Abstract
Multiple myeloma (MM) represents the malignant proliferation of terminally differentiated B cells, which, in many cases, is associated with the maintenance of high levels of the oncoprotein c-MYC. Overexpression of the histone methyltransferase MMSET (WHSC1/NSD2), due to t(4;14) chromosomal translocation, promotes the proliferation of MM cells along with global changes in chromatin; nevertheless, the precise mechanisms by which MMSET stimulates neoplasia remain incompletely understood. We found that MMSET enhances the proliferation of MM cells by stimulating the expression of c-MYC at the post-transcriptional level. A microRNA (miRNA) profiling experiment in t(4;14) MM cells identified miR-126* as an MMSET-regulated miRNA predicted to target c-MYC mRNA. We show that miR-126* specifically targets the 3′-untranslated region (3′-UTR) of c-MYC, inhibiting its translation and leading to decreased c-MYC protein levels. Moreover, the expression of this miRNA was sufficient to decrease the proliferation rate of t(4;14) MM cells. Chromatin immunoprecipitation analysis showed that MMSET binds to the miR-126* promoter along with the KAP1 corepressor and histone deacetylases, and is associated with heterochromatic modifications, characterized by increased trimethylation of H3K9 and decreased H3 acetylation, leading to miR-126* repression. Collectively, this study shows a novel mechanism that leads to increased c-MYC levels and enhanced proliferation of t(4;14) MM, and potentially other cancers with high MMSET expression.
Keywords: MMSET, t(4;14), multiple myeloma, proliferation, c-MYC, miR-126*
INTRODUCTION
Over the past decade, the molecular pathogenesis of multiple myeloma (MM) has become clearer with the identification of recurrent chromosomal translocations in ~40% of the cases. These anomalies are due to aberrant switch recombination that leads to the overexpression of a number of putative or known oncogenes such as MAF, MAFB and WHSC1/MMSET.1,2 More recently, whole genome sequencing in MM identified new recurrent point mutations in genes encoding coagulation cascade proteins, histone-modifying enzymes and RNA processing proteins.3 The underlying paradox of MM that may be explained by some of these genetic anomalies is the continued proliferation and self-renewal of a highly specialized cell. Normal plasma cell differentiation requires the BLIMP1/PRDM1 transcription factor, which stimulates proper differentiation of the postgerminal center B cell and represses the expression of c-MYC, a master regulator of cell growth.4 In contrast, malignant plasma cells continue to express c-MYC and enter the cell cycle while maintaining the specialized machinery required for production of high levels of immunoglobulin.
The MMSET gene is overexpressed in ~15% of MM cases because of the t(4;14) chromosomal translocation, which links the MMSET gene to the immunoglobulin promoter/enhancer. In most cases of t(4;14)-positive myeloma, the upregulation of MMSET is accompanied by the overexpression of FGFR3. Activation of FGFR3-mediated signaling would be expected to lead to activation of the mitogen-activated protein kinase cascade, serum response factors and ETS family of transcription factors, and eventually, stimulation of c-MYC transcription. However, in ~30% of cases of t(4;14)-associated myeloma, only MMSET is overexpressed.5,6 Given this background, it is reasonable to hypothesize that MMSET might also stimulate c-MYC expression. MMSET is a histone methyltransferase (HMT) that, in vitro, can methylate both histone H3 and histone H4, and is associated with transcriptional cofactors.7–11 In t(4;14) MM cells, MMSET overexpression correlates with elevated levels of H3K36 dimethylation (H3K36me2) and depressed levels of H3K27 trimethylation (H3K27me3) across the genome.11,12 These methylation patterns are associated with altered chromatin structure, gene expression profiles and cell growth. Nevertheless, the exact oncogenic mechanisms and genetic targets underlying MMSET activity are not well understood.
In efforts to identify the mechanisms by which MMSET enhances cell growth, we found that MMSET overexpression was associated with increased c-MYC protein, but not mRNA expression. This was related to the ability of MMSET to repress the expression of miR-126*, a microRNA (miRNA) that targets c-MYC. MMSET-mediated repression of this miRNA was associated with the recruitment of the KAP1 transcriptional corepressor, and a repressive histone modification, trimethylation of lysine 9 on histone H3 (H3K9me3). Collectively, these data point to a new mechanism by which c-MYC expression is elevated in t(4;14) MM, and start to elucidate the oncogenic mechanisms of MMSET in this disease.
MATERIALS AND METHODS
Cell culture
MM cell lines including the t(4;14)-positive cell lines, KMS11, KMS28BM, KMS26, H929, LP1, KMS34, OPM2, KMS18 and UTCM2, and the t(4;14)-negative cell lines, RPMI-8226, KMS12, FR4, XG6, U266, L363 and MM.M1 (gift from Michael Kuehl, National Cancer Institute, Bethesda, MD, USA), were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). The 293T cells (ATCC, Manassas, VA, USA) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS.
Loss- and gain-of-function systems
KMS11 cells were engineered to express an MMSET short hairpin RNA (shRNA) in a doxycycline (dox)-inducible manner.11,13 To obtain a KAP1 knockdown system, KMS11 cells were infected with a retroviral construct generated by cloning complementarily annealed oligonucleotides encoding a KAP1 shRNA sequence, into pSiren-ZsGreen (Clontech, Mountain View, CA, USA). The miR-126* expression vector was purchased from System Biosciences (Mountain View, CA, USA), and a mutant version of the construct was generated using the QuikChange Lightning Site-Directed Mutagenesis kit (Stratagene, Santa Clara, CA, USA). The c-MYC expression vector was described previously.14 The tagged MMSET and nuclear green fluorescent protein (GFP) constructs were prepared using pNTAP vector (Stratagene) (see Supplementary Data). Primers can be found in Supplementary Table 1.
Virus packaging and infections
Lentiviruses to overexpress miR-126* and the c-MYC 3′-untranslated region (UTR) luciferase reporter were generated by transfection of 293T cells with the plasmids described above, in addition to the packaging vectors psPAX2 and pMD2.G (Addgene, Cambridge, MA, USA),15 using Fugene 6 (Roche, Indianapolis, IN, USA). For infection of KMS11 cells, 1 ml of viral supernatant was added to 1 million cells in 1 ml of media, along with 6 µg/ml polybrene (Millipore, Billerica, MA, USA). Retroviruses harboring the KAP1 shRNA construct were produced as described above, and used to infect KMS11 cells in the presence of 4 µg/ml polybrene.
Proliferation assays
A total of 4000 cells were seeded in 96-well plates and the viability was assayed using the ATPlite luminescence assay system (PerkinElmer, Waltham, MA, USA). For miR-126*, the assays were performed at day 9 after infection of KMS11 cells with empty vector, wild-type or mutated miR-126*. For KAP1 knockdown, assays were performed at day 7 after infection with a KAP1 shRNA or control.
Immunoblotting and immunoprecipitation
Total cell lysates were prepared from cells resuspended in 0.1% NP-40 lysis buffer (Stratagene) supplemented with protease inhibitors (Roche), and subjected to three rounds of freezing/thawing. Nuclear fractions and immunoprecipitation were performed using the Nuclear Complex Co-IP Kit (Active Motif, Carlsbad, CA, USA). Proteins were separated using NuPAGE Bis-Tris Gel (Invitrogen, Grand Island, NY, USA), blotted with appropriate antibodies and detected using enhanced chemiluminescence (GE Healthcare, Piscataway, NJ, USA). Antibodies are listed in Supplementary Data.
Real-time PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and complementary DNA was generated using iScript (Bio-Rad, Hercules, CA, USA). Gene amplification was performed on the Roche Lightcycler 480 using the Quantitect SYBR Green PCR Mix (Qiagen). To determine miRNA expression levels, total RNA was isolated using the miRNeasy Mini Kit (Qiagen). The miRCURY LNA Universal RT microRNA PCR kit (Exiqon, Vedbaek, Denmark) was used for reverse transcription and miRNA amplification. Expression was calculated using the ΔΔCT method. GAPDH and U6 snRNA were used as internal controls. Primers for miR-126* (cat. 204584) and U6 snRNA (cat. 203907) were purchased from Exiqon. Other primer sequences are included in Supplementary Table 1.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) was performed as described previously.16 Ten million cells and 5 µg of each antibody (10 µl for H3-Ac) were utilized per ChIP. The same antibodies used for immunoblotting were utilized, as well as mouse IgG (ab18447; Abcam, Cambridge, MA, USA) and rabbit IgG (ab37415; Abcam) to serve as negative controls. PCR was performed using primers complimentary to the transcriptional start site (TSS) of miR-126/miR-126* or a region 10 kb upstream (see Supplementary Data).
Luciferase assays
For reporter assays, a c-MYC 3′-UTR vector was purchased from System Biosciences. A mutant c-MYC 3′-UTR construct was made using the QuikChange Lightning Site-Directed Mutagenesis kit (Stratagene). c-MYC 3′-UTR and miR-126* vectors were transduced into 293T cells using Fugene 6 (Roche). Briefly, 140 000 cells were transfected in 12-well plates with the indicated plasmids totaling 200 ng of DNA, and assayed after 3 days using the Luciferase Reporter Assay System (Promega, Madison, WI, USA) in a fluorescence plate reader (FLUOstar Optima, BMG Labtech, Cary, NC, USA).
Antagonizing miR-126* in MMSET-depleted KMS11 cells
KMS11 cells harboring an inducible MMSET shRNA were grown in the presence of dox for 7 days. The MMSET-depleted cells were transfected twice, on days 1 and 3, with 40 nM miR-126* inhibitor and control (Ambion, Grand Island, NY, USA), using HiPerFect (Qiagen).
RESULTS
MMSET stimulates cell growth and enhances c-MYC protein expression in t(4;14)+ MM cells
We previously reported that the t(4;14)-positive cell line KMS11 depends on its elevated MMSET expression to maintain proliferation, which markedly decreases upon dox-dependent shRNA-mediated MMSET knockdown11 (Supplementary Figure 1A). Cessation of MMSET knockdown by dox removal allowed the growth of KMS11 cells to resume within 3 days (Supplementary Figure 1B). These results, in accordance with previous data,11 indicate that loss of MMSET expression primarily leads to growth arrest rather than apoptosis.
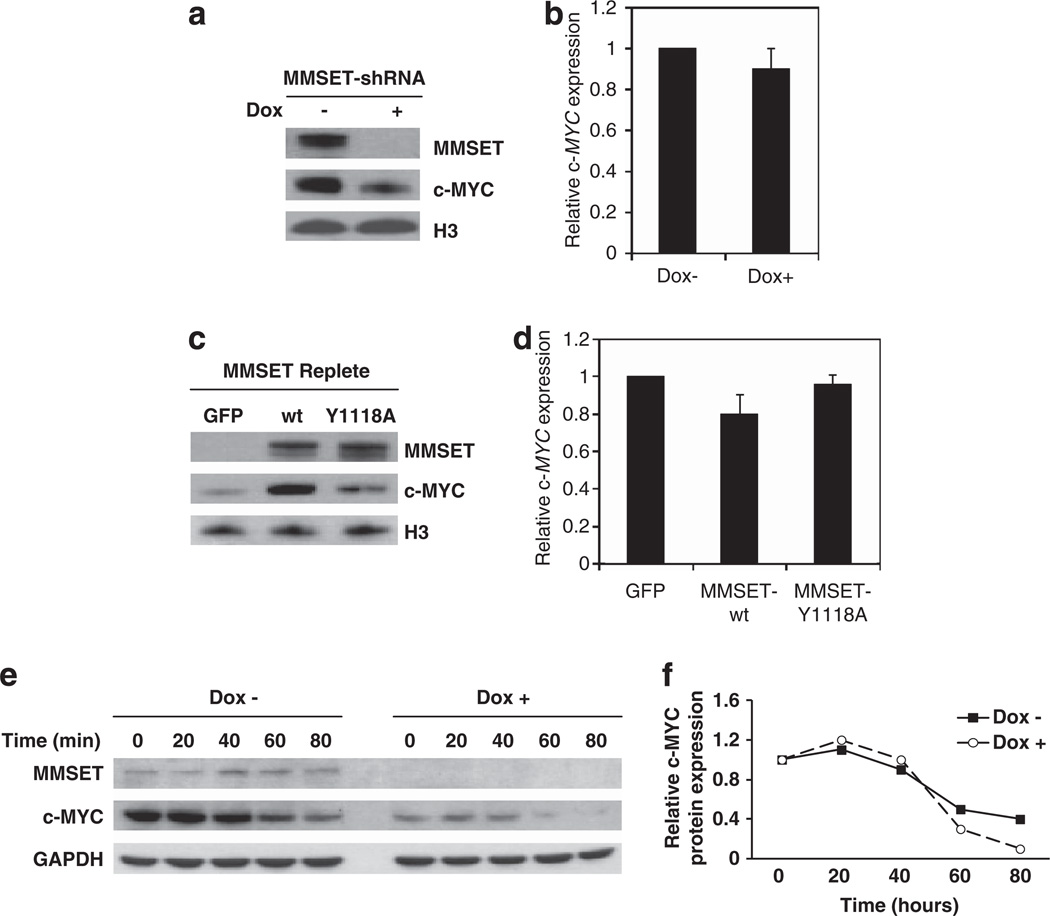
The formation of plasma cells from memory B cells requires upregulation of the BLIMP1/PRDM1 transcription factor that, in turn, represses c-MYC. MM is characterized by continued high expression of c-MYC in the presence of elevated BLIMP1 levels.13 Considering the pivotal role of c-MYC in the proliferation of MM cells, we hypothesized that it could mediate MMSET promotion of cell growth. Therefore, we first determined whether c-MYC expression was affected by MMSET. Depletion of MMSET in KMS11 cells was associated with decreased c-MYC protein levels (Figure 1a and Supplementary Figure 2); however, no significant change in c-MYC mRNA expression was detected (Figure 1b), indicating that MMSET affected c-MYC at the post-transcriptional level. Accordingly, overexpression of MMSET in the t(4;14)-negative RPMI-8226 cell line increased c-MYC at protein (Figure 1c) but not mRNA level (Figure 1d). The mutant MMSET-Y1118A, a protein defective for HMT activity,11 only mildly stimulated c-MYC protein expression, suggesting that the HMT activity of MMSET is an important factor in its ability to affect c-MYC. We next analyzed whether MMSET could affect the stability of c-MYC protein. KMS11 cells were treated with cycloheximide to abolish new mRNA translation, and c-MYC protein levels were determined by immunoblot at different time points after the addition of the drug, in the presence or absence of MMSET (Figure 1e). Although in the absence of MMSET the basal level of c-MYC was several fold lower than in the presence of MMSET, the kinetics of c-MYC degradation were similar to that observed in cells with high MMSET levels (Figure 1f). The decrease in c-MYC protein levels in the absence of MMSET without change in its half-life suggested that MMSET did not affect the stability of the protein, but rather promoted the translation of c-MYC mRNA.
Figure 1.
MMSET stimulates c-MYC protein but not mRNA expression in MM cells. (a) MMSET was depleted in t(4;14)-positive KMS11 cells using a dox-inducible shRNA. After nuclear fractionation, MMSET and c-MYC protein levels were assayed by immunoblot with histone H3 as a loading control. (b) Relative c-MYC mRNA levels of cells described in (a) were measured by real-time PCR and normalized to GAPDH mRNA levels. Values are represented relative to those observed in dox-negative cells (mean ± s.d. from three independent experiments). (c) The t(4;14)-negative RPMI-8226 myeloma cells were stably transduced with a retrovirus harboring wild-type or HMT inactive point mutant (Y1118A) MMSET, or a control retrovirus harboring GFP. After nuclear fractionation, MMSET and c-MYC protein levels were assayed by immunoblot with histone H3 as a loading control. (d) Relative c-MYC mRNA levels of cells described in (c) were measured by qPCR and normalized to GAPDH mRNA level. Values are represented relative to those obtained in cells transduced with the empty vector (mean ± s.d. from three experiments). (e) KMS11 cells cultured in the absence or presence of dox were treated with 10 mg/ml cycloheximide for the indicated times and lysates were immunoblotted for MMSET and c-MYC. GAPDH represents a loading control. (f) c-MYC protein levels obtained in (e) were quantified using ImageJ software (NIH, Bethesda, MD, USA), and represented as relative to those obtained at time 0.
MMSET represses miR-126* and leads to enhanced c-MYC protein levels in t(4;14)+ MM cells
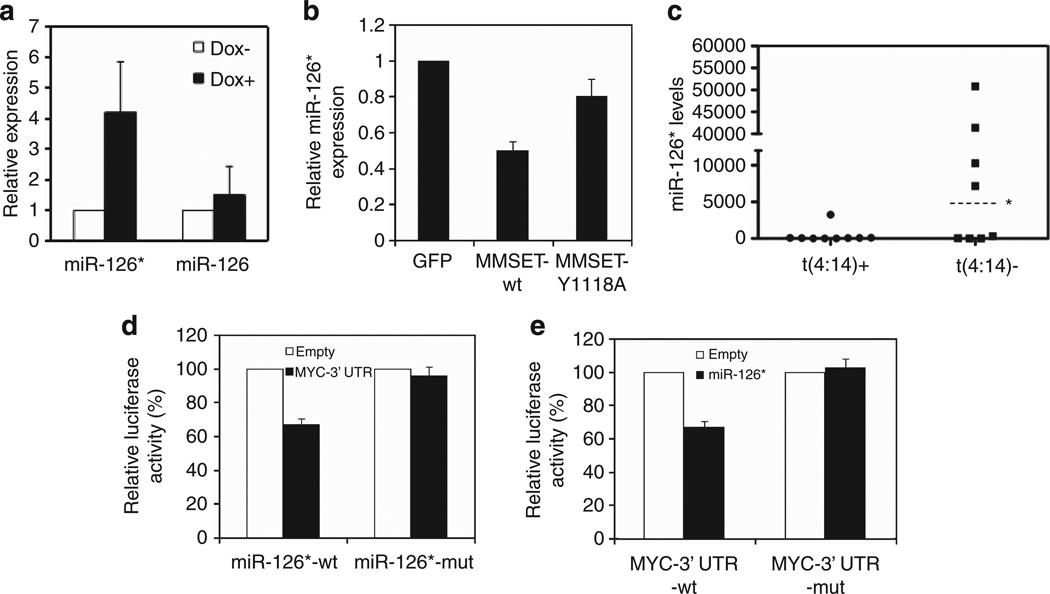
Among the several mechanisms that regulate mRNA translation, our study focused on the possibility that MMSET controlled c-MYC translation through the regulation of miRNAs. A miRNA microarray profiling experiment performed in the KMS11 loss-of-function system identified multiple miRNAs whose expression changed at least two fold in response to MMSET knockdown (Supplementary Figure 3A). Among the miRNAs repressed by MMSET, miR-126* and miR-1267 were predicted to target c-MYC (www.microrna.org). However, only miR-126* was validated as regulated in response to MMSET knockdown in KMS11 cells (Figure 2a). The expression of miR-126, the other miRNA generated from the same pre-miRNA, was modestly affected upon MMSET manipulation (Figure 2a). In a complementary experiment, ectopic expression of wild-type MMSET in t(4;14)-negative RPMI-8226 myeloma cells resulted in a 50% decrease in miR-126* levels. The mutant MMSET-Y1118A yielded a more modest 20% decrease in miR-126* expression (Figure 2b), in agreement with its less potent stimulation of c-MYC protein levels (Figure 1c). A panel of MM cell lines was then analyzed to determine whether basal levels of MMSET in these cells controlled miR-126* expression. Half of the t(4;14)-negative cell lines expressed high levels of miR-126*, whereas most of the t(4;14)-positive cell lines showed very low levels of the miRNA, consistent with the notion that MMSET could inhibit miR-126* expression in these cells (Figure 2c).
Figure 2.
MMSET represses miR-126*, a miRNA that targets c-MYC 3′-UTR. (a) MMSET was depleted in KMS11 cells using a dox-inducible shRNA for 7 days, and miR-126* and miR-126 levels were measured by quantitative real-time PCR (qPCR). U6 snRNA was used as a normalization control. Values are represented relative to that observed in dox-negative cells (mean ± s.d. from three independent experiments). (b) Relative miR-126* levels were measured by qPCR in t(4;14)-negative RPMI-8226 cells transduced with GFP, wild-type or mutant MMSET retroviruses. U6 snRNA was used as a normalization control. Values are represented relative to that observed in cells harboring the empty vector (mean ± s.d. from three independent experiments). (c) Relative miR-126* levels were measured by qPCR in 9 t(4;14)-positive and 8 t(4;14)-negative MM cell lines. U6 snRNA was used as a normalization control. The relative expression level of each cell line is calculated based on the lowest expression value. The median value (*) of the positive and negative cell lines are 64 and 3733, respectively. (d) A luciferase reporter gene linked to the c-MYC 3′-UTR was coexpressed in 293T cells with an expression vector for the wild-type or a mutant form of miR-126*. Luciferase activity is presented as percentage of the activity generated by the reporter gene when coexpressed with an empty vector. (e) Wild-type miR-126* was coexpressed in 293T cells with a luciferase reporter linked to the wild-type or a mutant c-MYC 3′-UTR. Luciferase activity is presented as percentage of the activity generated by the reporter gene when coexpressed with an empty vector.
To validate c-MYC as a real, and not only predicted, target of miR-126*, we evaluated its ability to alter the expression of a luciferase reporter containing the 3′-UTR of c-MYC. Transient transduction of 293T cells with the reporter construct along with a miR-126* expression vector showed a 40% decrease in luciferase activity (Figure 2d). Two-nucleotide mutations within the seed sequence of miR-126* or within the target sequence of the 3′-UTR of c-MYC (Supplementary Figure 3B) abolished the repression of the luciferase activity (Figures 2d and e), confirming the specificity of the interaction.
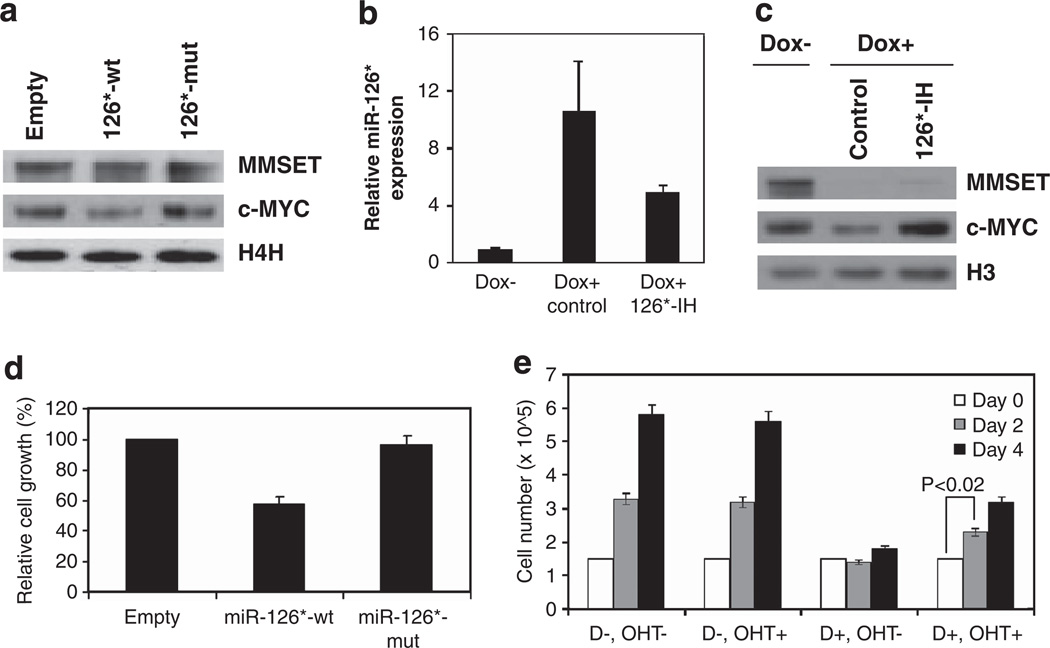
We next determined whether miR-126* targeting of the c-MYC 3′-UTR would affect its translation, leading to decreased c-MYC protein levels. KMS11 cells were infected with a lentiviral vector expressing wild-type or mutant miR-126*, and c-MYC protein levels were detected by immunoblot. Only wild-type but not mutant miR-126* decreased c-MYC protein levels (Figure 3a), in agreement with the results observed in the luciferase assays. To further confirm the ability of miR-126* to target c-MYC and abolish its translation, an antagomir specifically directed against miR-126* (miR-126* inhibitor) was transduced into KMS11 cells engineered to express MMSET shRNA (Figure 3b). The loss of c-MYC expression, associated with MMSET depletion and upregulation of miR-126*, was completely restored by introduction of the antagomir (Figure 3c). This experiment not only confirmed c-MYC as a target of miR-126*, but also indicated miR-126* as an important mediator of MMSET regulation of c-MYC.
Figure 3.
miR-126* decreases c-MYC protein levels and reduces the proliferation rate of MM cells. (a) KMS11 cells were stably transfected with an expression vector harboring wild-type or mutant miR-126*, or an empty vector. MMSET and c-MYC expression levels were assayed by immunoblot. Histone H4 represents a loading control. (b) KMS11 cells harboring an inducible MMSET shRNA were cultured in the presence of dox (dox+) for 7 days, and further grown for 6 days after transfection with a miR-126* inhibitor (126*-IH) or inhibitor control. KMS11 cells harboring an inducible MMSET shRNA were grown in parallel in the absence of dox (dox−). Relative miR-126* levels were determined by real-time PCR, normalized to U6 snRNA and represented as relative to that observed in cells grown in the absence of dox (mean ± s.d. of three independent experiments). (c) Nuclear extracts from cells described in (b) were subjected to immunoblot using an anti-MMSET or anti-c-MYC antibody. Histone H3 represents a loading control. (d) Growth of KMS11 cells transduced with wild-type or mutant miR-126*, or an empty vector, was determined by ATP production after 9 days of growth. The viable cell mass is displayed relative to cells transduced with the empty vector. (e) KMS11 cells harboring both an inducible MMSET shRNA and a c-MYC-ER expression vector were cultured in the absence or presence of dox (D− or D+) for 7 days, and further grown in the absence or presence of 200 nm 4-hydroxytamoxifen (OHT− or OHT+) up to 4 days. Live cells were counted by Trypan blue exclusion at indicated times. Values represent mean ± s.d. from three experiments.
We next analyzed whether miR-126* expression could recapitulate the effect of MMSET knockdown on cell growth. KMS11 cells infected with a lentiviral vector expressing wild-type miR-126* showed a 40% growth inhibition when compared with cells harboring a control vector, whereas the miR-126* mutant had no effect (Figure 3d). The effects of wild-type or mutant miR-126* on proliferation paralleled the c-MYC protein levels previously observed by immunoblot (Figure 3a). Finally, we determined whether enforced expression of a miRNA-resistant form of c-MYC could rescue the decrease in proliferation detected upon MMSET knockdown and miR-126* upregulation. KMS11 cells were infected with a retrovirus containing a conditional form of c-MYC (MYC-ER)18 that becomes translocated to the nucleus and activated upon treatment with 4-hydroxytamoxifen. c-MYC-ER is devoid of 3′-UTR of c-MYC and hence is unable to be affected by miR-126*. When MMSET levels were high, the myeloma cells grew with a doubling time of ~2 days, and the induction of exogenous c-MYC activity with 4-hydroxytamoxifen had no effect on cell growth (Figure 3e). In contrast, MMSET knockdown led to a cessation of cell growth that, upon induction of MYC-ER, was partially restored to a doubling time of 4 days (Figure 3e). Collectively, these data suggest that promotion of cell growth by MMSET is, at least, partially mediated by c-MYC overexpression driven by miRNA repression.
The KAP1 corepressor supports MMSET-mediated miR-126* repression
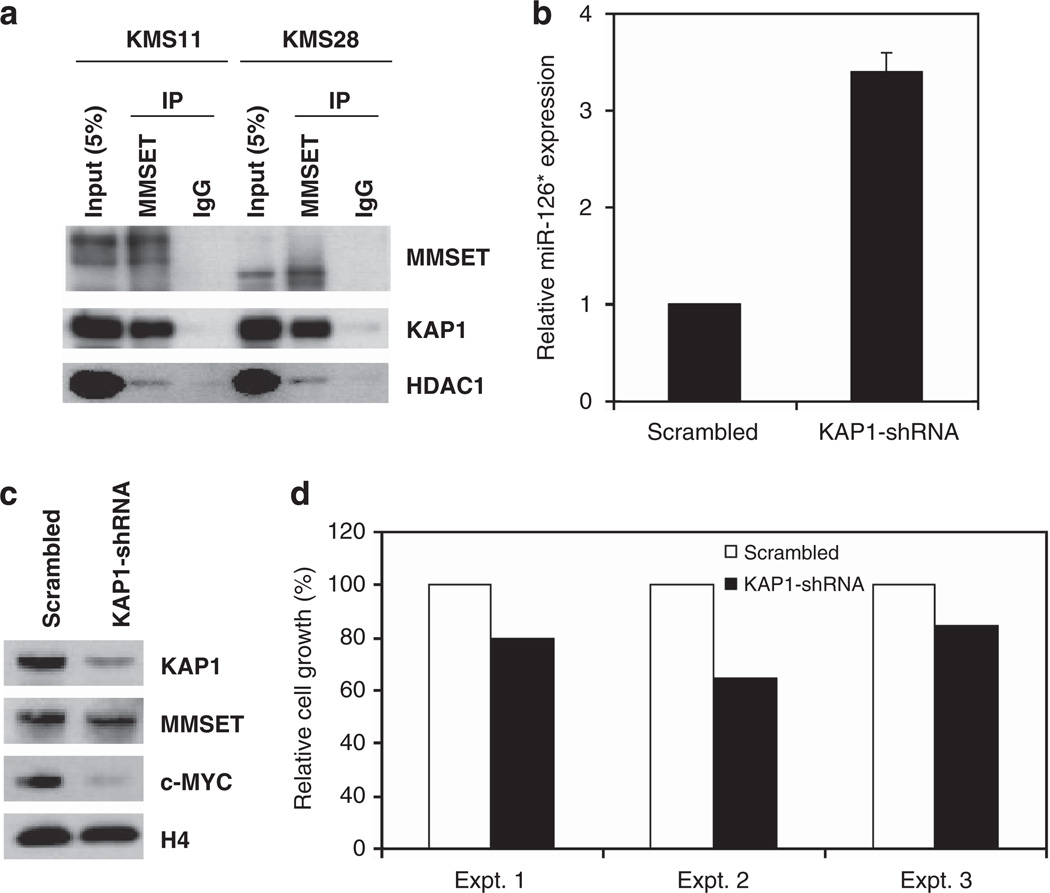
Although MMSET mediates methylation of H3K36, a histone modification associated with gene activation, it is clear from gene expression profiling and the ability of MMSET to interact with corepressors that the protein may also have a role in gene repression.7,8,11,19 To further investigate MMSET-mediated repression that could lead to the inactivation of miR-126/126* locus, we identified MMSET partner proteins. For this purpose, MMSET linked to an N-terminal streptavidin/calmodulin tag was expressed in 293 cells and affinity-purified, and associated proteins were identified by mass spectrometry (Supplementary Figure 4). A streptavidin/calmodulin-tagged nuclear GFP protein was purified in parallel as a control, in order to identify nonspecific binding proteins. Duplicate experiments identified 19 candidate proteins that associated with MMSET but not with tagged GFP (Supplementary Figure 4C). Among them, KAP1 (TRIM28), a previously identified corepressor that interacts with KRAB zinc-finger transcriptional repressors and the H3K9 methyltransferase SETDB1,20 was selected for further analysis. The interaction between MMSET and KAP1 was confirmed by co-immuno-precipitation in the t(4:14)-positive KMS11 and KMS28 MM cell lines, with HDAC1/MMSET interaction serving as a positive control7,19 (Figure 4a). MMSET was detected as a smaller species in KMS28 cells, because of the linkage of the immunoglobulin locus to a more 3′ location of the MMSET gene, which deletes part of the N-terminus of the protein.1 We next determined whether KAP1 was critical for the ability of MMSET to repress miR-126* and promote c-MYC protein levels. KMS11 cells transduced with an shRNA directed against KAP1 showed an almost 3.5-fold increase in miR-126* expression (Figure 4b), and a large decrease in c-MYC protein levels (Figure 4c). This effect was accompanied by a modest decrease in cell proliferation (Figure 4d) and no appreciable change in the level of MMSET (Figure 4c). These results indicated that KAP1 is a key factor in the repression of miR-126* expression and could mediate MMSET-dependent regulation of this miRNA.
Figure 4.
KAP1 interacts with MMSET and mediates miR-126* repression. (a) KAP1 interacts with MMSET. Nuclear extracts of KMS11 or KMS28 cells were subjected to immunoprecipitation (IP) with anti-MMSET antibody and immunoblotted with anti-MMSET, anti-KAP1 or anti-HDAC1 antibodies. IgG antibody was used as a control. (b) Relative miR-126* expression was determined by real-time PCR in KMS11 cells harboring a control or a KAP1 shRNA. U6 snRNA was used as a normalization control. Values are represented relative to that observed in cells transduced with the scramble shRNA (mean ± s.d. from three experiments). (c) Nuclear extracts from cells described in (b) were immunoblotted with the indicated antibodies. Histone H4 represents a loading control. (d) KMS11 cells were transduced with a KAP1 or scrambled shRNA and after 7 days cell mass was quantified by bioluminescent detection of cellular ATP. The viable cell mass is displayed relatively to control cells harboring the scramble shRNA.
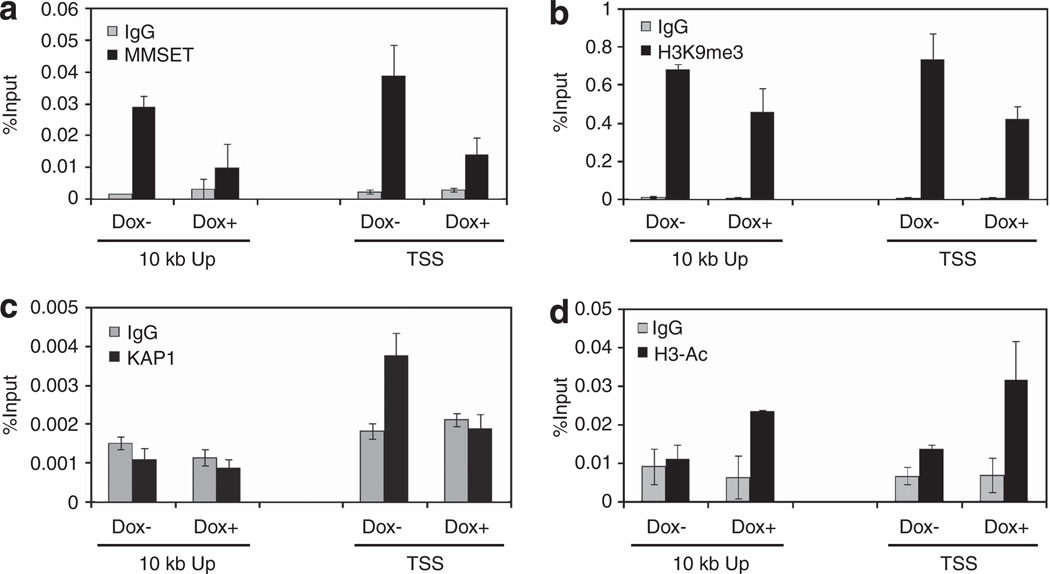
To determine whether KAP1 and MMSET might have a direct role in the repression of miR-126* expression, ChIP experiments were performed. In KMS11 cells, endogenous MMSET binding was detected at sequences near the predicted TSS of pre-miR-126*, as well as 10 kb upstream. shRNA-mediated knockdown of MMSET led to the loss of its binding at these sites (Figure 5a). The presence of MMSET at the miR-126* promoter was associated with the presence of H3K9me3, a mark enriched in inactive regions, which decreased upon MMSET depletion (Figure 5b). KAP1 was reported to bind to chromatin enriched in H3K9me3 sites21 and, accordingly, ChIP assays also showed that KAP1 was bound at the TSS of pre-miR-126* enriched for H3K9me3 (Figure 5c). Moreover, MMSET knockdown led to a complete loss of KAP1 enrichment at this locus (Figure 5c). In contrast, histone H3 acetylation, a mark indicative of gene activation, increased at both sites of pre-miR-126* upon MMSET knockdown (Figure 5d). H3K36 methylation, a modification altered globally by MMSET and associated with transcriptional elongation, was relatively low at both sites of premiR-126* and was unaffected by MMSET knockdown (data not shown).
Figure 5.
MMSET binding to the miR-126* promoter is associated with KAP1 presence and chromatin modifications favoring gene repression. Chromatin from MMSET shRNA-inducible KMS11 cells grown with or without dox for 7 days was immunoprecipitated with anti-MMSET (a), anti-H3K9me3 (b), anti-KAP1 (c) or anti-acetyl histone H3 (d) antibodies. Immunoglobulin G (IgG) antibody was used as a negative control. Precipitated DNA was analyzed by real-time PCR using primers amplifying regions near the predicted miR-126* TSS and 10 kb upstream (10 kb Up). Results are presented as percentage of total input DNA precipitated. Values represent mean ± s.d. from two experiments.
In a complementary experiment, we performed the same ChIP assays in the t(4;14)-negative RPMI-8226 cell line harboring GFP, wild-type MMSET or mutant MMSET-Y1118A. As expected, the ectopic expression of wild-type and mutant MMSET in RPMI-8226 cells led to its enrichment at both sites of pre-miR-126* (Supplementary Figure 5A). Although the presence of wild-type MMSET was associated with increased H3K9me3 and decreased H3 acetylation, mutant MMSET led to little change of these modifications when compared with the GFP control (Supplementary Figure 5B and C), correlating with the expression levels of miR-126* previously detected by quantitative real-time PCR (qPCR). Collectively, these data suggest that on certain loci such as that of miR-126/126*, MMSET can mediate transcriptional repression. Moreover, these results indicate that both MMSET and KAP1 bind the miR-126* locus, and that the presence of KAP1 at the locus depends on MMSET binding, suggesting that KAP1 may be recruited by MMSET and/or H3K9me3 to repress miR-126*.
HDACs support MMSET-mediated repression of miR-126*
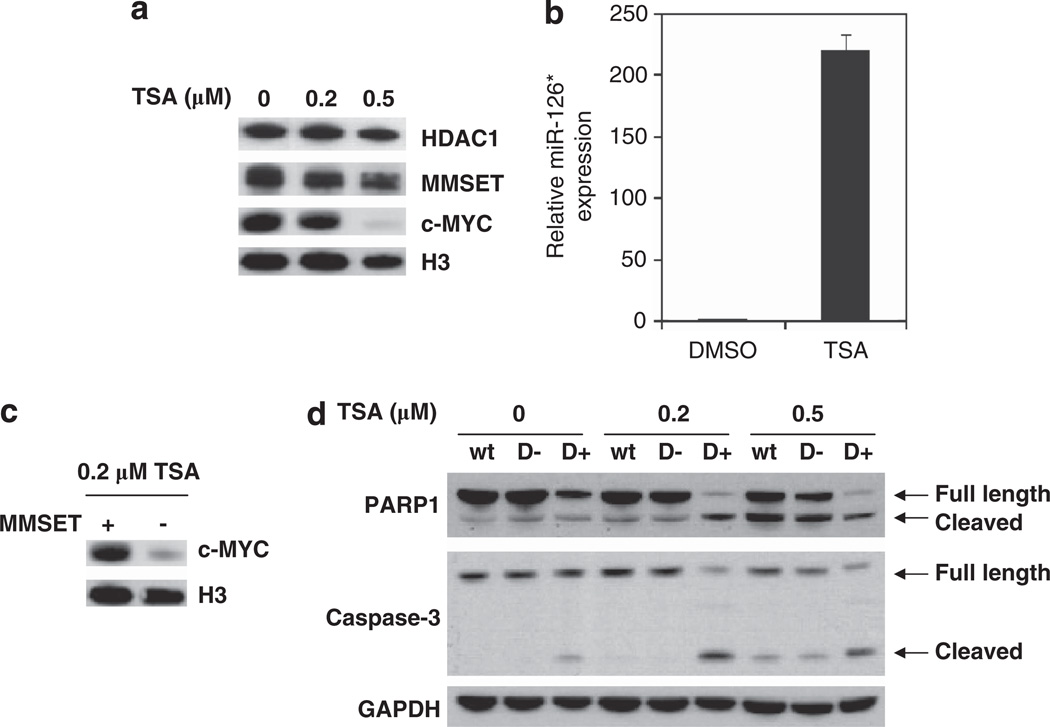
As histone deacetylases (HDACs) can interact with both MMSET7,8,19 and KAP120–22 to repress transcription (Figure 4a), we investigated the importance of HDACs in miR-126* and c-MYC expression in MM cells. ChIP assays in KMS11 cells detected the presence of HDAC1/2 on the miR-126* promoter (Supplementary Figure 6A), but this was unaffected by changes in the level of MMSET (data not shown). Therefore, we used the HDAC inhibitor trichostatin A (TSA) to determine if HDACs were required for miR-126* repression and c-MYC expression. When KMS11 cells were treated with 0.5 µm TSA for 24 h, c-MYC expression dramatically decreased and miR-126* levels significantly increased (Figures 6a and b). KMS11 cells transfected with a siRNA directed against HDAC1 also showed a decrease in c-MYC protein level (Supplementary Figure 6B). These results suggested that HDACs are involved in the repression of miR-126*, allowing for elevated c-MYC expression.
Figure 6.
HDACs promote miR-126* repression and collaborate with MMSET to mediate c-MYC expression. (a) KMS11 cells were treated with the indicated concentrations of TSA for 24 h. Nuclear extracts from these cells were immunoblotted for HDAC1, MMSET and c-MYC. Histone H3 was used as a loading control. (b) KMS11 cells were treated with 0.5 µm TSA or dimethylsulfoxide (DMSO) for 24 h, and relative miR-126* expression was determined by real-time PCR after normalization by U6 snRNA levels. Values are represent relative to that observed in cells treated with DMSO (mean ± s.d. from three experiments). (c) MMSET shRNA-inducible KMS11 cells were grown in the presence or absence of dox for 7 days, and further incubated with 0.2 µm TSA for 24 h. c-MYC protein levels were assayed by immunoblot with histone H3 as a loading control. (d) KMS11 cells described in (c) were treated with the indicated dose of TSA for 24 h. Parental KMS11 cells (wt) are presented as a control. Total cell lysates were subjected to immunoblot using an anti-PARP1 or anti-caspase-3 antibody. GAPDH represents a loading control.
To further explore the implications of the interplay between MMSET and HDACs, we treated KMS11 cells with different TSA concentrations before and after MMSET depletion. Although treatment of cells with 0.2 µm TSA on its own did not significantly alter c-MYC levels (Figure 6a), the combination of TSA treatment and shRNA-mediated depletion of MMSET strongly suppressed c-MYC expression (Figure 6c). This result suggested a collaboration between MMSET and HDACs in the regulation of c-MYC levels, potentially through miR-126*. To gain more insight into the combined biological effect of MMSET and HDACs in MM cells, we next analyzed apoptosis, a process promoted by HDAC inhibitors. Although low-dose TSA treatment or MMSET knockdown led to minimal apoptosis, combining TSA with MMSET depletion led to marked increase in apoptosis as measured by cleaved caspase-3 and PARP1 (Figure 6d). This suggests that combining HDAC inhibitors, already in clinical practice, with MMSET inhibitors, yet to be developed, may be a promising therapeutic strategy in t(4;14)-associated MM. This combination would potentially lead to an increase in the apoptosis of MM cells as well as to a reduced proliferation originated by a decreased c-MYC expression.
DISCUSSION
Overexpression of c-MYC represents a convergent mechanism of oncogenesis in MM, and is mediated in multiple ways. For example, overexpression of cyclin D1 or cyclin D3 by fusion to the immunoglobulin promoter/enhancer drives cell cycle entry through activation of the E2F transcription factor that promotes the expression of many genes, including c-MYC.23 Overexpression of the fibroblast growth factor receptor and activating mutations in RAS or B-RAF could induce c-MYC expression through the mitogen-activated protein kinase pathway. Another proposed mechanism for the continued high-level expression of c-MYC is the presence of an aberrantly spliced isoform of BLIMP1 in myeloma cells, which is unable to repress c-MYC transcription.24 IRF4, a key transcription factor in all myeloma cells, induces c-MYC, which in turn promotes IRF4 expression in a positive feedback loop.13 The drive for continued c-MYC expression is also typified in its frequent rearrangement, often to genes other than the immunoglobulin locus, particularly in late-stage MM and in virtually all established MM cell lines.2 Deregulated c-MYC expression in MM also occurs at the post-transcriptional level. The c-MYC gene contains an internal ribosome entry site in its 5′-UTR, frequently mutated in MM, allowing for enhanced translation through the actions of RNA-binding proteins.25 Furthermore, interleukin-6, a critical cytokine for myeloma growth, also stimulates c-MYC translation.26 In this work, we describe a novel mechanism to control the expression of c-MYC: the regulation of its translation by the HMT MMSET.
Many miRNAs are downregulated during transformation and tumor progression,27 suggesting they may act as tumor suppressors. The profile of miRNAs expressed in MM differs from that of normal plasma cells,28 and each subtype of MM has a specific miRNA signature,29,30 suggesting that deregulation of miRNA expression may have a relevant biological role in this disease. In the present study, we show that aberrant repression of miR-126* by MMSET leads to post-transcriptional enhancement of c-MYC expression, suggesting that miR-126* is a tumor suppressor. c-MYC is regulated by several miRNAs whose expression is suppressed in cancer. For example, the let-7 miRNA family, which targets c-MYC, is expressed at low levels in lung cancers.31 c-MYC overexpression in Burkitt’s lymphoma lacking rearrangement of the c-MYC gene was associated with downregulation of let-7c and miR-34b.32 miR-196b, which also represses c-MYC, is downregulated in acute lymphoblastic leukemia.33 Although none of the already described miRNAs targeting c-MYC was detected as regulated by MMSET in our miRNA profiling experiment, the possibility that some of these miRNAs collaborate with miR-126* to regulate c-MYC expression cannot be ruled out. Moreover, although this work focused on miRNA-mediated modulation of c-MYC mRNA translation as a mechanism by which MMSET promotes high c-MYC protein levels, it is also possible that MMSET could stimulate the translational machinery to increase the production of c-MYC and other key oncoproteins.
miR-126 and miR-126* are embedded within the EGFL7 gene. Traditionally, it was assumed that the passenger strand or miRNA* is rapidly degraded, whereas the guide strand is incorporated into the RISC (RNA-induced silencing complex) complex. Recent evidence indicates that miRNA* can also be functional and more abundant than previously thought. In fact, the strand of the miRNA that accumulates in a cell may vary according to species, cell type and developmental stage.17,34 Both miR-126 and miR-126* can be found associated with the RISC complex and have active roles in the control of gene expression. In myeloma cells, upon MMSET knockdown, both miR-126 and miR-126* increased, but the change in miR-126* was larger, suggesting that this miRNA was more readily incorporated into the RISC complex and protected from degradation. miR-126/126* was found to play a role in other cancers and hematopoiesis. For example, miR-126/126* is upregulated in core-binding factor-associated leukemia35 and controls hematopoietic differentiation of embryonic stem cells36 and zebrafish.37 A growth-suppressive function of miR-126 was found in breast38 and lung39 cancers, through the inhibition of vascular endothelial growth factor-mediated signaling. A possible oncogenic function of miR-126/126* was described in gastric cancer through suppression of Sox2 expression.40 Here, we identify a new tumor-suppressive action of miR-126* through inhibition of c-MYC expression.
miRNAs may be silenced in cancer because of deletion of chromosome segments,41 aberrant DNA hypermethylation and silencing of miRNA host gene transcription42 or by oncogenic transcriptional regulators. For example, the AML1-ETO fusion protein assembles a heterochromatin complex including DNA methyltransferase on the miR-223 promoter.43 Here, we show that an overexpressed HMT can repress a miRNA. The relevance of miR-126* regulation by MMSET was highlighted by the fact that most MMSET-overexpressing myeloma cell lines had undetectable levels of miR-126*. In some of the t(4;14)-negative cell lines, however, the low levels of MMSET did not appear to be sufficient to promote the expression of miR-126*, suggesting that other mechanisms may repress this miRNA. For example, in acute myeloid leukemia, miR-126/126* expression was associated with promoter DNA methylation status,44,45 and in breast cancer cells oncogenic Src repressed miR-126/126* expression.46 Therefore, MMSET may be a major but not the only influence on the expression of this important miRNA pair in MM.
MMSET overexpression was associated with heterochromatic modifications of the miR-126* promoter, characterized by increased H3K9me3 and decreased histone H3 acetylation. This contrasts with the action of MMSET on many other loci, defined by increased H3K36 methylation, decreased H3K27 methylation and gene activation.11,47 On the miR-126/126* promoter, increased MMSET binding was not associated with increased H3K36 methylation. Why this and other regions of the genome (data not shown) are protected from an otherwise global change in histone methylation remains to be understood.
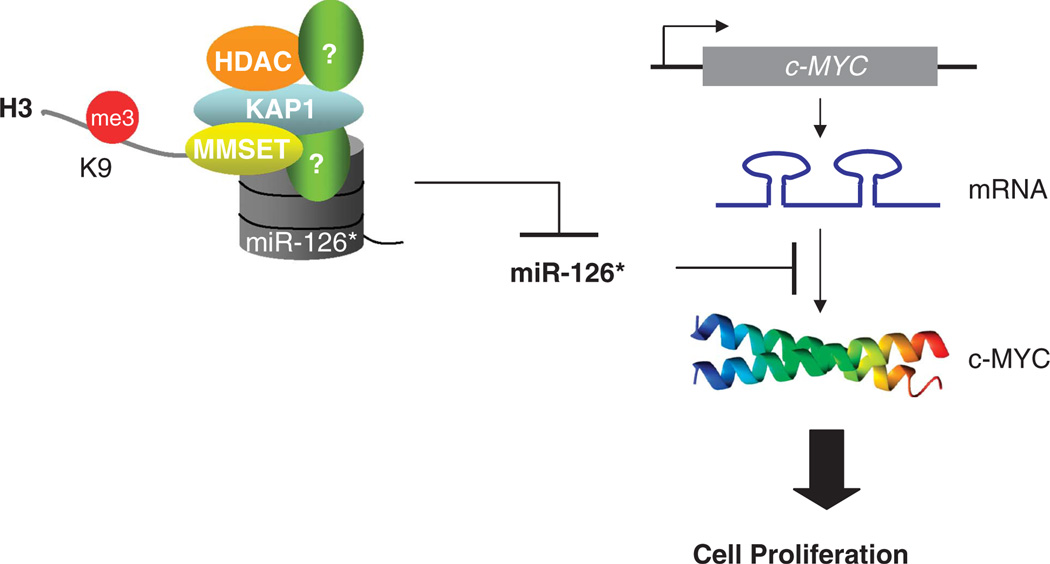
Nevertheless, at the miR-126/126* promoter, repression in response to MMSET was associated with KAP1, a critical cofactor of KRAB zinc-finger proteins.48 KAP1 recruits the NuRD-HDAC complex and the H3K9-specific HMT SETDB1 to induce heterochromatin formation.22 Genome-wide analysis of KAP1 binding showed that KAP1 target promoters were enriched for H3K9me3.21 Consistent with this, we show that KAP1 and H3K9me3 were enriched at the miR-126* promoter in the presence of MMSET. KAP1 recruitment to this promoter may be triggered by the enrichment in H3K9me3; alternatively, KAP1 may be attracted to the miR-126/126* locus by MMSET, as suggested by the direct interaction between both factors observed. Previous studies showed that HDAC1 and/or 2, which are components of the KAP-1-NuRD complex,22 can interact with MMSET and play a role in its ability to repress transcription.7,19 Taken together, in t(4;14) MM cells, MMSET in combination with KAP1 and HDACs inhibits miR-126* transcription through heterochromatic histone modifications, helping to maintain high c-MYC protein levels and cellular proliferation (Figure 7).
Figure 7.
A model for MMSET regulation of miR-126* and c-MYC expression. MMSET, in collaboration with KAP1 and HDAC proteins among others, binds the miR-126/126* locus, promoting chromatin modifications that lead to gene repression. Decreased levels of miR-126* release the 3′-UTR region of c-MYC mRNA, allowing for an increased translation of the transcript. As a result, elevated c-MYC protein expression promotes the proliferation of t(4;14) MM cells.
HDAC inhibitors decrease proliferation and induce apoptosis of cancer cells.49,50 The HDAC inhibitor vorinostat/SAHA (suberoylanilidehydroxamic acid) is approved for the treatment of cutaneous T-cell lymphoma, and other HDAC inhibitors have entered clinical trials in lymphomas, MM and acute myeloid leukemia.51,52 HDAC inhibitors have been combined with other antitumor agents to elucidate a minimal effective dose, to reduce side effects, and to maximize treatment efficacy.51,53 In this study, the combination of MMSET depletion and an HDAC inhibitor markedly increased apoptosis in t(4;14)+ MM cells. This may be because of global changes in the structure and function of the chromatin in response to the loss of MMSET,11 and stimulation of DNA damage, a known effect of HDAC treatment,54 as well as effects on the expression of miR-126* and other genes at the transcriptional and post-transcriptional level.54 These results suggest that combining inhibitors of MMSET and HDAC function may represent a future therapeutic strategy for t(4;14)-associated myeloma.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Louis Staudt for providing the MMSET-shRNA cells and Dr Marcus Peter for helpful suggestions. This work was supported by NIH Grant CA123204, a Leukemia and Lymphoma Specialized Center of Research grant, a NIH Physical Science Oncology Center Grant U54CA143869 and Epizyme, Inc. (to JDL). EM-G was supported by a European Hematology Association Fellowship, RP by NRSA HL099177 and TE by an Alfonso Martin Escudero fellowship.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 2.Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr. 2008;39:25–31. doi: 10.1093/jncimonographs/lgn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 5.Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101:1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- 6.Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- 7.Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JY, Kee HJ, Choe NW, Kim SM, Eom GH, Baek HJ, et al. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol Cell Biol. 2008;28:2023–2034. doi: 10.1128/MCB.02130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brito JL, Walker B, Jenner M, Dickens NJ, Brown NJ, Ross FM, et al. MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica. 2009;94:78–86. doi: 10.3324/haematol.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McConnell MJ, Chevallier N, Berkofsky-Fessler W, Giltnane JM, Malani RB, Staudt LM, et al. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol Cell Biol. 2003;23:9375–9388. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Hum Genet. 2007;Chapter 12(Unit 12):10. doi: 10.1002/0471142905.hg1210s54. [DOI] [PubMed] [Google Scholar]
- 16.Kim MK, Mason JM, Li CM, Berkofsky-Fessler W, Jiang L, Choubey D, et al. A pathologic link between Wilms tumor suppressor gene, WT1, and IFI16. Neoplasia. 2008;10:69–78. doi: 10.1593/neo.07869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuchenbauer F, Mah SM, Heuser M, McPherson A, Ruschmann J, Rouhi A, et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood. 2011;118:3350–3358. doi: 10.1182/blood-2010-10-312454. [DOI] [PubMed] [Google Scholar]
- 18.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todoerti K, Ronchetti D, Agnelli L, Castellani S, Marelli S, Deliliers GL, et al. Transcription repression activity is associated with the type I isoform of the MMSET gene involved in t(4;14) in multiple myeloma. Br J Haematol. 2005;131:214–218. doi: 10.1111/j.1365-2141.2005.05741.x. [DOI] [PubMed] [Google Scholar]
- 20.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ., 3rd Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J Biol Chem. 2009;284:35670–35680. doi: 10.1074/jbc.M109.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oswald F, Lovec H, Moroy T, Lipp M. E2F-dependent regulation of human MYC: trans-activation by cyclins D1 and A overrides tumour suppressor protein functions. Oncogene. 1994;9:2029–2036. [PubMed] [Google Scholar]
- 24.Morgan MA, Magnusdottir E, Kuo TC, Tunyaplin C, Harper J, Arnold SJ, et al. Blimp-1/Prdm1 alternative promoter usage during mouse development and plasma cell differentiation. Mol Cell Biol. 2009;29:5813–5827. doi: 10.1128/MCB.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cobbold LC, Wilson LA, Sawicka K, King HA, Kondrashov AV, Spriggs KA, et al. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene. 2010;29:2884–2891. doi: 10.1038/onc.2010.31. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Frost P, Hoang B, Benavides A, Gera J, Lichtenstein A. IL-6-induced enhancement of c-Myc translation in multiple myeloma cells: critical role of cytoplasmic localization of the rna-binding protein hnRNP A1. J Biol Chem. 2011;286:67–78. doi: 10.1074/jbc.M110.153221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 28.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez NC, Sarasquete ME, Misiewicz-Krzeminska I, Delgado M, De Las Rivas J, Ticona FV, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 2010;24:629–637. doi: 10.1038/leu.2009.274. [DOI] [PubMed] [Google Scholar]
- 30.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–e26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 31.Wong TS, Man OY, Tsang CM, Tsao SW, Tsang RK, Chan JY, et al. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J Cancer Res Clin Oncol. 2011;137:415–422. doi: 10.1007/s00432-010-0898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leucci E, Cocco M, Onnis A, De Falco G, van Cleef P, Bellan C, et al. MYC translocation-negative classical Burkitt lymphoma cases: an alternative pathogenetic mechanism involving miRNA deregulation. J Pathol. 2008;216:440–450. doi: 10.1002/path.2410. [DOI] [PubMed] [Google Scholar]
- 33.Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem. 2010;340:97–106. doi: 10.1007/s11010-010-0406-9. [DOI] [PubMed] [Google Scholar]
- 34.Biasiolo M, Sales G, Lionetti M, Agnelli L, Todoerti K, Bisognin A, et al. Impact of host genes and strand selection on miRNA and miRNA* expression. PLoS One. 2011;6:e23854. doi: 10.1371/journal.pone.0023854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Chen J. In vitro functional study of miR-126 in leukemia. Methods Mol Biol. 2011;676:185–195. doi: 10.1007/978-1-60761-863-8_13. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Gschweng E, Van Handel B, Cheng D, Mikkola HK, Witte ON. Regulated expression of microRNAs-126/126* inhibits erythropoiesis from human embryonic stem cells. Blood. 2011;117:2157–2165. doi: 10.1182/blood-2010-08-302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabher C, Payne EM, Johnston AB, Bolli N, Lechman E, Dick JE, et al. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia. 2011;25:506–514. doi: 10.1038/leu.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157–164. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 39.Miko E, Margitai Z, Czimmerer Z, Varkonyi I, Dezso B, Lanyi A, et al. miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett. 2011;585:1191–1196. doi: 10.1016/j.febslet.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 40.Otsubo T, Akiyama Y, Hashimoto Y, Shimada S, Goto K, Yuasa Y. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One. 2011;6:e16617. doi: 10.1371/journal.pone.0016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouhi A, Mager DL, Humphries RK, Kuchenbauer F. MiRNAs, epigenetics, and cancer. Mamm Genome. 2008;19:517–525. doi: 10.1007/s00335-008-9133-x. [DOI] [PubMed] [Google Scholar]
- 43.Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Cammarata G, Augugliaro L, Salemi D, Agueli C, La Rosa M, Dagnino L, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Shen Y, Ichikawa H, Antes T, Goldberg GS. Regulation of miRNA expression by Src and contact normalization: effects on nonanchored cell growth and migration. Oncogene. 2009;28:4272–4283. doi: 10.1038/onc.2009.278. [DOI] [PubMed] [Google Scholar]
- 47.Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ, et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2012 doi: 10.1038/onc.2012.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alter MD, Hen R. Putting a KAP on transcription and stress. Neuron. 2008;60:733–735. doi: 10.1016/j.neuron.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Schrump DS. Cytotoxicity mediated by histone deacetylase inhibitors in cancer cells: mechanisms and potential clinical implications. Clin Cancer Res. 2009;15:3947–3957. doi: 10.1158/1078-0432.CCR-08-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stimson L, La Thangue NB. Biomarkers for predicting clinical responses to HDAC inhibitors. Cancer Lett. 2009;280:177–183. doi: 10.1016/j.canlet.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 51.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 52.Mitsiades CS, Davies FE, Laubach JP, Joshua D, San Miguel J, Anderson KC, et al. Future directions of next-generation novel therapies, combination approaches, and the development of personalized medicine in myeloma. J Clin Oncol. 2011;29:1916–1923. doi: 10.1200/JCO.2010.34.0760. [DOI] [PubMed] [Google Scholar]
- 53.Gronbaek K, Hother C, Jones PA. Epigenetic changes in cancer. APMIS. 2007;115:1039–1059. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- 54.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;280:125–133. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.