. The genetics of inbreeding depression. Nature Reviews Genetics. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- Christiansen FB. Sufficient conditions for protected polymorphism in a subdivided population. American Naturalist. 1974;108:157–166. [Google Scholar]
- Clarke BC. The evolution of genetic diversity. Proceedings of the Royal Society B: Biological Sciences. 1979;205:453–474. doi: 10.1098/rspb.1979.0079. [DOI] [PubMed] [Google Scholar]
- Clausen J, Keck DD, Hiesey WM. Experimental studies on the nature of species. I. The effect of varied environments on western American plants. Washington, DC, USA: Carnegie Institute; 1940. [Google Scholar]
- Clausen J, Keck DD, Hiesey WM. Experimental studies on the nature of species. III. Environmental responses of climatic races of Achillea. Washington, DC, USA: Carnegie Institute; 1948. [Google Scholar]
- Crow JF. Muller, Dobzhansky, and overdominance. Journal of Historical Biology. 1987;20:351–380. [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. New York, NY, USA: Harper and Row; 1970. [Google Scholar]
- Darwin C. The effects of cross- and self-fertilisation in the vegetable kingdom. London, UK: John Murray; 1876. [Google Scholar]
- Darwin C. The different forms of flowers on plants of the same species. London, UK: John Murray; 1877. [Google Scholar]
- Deakin MAB. Sufficient conditions for genetic polymorphism. American Naturalist. 1966;100:690–692. [Google Scholar]
- Decaestecker E, Gaba S, Raeymaekers JAM, Stoks R, Van Kerckhoven L, Ebert D, De Meester L. Host–parasite “Red Queen” dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- Delannay X, Gouyon PH, Valdeyron G. Mathematical study of the evolution of gynodioecy with cytoplasmic inheritance under the effect of a nuclear restorer gene. Genetics. 1981;99:169–181. [PMC free article] [PubMed] [Google Scholar]
- Dempster E. Maintenance of genetic heterogeneity. Cold Spring Harbor Symposia on Quantitative Biology. 1955;20:25–32. doi: 10.1101/sqb.1955.020.01.005. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. New York, NY, USA: Columbia University Press; 1951. [Google Scholar]
- Dobzhansky TH. A review of some fundamental concepts and problems of population genetics. Cold Spring Harbor Symposium Quantitative Biology. 1955;20:1–15. doi: 10.1101/sqb.1955.020.01.003. [DOI] [PubMed] [Google Scholar]
- East EM. Inbreeding in corn. Reports of the Connecticut Agricultural Experiments Station. 1908;1907:419–428. [Google Scholar]
- Eckert CG, Manicacci D, Barrett SCH. Frequency-dependent selection on morph ratios in tristylous Lythrum salicaria (Lythracae) Heredity. 1996;77:581–588. [Google Scholar]
- Edwards A. Natural selection and the sex ratio: Fisher’s sources. American Naturalist. 1998;151:564–569. doi: 10.1086/286141. [DOI] [PubMed] [Google Scholar]
- Emerson S. The genetics of self-incompatibility in Oenothera organensis. Genetics. 1938;23:190–202. doi: 10.1093/genetics/23.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK, Lorence DH. Heterodichogamy of a novel type in Hernandia (Hernandiaceae) and its structural basis. International Journal of Plant Sciences. 2004;165:753–763. [Google Scholar]
- Ennos RA. Maintenance of genetic variation in plant populations. Evolutionary Biology. 1983;16:129–155. [Google Scholar]
- Epperson BK, Clegg MT. Spatial auto-correlation analysis of flower color polymorphisms within substructured populations of morning glory (Ipomoea purpurea) American Naturalist. 1986;128:840–858. [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to quantitative genetics. Essex, UK: Prentice Hall; 1996. [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford, UK: Clarendon Press; 1930. [Google Scholar]
- Fisher RA. The theoretical consequences of polyploid inheritance for the mid style form of Lythrum salicaria. Annals of Eugenics. 1941;11:31–38. [Google Scholar]
- Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- Ford EB. Ecological genetics. 3. London, UK: Chapman and Hall; 1971. [Google Scholar]
- Frank SA. The evolutionary dynamics of cytoplasmic male sterility. American Naturalist. 1989;133:345–376. [Google Scholar]
- Frey FM. Opposing natural selection from herbivores and pathogens may maintain floral-color polymorphism in Claytonia virginica (Portulaceceae) Evolution. 2004;58:2426–2437. doi: 10.1111/j.0014-3820.2004.tb00872.x. [DOI] [PubMed] [Google Scholar]
- Galloway LF, Fenster CB. Population differentiation in an annual legume: local adaptation. Evolution. 2000;54:1173–1181. doi: 10.1111/j.0014-3820.2000.tb00552.x. [DOI] [PubMed] [Google Scholar]
- Gardner CO. Estimates of genetic parameters in cross-fertilizing plants and their implications in plant breeding. National Academy of Sciences – National Research Council Publication. 1963;982:225–252. [Google Scholar]
- Gigord LDB, Macnair MR, Smithson A. Negative frequency-dependent selection maintains a dramatic flower color polymorphism in the rewardless orchid Dactylorhiza sambucina (L.) Soo. Proceedings of the National Academy of Sciences, USA. 2001;98:6253–6255. doi: 10.1073/pnas.111162598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH, Turelli M. Genotype–environment interactions and the maintenance of polygenic variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igić B. Species selection maintains self-incompatibility. Science. 2010;330:493–495. doi: 10.1126/science.1194513. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Holsinger KE. Maintenance of polygenic variation in spatially structured populations: roles for local mating and genetic redundancy. Evolution. 1992;46:412–429. doi: 10.1111/j.1558-5646.1992.tb02048.x. [DOI] [PubMed] [Google Scholar]
- Gomez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- Gouyon PH, Vichot F, van Damme JMM. Nuclear cytoplasmic male sterility: single-point equilibria versus limit cycles. American Naturalist. 1991;137:498–514. [Google Scholar]
- Grant PR, Grant BR. Predicting microevolutionary responses to directional selection on heritable variation. Evolution. 1995;49:241–251. doi: 10.1111/j.1558-5646.1995.tb02236.x. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. A mathematical theory of natural and artificial selection. Part V. Selection and mutation. Proceedings of the Cambridge Philosophical Society. 1927;23:838–844. [Google Scholar]
- Haldane JBS. Disease and evolution. Ricerca Scientifica. 1949;19:68–76. [Google Scholar]
- Haldane JBS. The statics of evolution. In: Huxley J, Hardy AC, Ford EB, editors. Evolution as a process. London, UK: Allen and Unwin; 1954. pp. 109–121. [Google Scholar]
- Haldane JBS, Jayakar SD. Polymorphism due to selection of varying direction. Journal of Genetics. 1963;58:237–242. [Google Scholar]
- Hedrick PW. Genetic variation in a heterogeneous environment. II. Temporal heterogeneity and directional selection. Genetics. 1976;84:145–157. doi: 10.1093/genetics/84.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: the age of genomics. Annual Review of Ecology, Evolution, and Systematics. 2006;37:67–93. [Google Scholar]
- Houliston GJ, Olson MS. Nonneutral evolution of organelle genes in Silene vulgaris. Genetics. 2006;174:1983–1994. doi: 10.1534/genetics.106.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. Genetics and the understanding of selection. Nature Reviews Genetics. 2009;10:83–93. doi: 10.1038/nrg2506. [DOI] [PubMed] [Google Scholar]
- Ioerger TR, Clark AG, Kao T-H. Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proceedings of the National Academy of Sciences, USA. 1990;87:9732–9735. doi: 10.1073/pnas.87.24.9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. A classification of selection-migration structures and conditions for protected polymorphism. In: Hecht MK, Wallace B, Prance GT, editors. Evolutionary Biology. New York, NY, USA: Plenum; 1982. pp. 61–204. [Google Scholar]
- Kelly CA. Spatial and temporal variation in selection on correlated life-history traits and plant size in Chamaecrista fasciculata. Evolution. 1992;46:1658–1673. doi: 10.1111/j.1558-5646.1992.tb01160.x. [DOI] [PubMed] [Google Scholar]
- Kelly JK. Geographical variation in selection, from phenotypes to molecules. American Naturalist. 2006;167:481–495. doi: 10.1086/501167. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. New York, NY, USA: Cambridge University Press; 1983. [Google Scholar]
- Kondrashov AS, Yampolsky LY. High genetic variability under the balance between symmetric mutation and fluctuating stabilizing selection. Genetical Research. 1996;68:157–164. [Google Scholar]
- Koskella B. Phage-mediated selection on microbiota of a long-lived host. Current Biology. 2013;23:1256–1260. doi: 10.1016/j.cub.2013.05.038. [DOI] [PubMed] [Google Scholar]
- Kruckeberg AR, Walker RB, Leviton AE, editors. Genecology and ecogeographic races. San Francisco, CA, USA: Pacific division AAAS; 1995. [Google Scholar]
- Lahiani E, Dufaÿ M, Castric V, Le Cadre S, Charlesworth D, Van Rossum F, Touzet P. Disentangling the effects of mating systems and mutation rates on cytoplasmic diversity in gynodioecious Silene nutans and dioecious S. otites. Heredity. 2013;11:157–164. doi: 10.1038/hdy.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L, Burdon JJ, Dodds PN, Thrall PH. Spatial variation in disease resistance: from molecules to metapopulations. Journal of Ecology. 2011;99:96–112. doi: 10.1111/j.1365-2745.2010.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latter BDH. Natural selection for an intermediate optimum. Australian Journal of Biological Science. 1960;13:30–35. [Google Scholar]
- Leimu R, Fischer M. A meta-analysis of local adaptation in plants. PLoS ONE. 2008;3:e4010. doi: 10.1371/journal.pone.0004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H. Genetic equilibrium when more than one ecological niche is available. American Naturalist. 1953;87:331–333. [Google Scholar]
- Lewis D. Male sterility in natural populations of hermaphrodite plants. New Phytologist. 1941;40:56–63. [Google Scholar]
- Lewontin RC. The genetic basis of evolutionary change. New York, NY, USA: Columbia University Press; 1974. [Google Scholar]
- Lloyd DG. The maintenance of gynodioecy and androdioecy in angiosperms. Genetica. 1975;45:325–339. [Google Scholar]
- Lloyd DG, Webb CJ. The avoidance of interference between the presentation of pollen and stigmas in angiosperms. I. Dichogamy. New Zealand Journal of Botany. 1986;24:135–162. [Google Scholar]
- Lynch M, Walsh B. Genetics and analysis of quantitative characters. Sunderland, MA, USA: Sinauer Associates; 1998. [Google Scholar]
- Maynard Smith J. Evolutionary genetics. Oxford, UK: Oxford University Press; 1989. [Google Scholar]
- McCauley DE. Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytologist. 2013 doi: 10.1111/nph.12431. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nature Reviews Genetics. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Mokkonen M, Kokko H, Koskela E, Lehtonen J, Mappes T, Martiskainen H, Mills SC. Negative frequency-dependent selection of sexually antagonistic alleles in Myodes glareolus. Science. 2011;334:972–974. doi: 10.1126/science.1208708. [DOI] [PubMed] [Google Scholar]
- Moll RH, Lindsey MF, Robinson HF. Estimates of genetic variances and level of dominance in Maize. Genetics. 1964;49:411–423. doi: 10.1093/genetics/49.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin, Germany: Springer; 2001. [Google Scholar]
- Renner SS. How common is heterodichogamy? Trends in Ecology and Evolution. 2001;16:595–597. [Google Scholar]
- Schemske DW. Population structure and local selection in Impatiens pallida (Balsaminaceae), a selfing annual. Evolution. 1984;38:817–832. doi: 10.1111/j.1558-5646.1984.tb00354.x. [DOI] [PubMed] [Google Scholar]
- Schiestl FP. On the success of a swindle: pollination by deception in orchids. Naturwissenschaften. 2005;92:255–264. doi: 10.1007/s00114-005-0636-y. [DOI] [PubMed] [Google Scholar]
- Schmitt J, Gamble SE. The effect of distance from parental site on offspring performance and inbreeding depression in Impatiens capensis: a test of the local adaptation hypothesis. Evolution. 1990;44:2022–2030. doi: 10.1111/j.1558-5646.1990.tb04308.x. [DOI] [PubMed] [Google Scholar]
- Shull GH. The composition of a field of Maize. Reports of the American Breeders Association. 1908;4:296–301. [Google Scholar]
- Shykoff JA, Kolokotronis S-O, Collin CL, López-Villavicencio M. Effects of male sterility on reproductive traits in gynodioecious plants: a meta-analysis. Oecologia. 2003;135:1–9. doi: 10.1007/s00442-002-1133-z. [DOI] [PubMed] [Google Scholar]
- Silander JA. Microevolution and clone structure in Spartina patens. Science. 1979;203:658–660. doi: 10.1126/science.203.4381.658. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Spatial patterns in the distribution of polygenic characters. Journal of Theoretical Biology. 1978;70:213–228. doi: 10.1016/0022-5193(78)90348-x. [DOI] [PubMed] [Google Scholar]
- Sprengel CK. Das entdeckte Geheimniss in der Natur im Bau und in der Befruchtung der Blumen. Translation. In: Lloyd DG, Barrett SCH, editors. Floral biology: studies of floral evolution in animal-pollinated plants. New York, NY, USA: Chapman & Hall; 1793. pp. 3–43. [Google Scholar]
- Städler T, Delph LF. Ancient mitochondrial haplotypes and evidence for intragenic recombination in a gynodioecious plant. Proceedings of the National Academy of Sciences, USA. 2002;99:11 730–11 735. doi: 10.1073/pnas.182267799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Stewart SC, Schoen DJ. Pattern of phenotypic viability and fecundity selection in a natural population of Impatiens pallida. Evolution. 1987;41:1290–1301. doi: 10.1111/j.1558-5646.1987.tb02467.x. [DOI] [PubMed] [Google Scholar]
- Stratton DA, Bennington CC. Fine-grained spatial and temporal variation in selection does not maintain genetic variation in Erigeron annuus. Evolution. 1998;52:678–691. doi: 10.1111/j.1558-5646.1998.tb03693.x. [DOI] [PubMed] [Google Scholar]
- Subramaniam B, Rausher MD. Balancing selection on a floral polymorphism. Evolution. 2000;54:691–695. doi: 10.1111/j.0014-3820.2000.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Tack AJM, Thrall PH, Barrett LG, Burdon JJ, Laine A-L. Variation in infectivity and aggressiveness in space and time in wild host-pathogen systems: causes and consequences. Journal of Evolutionary Biology. 2012;25:1918–1936. doi: 10.1111/j.1420-9101.2012.02588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall PH, Laine A-L, Ravensdale M, Nemri A, Dodds PN, Barrett LG, Burdon JJ. Rapid genetic changes underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecology Letters. 2012;15:425–435. doi: 10.1111/j.1461-0248.2012.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Araki H, Stahl EA, Bergelson J, Kreitman M. Signature of balancing selection in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2002;99:11525–11530. doi: 10.1073/pnas.172203599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsor SJ, Kalisz S, Fisher J, Holtsford TP. A life-history based study of population genetic structure: seed bank to adults in Plantago lanceolata. Evolution. 1993;47:833–843. doi: 10.1111/j.1558-5646.1993.tb01237.x. [DOI] [PubMed] [Google Scholar]
- Touzet P, Delph LF. The effect of breeding system on polymorphism in mitochondrial genes of Silene. Genetics. 2009;181:631–644. doi: 10.1534/genetics.108.092411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter MV, Spencer HG. Frequency-dependent selection and the maintenance of genetic variation: exploring the parameter space of the multiallelic pairwise interaction model. Genetics. 2007;176:1729–1740. doi: 10.1534/genetics.107.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M. Heritable genetic variation via mutation-selection balance: Lerch’s zeta meets the abdominal bristle. Theoretical Population Biology. 1984;25:138–193. doi: 10.1016/0040-5809(84)90017-0. [DOI] [PubMed] [Google Scholar]
- Turkington R, Harper JL. The growth, distribution and neighbour relationships of Trifolium repens in a permanent pasture: IV. Fine-scale biotic differentiation. Journal of Ecology. 1979;67:245–254. [Google Scholar]
- Wang X, Zhang P, Du Q, He H, Zhao L, Ren Y, Endress PK. Heterodichogamy in Kingdonia (Circaeasteraceae, Ranunculales) Annals of Botany. 2012;109:1125–1132. doi: 10.1093/aob/mcs041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waser NM, Price MV. Reciprocal transplant experiments with Delphinium nelsonii (Ranunculaceae): evidence for local adaptation. American Journal of Botany. 1985;72:1726–1732. [Google Scholar]
- Wright S. Evolution in mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations, the theory of gene frequencies. Vol. 2. Chicago, IL, USA: University of Chicago Press; 1969. [Google Scholar]


