Abstract
We introduce vector diffusion maps (VDM), a new mathematical framework for organizing and analyzing massive high-dimensional data sets, images, and shapes. VDM is a mathematical and algorithmic generalization of diffusion maps and other nonlinear dimensionality reduction methods, such as LLE, ISOMAP, and Laplacian eigenmaps. While existing methods are either directly or indirectly related to the heat kernel for functions over the data, VDM is based on the heat kernel for vector fields. VDM provides tools for organizing complex data sets, embedding them in a low-dimensional space, and interpolating and regressing vector fields over the data. In particular, it equips the data with a metric, which we refer to as the vector diffusion distance. In the manifold learning setup, where the data set is distributed on a low-dimensional manifold ℳd embedded in ℝp, we prove the relation between VDM and the connection Laplacian operator for vector fields over the manifold.
1 Introduction
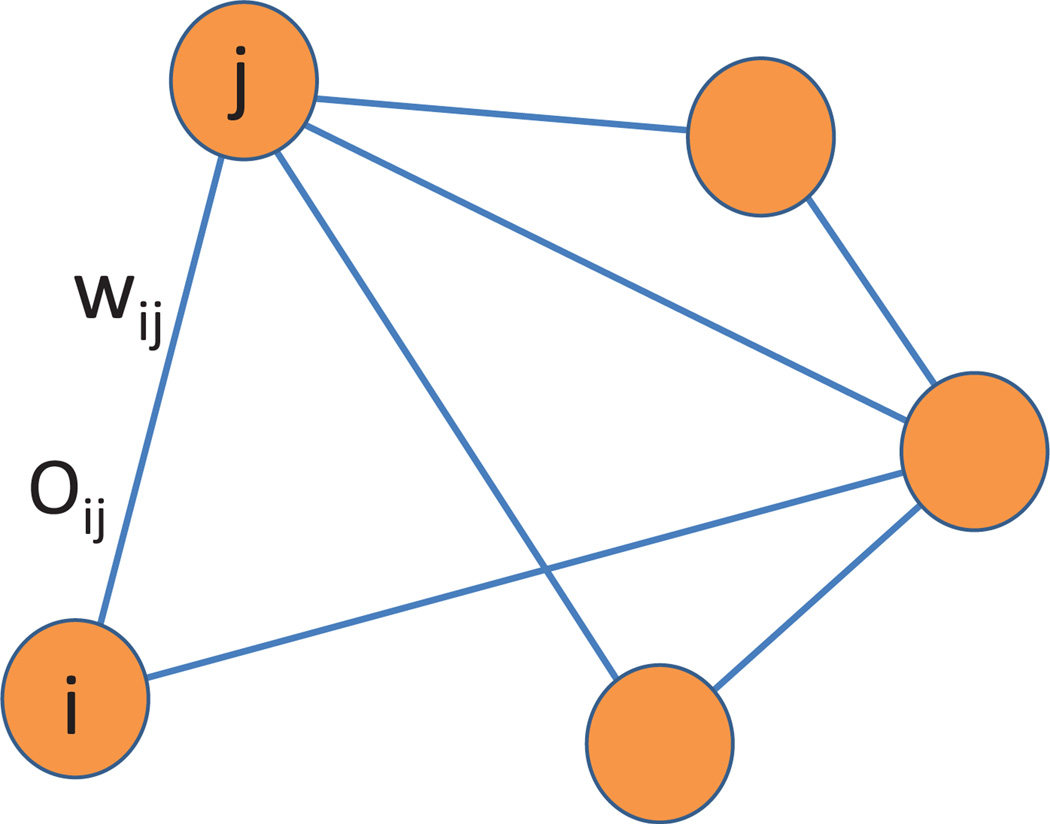
A popular way to describe the affinities between data points is using a weighted graph, whose vertices correspond to the data points, edges that connect data points with large enough affinities, and weights that quantify the affinities. In the past decade we have witnessed the emergence of nonlinear dimensionality reduction methods, such as locally linear embedding (LLE) [33], ISOMAP [39], Hessian LLE [12], local tangent space alignment (LTSA) [42], Laplacian eigenmaps [2], and diffusion maps [9]. These methods use the local affinities in the weighted graph to learn its global features. They provide invaluable tools for organizing complex networks and data sets, embedding them in a low-dimensional space, and studying and regressing functions over graphs. Inspired by recent developments in the mathematical theory of cryo-electron microscopy [18, 37] and synchronization [10, 35], in this paper we demonstrate that in many applications, the representation of the data set can be vastly improved by attaching to every edge of the graph not only a weight but also a linear orthogonal transformation (see Figure 1.1).
Figure 1.1.
In VDM, the relationships between data points are represented as a weighted graph, where the weights wij are accompanied by linear orthogonal transformations Oij.
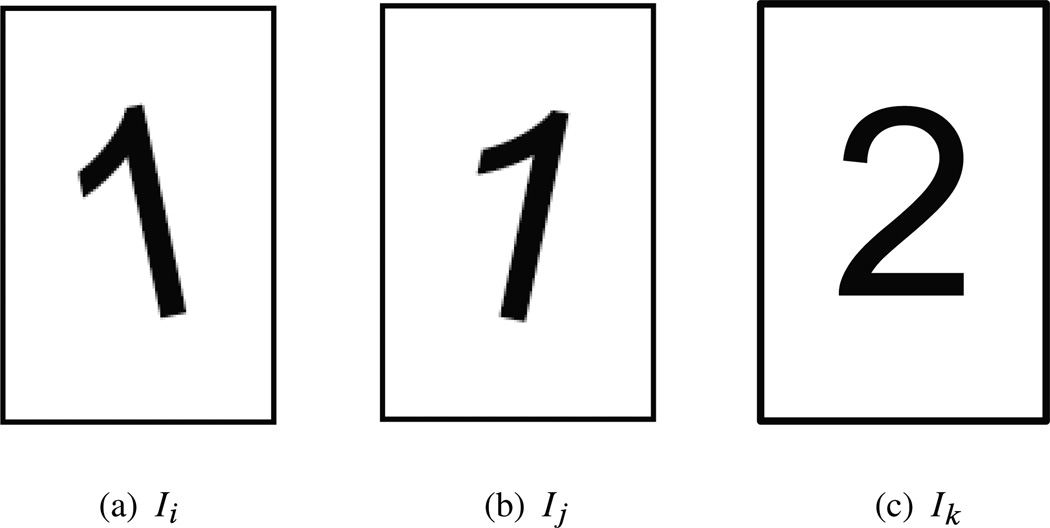
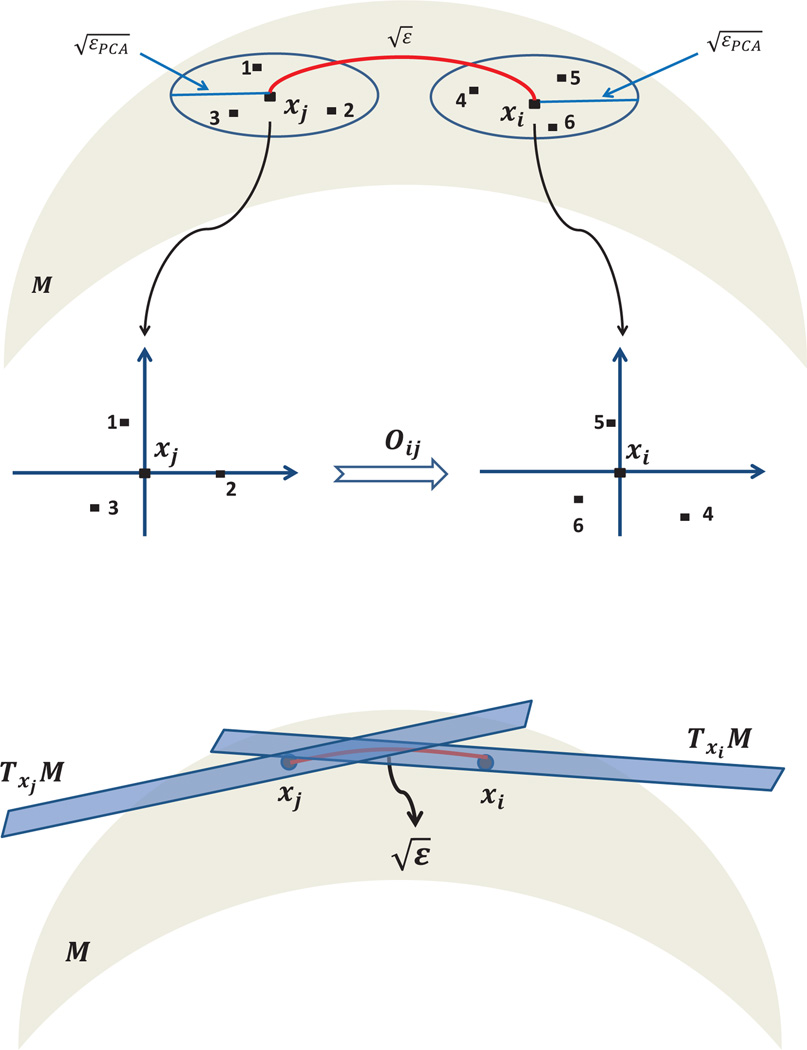
Consider, for example, a data set of images, or small patches extracted from images (see, e.g., [8, 27]). While weights are usually derived from the pairwise comparison of the images in their original representation, we instead associate the weight wij to the similarity between image i and image j when they are optimally rotationally aligned. The dissimilarity between images when they are optimally rotationally aligned is sometimes called the rotationally invariant distance [31]. We further define the linear transformation Oij as the 2 × 2 orthogonal transformation that registers the two images (see Figure 1.2). Similarly, for data sets consisting of three-dimensional shapes, Oij encodes the optimal 3 × 3 orthogonal registration transformation. In the case of manifold learning, the linear transformations can be constructed using local principal component analysis (PCA) and alignment, as discussed in Section 2.
Figure 1.2.
An example of a weighted graph with orthogonal transformations: Ii and Ij are two different images of the digit 1, corresponding to nodes i and j in the graph. Oij is the 2 × 2 rotation matrix that rotationally aligns Ij with Ii and wij is some measure for the affinity between the two images when they are optimally aligned. The affinity wij is large, because the images Ii and OijIj are actually the same. On the other hand, Ik is an image of the digit 2, and the discrepancy between Ik and Ii is large even when these images are optimally aligned. As a result, the affinity wik would be small, perhaps so small that there is no edge in the graph connecting nodes i and k. The matrix Oik is clearly not as meaningful as Oij. If there is no edge between i and k, then Oik is not represented in the weighted graph.
In this paper, the linear transformations relating data points are restricted to orthogonal transformations in O(d). Transformations belonging to other matrix groups, such as translations and dilations, are not treated here.
While diffusion maps and other nonlinear dimensionality reduction methods are either directly or indirectly related to the heat kernel for functions over the data, our VDM framework is based on the heat kernel for vector fields. We construct this kernel from the weighted graph and the orthogonal transformations. Through the spectral decomposition of this kernel, VDM defines an embedding of the data in a Hilbert space. In particular, it defines a metric for the data, that is, distances between data points that we call vector diffusion distances. For some applications, the vector diffusion metric is more meaningful than currently used metrics, since it takes into account the linear transformations, and as a result, it provides a better organization of the data. In the manifold learning setup, we prove a convergence theorem illuminating the relation between VDM and the connection Laplacian operator for vector fields over the manifold.
The paper is organized in the following way: First, Table 1.1 summarizes the notation used throughout this paper. In Section 2 we describe the manifold learning setup and a procedure to extract the orthogonal transformations from a point cloud scattered in a high-dimensional euclidean space using local PCA and alignment. In Section 3 we specify the vector diffusion mapping of the data set into a finite-dimensional Hilbert space. At the heart of the vector diffusion mapping construction lies a certain symmetric matrix that can be normalized in slightly different ways. Different normalizations lead to different embeddings, as discussed in Section 4. These normalizations resemble the normalizations of the graph Laplacian in spectral graph theory and spectral clustering algorithms. In the manifold learning setup, it is known that when the point cloud is uniformly sampled from a low-dimensional Riemannian manifold, then the normalized graph Laplacian approximates the Laplace-Beltrami operator for scalar functions. In Section 5 we formulate a similar result, stated as Theorem 5.3, for the convergence of the appropriately normalized vector diffusion mapping matrix to the connection Laplacian operator for vector fields.1
TABLE 1.1.
Summary of symbols used throughout the paper.
| Symbol | Meaning |
|---|---|
| p | Dimension of the ambient euclidean space |
| d | Dimension of the low-dimensional Riemannian manifold |
| ℳd | d-dimensional Riemannian manifold embedded in ℝp |
| ι | Embedding of ℳd into ℝp |
| g | Metric of ℳ induced from ℝp |
| dV | Volume form associated with the metric g |
| n | Number of data points sampled from ℳd |
| x1 …,xn | Points sampled from ℳd |
| expx | Exponential map at x |
| Δ | Laplace-Beltrami operator |
| Tℳ | Tangent bundle of ℳ |
| Txℳ | Tangent space to ℳ at x |
| X | Vector field |
| Ck(Tℳ) | Space of kth continuously differentiable vector fields, k = 1,2,… |
| L2(Tℳ) | Space of squared-integrable vector fields |
| Px,y | Parallel transport from y to x along the geodesic linking them |
| ∇ | Connection of the tangent bundle |
| ∇2 | Connection (rough) Laplacian |
| et∇2 | Heat kernel associated with the connection Laplacian |
| ℛ | Riemannain curvature tensor |
| Ric | Ricci curvature |
| s | Scalar curvature |
| Π | Second fundamental form of the embedding ι |
| K, KPCA | Kernel functions |
| ∊,∊PCA | Bandwidth parameters of the kernel functions |
The proof of Theorem 5.3 appears in Appendix B. We verify Theorem 5.3 numerically for spheres of different dimensions, as reported in Section 6 and Appendix C. We also use other surfaces to perform numerical comparisons between the vector diffusion distance, the diffusion distance, and the geodesic distance. In Section 7 we briefly discuss out-of-sample extrapolation of vector fields via the Nyström extension scheme. The role played by the heat kernel of the connection Laplacian is discussed in Section 8. We use the well-known short-time asymptotic expansion of the heat kernel to show the relationship between vector diffusion distances and geodesic distances for nearby points. In Section 9 we briefly discuss the application of VDM to cryo-electron microscopy, as a prototypical multireference rotational alignment problem. We conclude in Section 10 with a summary followed by a discussion of some other possible applications and extensions of the mathematical framework.
2 Data Sampled from a Riemannian Manifold
One of the main objectives in the analysis of a high-dimensional large data set is to learn its geometric and topological structure. Even though the data itself is parametrized as a point cloud in a high-dimensional ambient space ℝpthe correlation between parameters often suggests the popular “manifold assumption” that the data points are distributed on (or near) a single low-dimensional Riemannian manifold ℳd embedded in ℝp, where d is the dimension of the manifold and d ≪ p. Suppose that the point cloud consists of n data points x1; x2, …, xn that are viewed as points in ℝp but are restricted to the manifold.
We now describe how the orthogonal transformations Oij can be constructed from the point cloud using local PCA and alignment.
Local PCA. For every data point xi we suggest estimating a basis to the tangent plane Txiℳ to the manifold at xi using the following procedure, which we refer to as local PCA. We fix a scale parameter ∊PCA > 0 and define 𝒩xi, ∊PCA as the neighbors of xi inside a ball of radius centered at xi:
Denote the number of neighboring points of xi by Ni,2 that is, Ni |𝒩xi, ∊PCA|, and denote the neighbors of xi by xi1, xi2, …, xiNi. We assume that ∊PCA is large enough so that Ni ≥ d, but at the same time ∊PCA is small enough such that Ni ≪ n. At this point we assume d is known; methods to estimate it will be discussed later in this section. In Theorem B.1 we show that a satisfactory choice for ∊PCA is given by ∊PCA = O(n−2/(d+1)), so that Ni = O(n1/(d+1)). In fact, it is even possible to choose ∊PCA = O (n−2/(d+2)) if the manifold does not have a boundary.
Observe that the neighboring points are located near Txiℳ, where deviations are possible due to curvature. Define Xi to be a p × Ni matrix whose jth column is the vector xij – xi that is,
In other words, Xi is the data matrix of the neighbors shifted to be centered at the point xi. Notice that while it is more common to shift the data for PCA by the mean , here we shift the data by xi. Shifting the data by µi is also possible for all practical purposes, but has the slight disadvantage of complicating the proof for the convergence of the local PCA step (see Appendix B.1).
The local covariance matrix corresponding to the neighbors of xi is . Among the neighbors, those that are further away from xi contribute the most to the covariance matrix. However, if the manifold is not flat at xi then we would like to give more emphasis to the nearby points, so that the tangent space estimation is more accurate. In order to give more emphasis to nearby points, we weigh the contribution of each point by a monotonically decreasing function of its distance from xi. Let KPCA be a C2 positive monotonic decreasing function with support on the interval [0, 1], for example, the Epanechnikov kernel KPCA(u) = (1–u2)χ[0,1], where χ is the indicator function. Let Di be an Ni × Ni diagonal matrix with
and define the p × Ni matrix Bi as
| (2.1) |
The local weighted covariance matrix at xi, which we denote by Ξi is
| (2.2) |
Since KPCA is supported on the interval [0, 1] the covariance matrix Ξi can also be represented as
| (2.3) |
The definition of Di(j, j) above is via the square root of the kernel, so it appears linearly in the covariance matrix. We denote the singular values of Bi by σi;1 ≥ σi;2≥⋯ ≥σi,Ni. The eigenvalues of the p × p local covariance matrix Ξi equal the squared singular values. Since the ambient space dimension p is typically large, it is usually more efficient to compute the singular values and singular vectors of Bi rather than the eigendecomposition of Ξi.
Suppose that the singular value decomposition (SVD) of Bi is given by
The columns of the p × Ni matrix Ui are orthonormal and are known as the left singular vectors
We define the p × d matrix Oi by the first d left singular vectors (corresponding to the largest singular values):
| (2.4) |
The d columns of Oi are orthonormal, i.e., . The columns of Oi represent an orthonormal basis to a d-dimensional subspace of ℝp. This basis is a numerical approximation to an orthonormal basis of the tangent plane Txiℳ. The order of the approximation (as a function of ∊PCA and n, where n is the number of the data points) is established later in Appendix B, using the fact that the columns of Oi are also the eigenvectors (corresponding to the d largest eigenvalues) of the p × p; weighted covariance matrix. Ξi. We emphasize that the covariance matrix is never actually formed due to its excessive storage requirements, and all computations are performed with the matrix Bi.
In cases where the intrinsic dimension d is not known in advance, the following procedure can be used to estimate d. The underlying assumption is that data points lie exactly on the manifold, without any noise contamination. We remark that in the presence of noise, other procedures (e.g., [28]) could give a more accurate estimation.
Because our previous definition for the neighborhood size parameter ∊PCA involves d, we are not allowed to use it when trying to estimate d. Instead, we can take the neighborhood size ∊PCA to be any monotonically decreasing function of n that satisfies . This condition ensures that the number of neighboring points increases indefinitely in the limit of an infinite number of sampling points. One can choose, for example, ∊PCA = 1/log(n), though other choices are also possible.
Notice that if the manifold is flat, then the neighboring points in 𝒩xi, ∊PCA are located exactly on Txiℳ and as a result rank (Xi) = rank (Bi) = d and Bi has exactly d nonvanishing singular values (i.e., σi;d+1 = σi,d+2 = ⋯ = σi,Ni = 0). In such a case, the dimension can be estimated as the number of nonzero singular values. For nonflat manifolds, due to curvature, there may be more than d nonzero singular values. Clearly, as n goes to infinity, these singular values approach 0, since the curvature effect disappears. A common practice is to estimate the dimension as the number of singular values that account for a high enough percentage of the variability of the data. That is, one sets a threshold γ between 0 and 1 (usually closer to 1 than to 0), and estimates the dimension as the smallest integer di for which
For example, setting γ = 0:9 means that di singular values account for at least 90% variability of the data, while di − 1 singular values account for less than 90%. We refer to the smallest integer di as the estimated local dimension of ℳ at xi. From the previous discussion it follows that this procedure produces an accurate estimation of the dimension at each point as n goes to infinity. One possible way to estimate the dimension of the manifold would be to use the mean of the estimated local dimensions d1; d2; …, dn, that is, (and then round it to the closest integer). The mean estimator minimizes the sum of squared errors . We estimate the intrinsic dimension of the manifold by the median value of all the di’s; that is, we define the estimator d̂ for the intrinsic dimension d as
In all proceeding steps of the algorithm we use the median estimator d̂, but in order to facilitate the notation we write d instead of y d̂.
Alignment Suppose xi and xj are two nearby points whose euclidean distance satisfies , where ∊ > 0 is a scale parameter different from the scale parameter ∊PCA. In fact, ∊ is much larger than ∊PCA as we later choose ∊ = O(n−2/(d+4)), while, as mentioned earlier, ∊PCA = O(n−2/(d+1)) (manifolds with boundary) or ∊PCA = O(n−2/(d+2)) (manifolds with no boundary). In any case, ∊ is small enough so that the tangent spaces Txiℳ and Txjℳ are also close (in the sense that their Grassmannian distance given approximately by the operator norm is small). Therefore, the column spaces of Oi and Oj are almost the same. If the subspaces were to be exactly the same, then the matrices Oi and Oj would differ by a d × d orthogonal transformation Oij satisfying OiOij = Oj, or equivalently . In that case, would be the matrix representation of the operator that transport vectors from Txjℳ to Txjℳ, viewed as copies of ℝd. The subspaces, however, are usually not exactly the same, due to curvature. As a result, the matrix is not necessarily orthogonal, and we define Oij as its closest orthogonal matrix, i.e.,
| (2.5) |
where ∥·∥HS is the Hilbert-Schmidt (HS) norm (given by for any real matrix A) and O(d) is the set of orthogonal d × d matrices. This minimization problem has a simple solution3 [1, 13, 21, 25] via the SVD of . Specifically, if
is the SVD of , then Oij is given by
We refer to the process of finding the optimal orthogonal transformation between bases as alignment. Later in Appendix B we show that the matrix Oij is an approximation to the parallel transport operator4 from Txjℳ to Txiℳ whenever xi and xj are nearby.
Note that not all bases are aligned; only the bases of nearby points are aligned. We set E to be the edge set of the undirected graph over n vertices that correspond to the data points, where an edge between i and j exists if and only if their corresponding bases are aligned by the algorithm5 (or equivalently, if and only if . The weights wij are defined6 using a kernel function K as
| (2.6) |
where we assume that K is supported on the interval [0, 1]. For example, the Gaussian kernel K(u) = exp {−u2}χ[0,1] leads to weights of the form and 0 otherwise. Notice that the kernel K used for the definition of the weights wij could be different from the kernel KPCA used for the previous step of local PCA.
3 Vector Diffusion Mapping
We construct the following matrix S:
| (3.1) |
That is, S is a block matrix, with n × n blocks, each of which is of size d × d. Each block is either a d × d orthogonal transformation Oij multiplied by the scalar weight wij or a zero d × d matrix. (As mentioned in a previous footnote, the edge set does not contain self-loops, so wii = 0 and S(i, i) = 0d × d.) The matrix S is symmetric since and wij = wji, and its overall size is nd × nd. We define a diagonal matrix D of the same size, where the diagonal blocks are scalar matrices given by
| (3.2) |
and
| (3.3) |
is the weighted degree of node i. The matrix D−1S can be applied to vectors v of length nd, which we regard as n vectors of length d, such that v(i) is a vector in ℝd viewed as a vector in Txiℳ. The matrix D−1S is an averaging operator for vector fields, since
| (3.4) |
This implies that the operator D−1S : (ℝd)n → (ℝd)n transports vectors from the tangent spaces Txjℳ (that are nearby to Txiℳ) to Txiℳ and then averages the transported vectors in Txiℳ.
Notice that diffusion maps and other nonlinear dimensionality reduction methods make use of the weight matrix but not of the transformations Oij. In diffusion maps, the weights are used to define a discrete random walk over the graph, where the transition probability aij in a single time step from node i to node j is given by
| (3.5) |
The Markov transition matrix can be written as
| (3.6) |
where 𝒟 is an n × n diagonal matrix with
| (3.7) |
While A is the Markov transition probability matrix in a single time step, At is the transition matrix for t steps. In particular, At (i, j) sums the probabilities of all paths of length t that start at i and end at j. Coifman and Lafon [9, 26] showed that At can be used to define an inner product in a Hilbert space. Specifically, the matrix A is similar to the symmetric matrix 𝒟−1/2W 𝒟−1/2 through A = 𝒟−1/2(𝒟−1/2W𝒟−1/2)𝒟1/2. It follows that A has a complete set of real eigenvalues and eigenvectors , respectively, satisfying Aφl = µlφl. Their diffusion mapping Φt is given by
| (3.8) |
where φl(i) is the ith entry of the eigenvector φl. The mapping Φt satisfies
| (3.9) |
where 〈·,·〉 is the usual dot product over euclidean space. The metric associated to this inner product is known as the diffusion distance. The diffusion distance dDM,t (i, j) between i and j is given by
| (3.10) |
Thus, the diffusion distance between i and j is the weighted-ℓ2 proximity between the probability clouds of random walkers starting at i and j after t steps.
In the VDM framework, we define the affinity between i and j by considering all paths of length t connecting them, but instead of just summing the weights of all paths, we sum the transformations. A path of length t from j to i is some sequence of vertices j0, j1, …, jt with j0 = j and jt = i, and its corresponding orthogonal transformation is obtained by multiplying the orthogonal transformations along the path in the following order:
| (3.11) |
Every path from j to i may therefore result in a different transformation. This is analogous to the parallel transport operator from differential geometry that depends on the path connecting two points whenever the manifold has curvature (e.g., the sphere). Thus, when adding transformations of different paths, cancellations may happen.
We would like to define the affinity between i and j as the consistency between these transformations, with higher affinity expressing more agreement among the transformations that are being averaged. To quantify this affinity, we consider again the matrix D−1S, which is similar to the symmetric matrix
| (3.12) |
through D−1S = D−1/2S̃D1/2, and define the affinity between i and j as
that is, as the squared HS norm of the d × d matrix S̃2t (i, j), which takes into account all paths of length 2t, where t is a positive integer. In a sense, measures not only the number of paths of length 2t connecting i and j but also the amount of agreement between their transformations. That is, for a fixed number of paths, is larger when the path transformations are in agreement and is smaller when they differ.
Since S̃ is symmetric, it has a complete set of eigenvectors v1; v2; …, vnd and eigenvalues λ1, λ2; …, λnd. We order the eigenvalues in decreasing order of magnitude |λ1|≥ |λ2|≥…≥|λnd|. The spectral decompositions of S̃ and S̃2t are given by
| (3.13) |
where vl(i) ∈ ℝd for i = 1, 2, …, n and l = 1, 2, …, nd. The HS norm of S̃2t (i, j) is calculated using the trace:
| (3.14) |
It follows that the affinity is an inner product for the finite-dimensional Hilbert space ℝ(nd)2 via the mapping Vt:
| (3.15) |
That is,
| (3.16) |
Note that in the manifold learning setup, the embedding i ↦ Vt(i) is invariant to the choice of basis for Txiℳ because the dot products 〈vl(i), vr(i)〉 are invariant to orthogonal transformations. We refer to Vt as the vector diffusion mapping.
From the symmetry of the dot products 〈vl (i), vr (i)〉, it is clear that is also an inner product for the finite-dimensional Hilbert space ℝnd(nd+1)/2 corresponding to the mapping
We define the symmetric vector diffusion distance dVDM,t (i, j) between nodes i and j as
| (3.17) |
The matrices I − S̃ and I + S̃ are positive semidefinite due to the following identity:
| (3.18) |
for any v ∈ ℝnd. As a consequence, all eigenvalues λl of S̃ reside in the interval [−1; 1]. In particular, for large enough t, most terms of the form (λlλr)2t in (3.14) are close to 0, and can be well approximated by using only the few largest eigenvalues and their corresponding eigenvectors. This lends itself to an efficient approximation of the vector diffusion distances dVDM;t(i, j) of (3.17), and it is not necessary to raise the matrix S̃ to its 2t power (which usually results in dense matrices). Thus, for any δ > 0, we define the truncated vector diffusion mapping that embeds the data set in ℝm2 (or equivalently, but more efficiently, in ℝm(m+1)/2) using the eigenvectors v1, v2; …, vm as
| (3.19) |
where m = m(t, δ) is the largest integer for which
We remark that Vt is defined through rather than , because we cannot guarantee that in general all eigenvalues of S̃ are nonnegative. In Section 8, we show that in the continuous setup of the manifold learning problem all eigenvalues are nonnegative. We anticipate that for most practical applications that correspond to the manifold assumption, all negative eigenvalues (if any) would be small in magnitude (say, smaller than δ). In such cases, one can use any real t > 0 for the truncated vector diffusion map .
4 Normalized Vector Diffusion Mappings
It is also possible to obtain slightly different vector diffusion mappings using different normalizations of the matrix S. These normalizations are similar to the ones used in the diffusion map framework [9]. For example, notice that
| (4.1) |
are the right eigenvectors of D−1S, that is, D−1Swl = λlwl. We can thus define another vector diffusion mapping, denoted , as
| (4.2) |
From (4.1) it follows that and Vt satisfy the relations
| (4.3) |
and
| (4.4) |
As a result,
| (4.5) |
In other words, the Hilbert-Schmidt norm of the matrix D−1S leads to an embedding of the data set in a Hilbert space only upon proper normalization by the vertex degrees (similar to the normalization by the vertex degrees in (3.9) and (3.10) for the diffusion map). We define the associated vector diffusion distances as
| (4.6) |
We comment that the normalized mappings that map the data points to the unit sphere are equivalent, that is,
| (4.7) |
This means that the angles between pairs of embedded points are the same for both mappings. For a diffusion map, it has been observed that in some cases the distances
are more meaningful than ∥Φt(i)–Φt(j)∥ (see, for example, [17]). This may also suggest the usage of the distances
in the VDM framework.
Another important family of normalized diffusion mappings is obtained by the following procedure. Suppose 0 ≤ α ≤ 1, and define the symmetric matrices Wα and Sα as
| (4.8) |
and
| (4.9) |
We define the weighted degrees degα(1), degα(2), …, degα(n) corresponding to Wα by
the n × n diagonal matrix Dα as
| (4.10) |
and the n × n block diagonal matrix Dα (with blocks of size d × d) as
| (4.11) |
We can then use the matrices Sα and Dα (instead of S and D) to define the vector diffusion mappings . Notice that for α = 0 we have S0 = S and D0 = D, so that V0,t = Vt and . The case α = 1 turns out to be especially important as discussed in the next section.
5 Convergence to the Connection Laplacian
For diffusion maps, the discrete random walk over the data points converges to a continuous diffusion process over that manifold in the limit n → ∞ and ∊ → 0. This convergence can be stated in terms of the normalized graph Laplacian L given by
In the case where the data points are sampled independently from the uniform distribution over ℳd the graph Laplacian converges pointwise to the Laplace-Beltrami operator, as we have the following proposition [3, 20, 26, 34]: If f: ℳd → ℝ is a smooth function (e.g., f ∈ C3(ℳ)), then with high probability
| (5.1) |
where Δℳ is the Laplace-Beltrami operator on ℳd. The error consists of two terms: a bias term O(∊) and a variance term that decreases as but also depends on ∊. Balancing the two terms may lead to an optimal choice of the parameter ∊ as a function of the number of points n. In the case of uniform sampling, Belkin and Niyogi [4] have shown that the eigenvectors of the graph Laplacian converge to the eigenfunctions of the Laplace-Beltrami operator on the manifold, which is stronger than the pointwise convergence given in (5.1).
In the case where the data points are independently sampled from a probability density function p(x) whose support is a d-dimensional manifold ℳd and satisfies some mild conditions, the graph Laplacian converges pointwise to the Fokker-Planck operator as stated in following proposition [3, 20, 26, 34]: If f ∈ C3(ℳ), then with high probability
| (5.2) |
where the potential term U is given by U(x) = −2 log p(x). The error is interpreted in the same way as in the uniform sampling case. In [9] it is shown that it is also possible to recover the Laplace-Beltrami operator for nonuniform sampling processes using W1 and 𝒟1 (that correspond to α = 1 in (4.8) and (4.11)). The matrix converges to the Laplace-Beltrami operator independently of the sampling density function p(x).
For VDM, we prove in Appendix B the following theorem, Theorem 5.3, that states that the matrix , where 0 ≤ α ≤ 1, converges to the connection Laplacian operator (defined via the covariant derivative; see Appendix A and [32]) plus some potential terms depending on p(x). In particular, converges to the connection Laplacian operator, without any additional potential terms. Using the terminology of spectral graph theory, it may thus be appropriate to call the connection Laplacian of the graph.
The main content of Theorem 5.3 specifies the way in which VDM generalizes diffusion maps: while diffusion mapping is based on the heat kernel and the Laplace-Beltrami operator over scalar functions, VDM is based on the heat kernel and the connection Laplacian over vector fields. While for diffusion maps the computed eigenvectors are discrete approximations of the Laplacian eigenfunctions, for VDM the lth eigenvector vl of is a discrete approximation of the lth eigenvector field Xl of the connection Laplacian ▽2 over ℳ, which satisfies ▽2Xl= −λlXl for some λ1 ≥ 0.
In the formulation of Theorem 5.3, as well as in the remainder of the paper, we slightly change the notation used so far in the paper, as we denote the sampled data points in ℳd by x1; x2; …, xn, and the observed data points in ℝp by ι(x1), ι(x2), …, ι(xn), where ι : ℳ ↪ ℝp is the embedding of the Riemannian manifold ℳ in ℝp. Furthermore, we denote by ι*Txiℳ the d-dimensional subspace of ℝp that is the embedding of Txiℝ in ℝp. It is important to note that in the manifold learning setup, the manifold ℳ, the embedding ι, and the points x1, x2; …, xn ∈ ℳ are assumed to exist but cannot be directly observed.
Theorems 5.3, 5.5, and 5.6, stated later in this section, and their proofs in Appendix B all share the following assumption:
ASSUMPTION 5.1.
ι : ℳ ↪ ℳp is a smooth d-dimensional compact Riemannian manifold embedded in ℳp, with metric g induced from the canonical metric on ℳp.
When ∂ ℳ ≠, we denote ℳt = {x ∈ ℳ : miny∈∂ℳd(x, y) ≤ t}, where t > 0 and d(x; y) is the geodesic distance between x and y.
The data points x1, x2; …, xn are independent samples from ℳ according to the probability density function p ∈ C3 (ℳ) supported on ℳ ⊂ ℝp, where p is uniformly bounded from below and above, that is, 0 < pm ≤ p(x) ≤ pM < ∞.
K ∈ C2([0, 1)) is a positive function. Also, ml := ∫ℝd ∥x∥lK(∥x∥)dx and . We assume m0 = 1.
The vector field X is in C3(Tℳ).
Denote τ to be the largest number having the following property: the open normal bundle about ℳ of radius r is embedded in ℝp for every r < τ [30]. This condition holds automatically since ℳ is compact. In all theorems, we assume that . In [30], 1/τ is referred to as the “condition number” of ℳ.
To ease notation, in what follows we use the same notation ▽ to denote different connections on different bundles whenever there is no confusion and the meaning is clear from the context.
DEFINITION 5.2. For ∊ > 0, define
We define the empirical probability density function by
and for 0 ≤ α ≤ 1 define the α-normalized kernel K∊, α by
For 0 ≤ α ≤ 1, we define
THEOREM 5.3. In addition to Assumption 5.1, suppose ℳ is closed and ∊PCA = O(n−2/(d+2)). For all xi with high probability (w.h.p.)
| (5.3) |
where is an orthonormal basis for a d-dimensional subspace of ℝp determined by local PCA (i.e., the columns of Oi), is an orthonormal basis for ι*Txiℳ, Oij is the optimal orthogonal transformation determined by the alignment procedure, and , where El is an orthonormal basis for Txiℳ.
When ∊PCA = O(n−2/(d+1)), then the same almost surely convergence results above hold but with a slower convergence rate.
COROLLARY 5.4. For ∊ = O(n−2/(d+4)) almost surely,
| (5.4) |
and in particular
| (5.5) |
When the manifold is compact with boundary, (5.3) does not hold at the boundary. However, we have the following result for the convergence behavior near the boundary:
THEOREM 5.5. Suppose the boundary ∂ℳ is smooth and that Assumption 5.1 applies. Choose ∊PCA = O(n−2/d+1)). When we have
| (5.6) |
where x0 = argminy∈∂ℳ d(xi, y), are constants defined in (B.93) and (B.94)and ∂d is the normal direction to the boundary at x0.
For the choice ∊ = O(n−2/(d+4)) (as in Corollary 5.4), the error appearing in (5.6) is O(∊3/4), which is asymptotically smaller than , which is the order of . A consequence of Theorem 5.3, Theorem 5.5, and the above discussion about the error terms is that the eigenvectors of are discrete approximations of the eigenvector fields of the connection Laplacian operator with homogeneous Neumann boundary condition that satisfy
| (5.7) |
We remark that the Neumann boundary condition also emerges for the choice ∊PCA = O(n−2/(d+2)). This is due to the fact that the error in the local PCA term is , which is asymptotically smaller than O(∊1/2 = O(n−1/(d+4)) error term.
Finally, Theorem 5.6 details the way in which the algorithm approximates the continuous heat kernel of the connection Laplacian:
THEOREM 5.6. Under Assumption 5.1, for any t > 0, the heat kernel et▽2 can be approximated on L2(Tℳ) by , that is,
6 Numerical Simulations
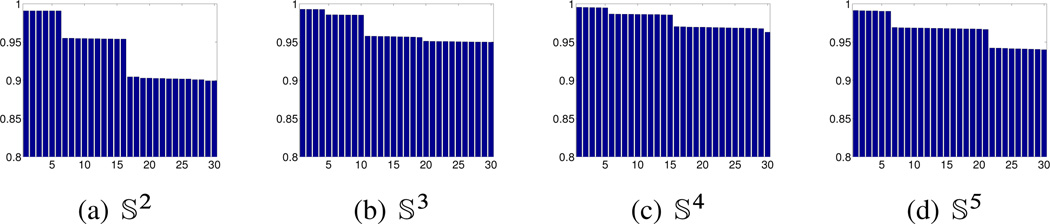
In all numerical experiments reported in this section, we use the normalized vector diffusion mapping corresponding to α = 1 in (4.9) and (4.10); that is, we use the eigenvectors of to define the VDM. In all experiments we used the kernel function K (u) = e−5u2χ[0,1] for the local PCA step as well as for the definition of the weights wij. The specific choices for ∊ and ∊PCA are detailed below. We remark that the results are not very sensitive to these choices; that is, similar results are obtained for a wide regime of parameters. The purpose of the first experiment is to numerically verify Theorem 5.3 using spheres of different dimensions. Specifically, we sampled n = 8000 points uniformly from 𝕊d embedded in ℝd+1 for d = 2, 3, 4, 5. Figure 6.1 shows bar plots of the largest 30 eigenvalues of the matrix for ∊PCA = 0:1 when d = 2, 3, 4 and ∊PCA = 0:2 when d = 5, and . It is noticeable that the eigenvalues have numerical multiplicities greater than 1. Since the connection Laplacian commutes with rotations, the dimensions of its eigenspaces can be calculated using representation theory (see Appendix C). In particular, our calculation predicted the following dimensions for the eigenspaces of the largest eigenvalues:
Figure 6.1.
Bar plots of the largest 30 eigenvalues of for n = 8000 points uniformly distributed over spheres of different dimensions.
These dimensions are in full agreement with the bar plots shown in Figure 6.1.
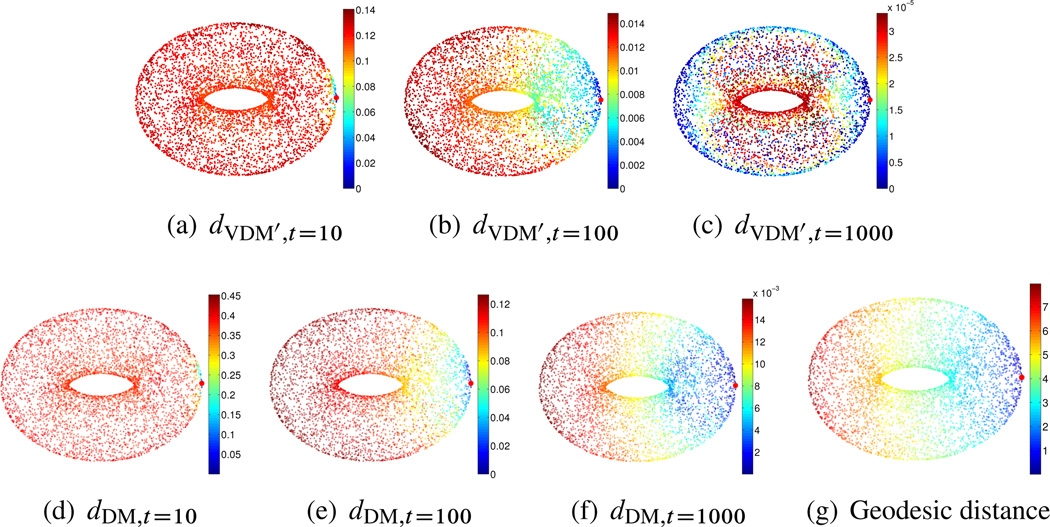
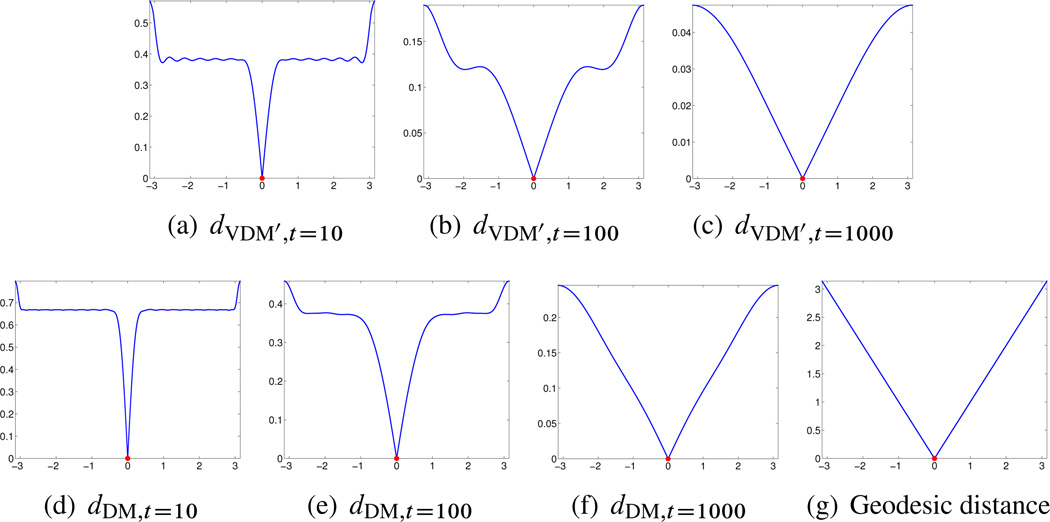
In the second set of experiments, we numerically compare the vector diffusion distance, the diffusion distance, and the geodesic distance for different compact manifolds with and without boundaries. The comparison is performed for the following four manifolds: (1) the sphere 𝕊2 embedded in ℝ3; (2) the torus 𝕋2 embedded in ℝ3; (3) the interval [−π, π] in ℝ and (4) the square [0, 2π] × [0, 2π] in ℝ2. For both VDM and DM we truncate the mappings using δ = 0.2; see (3.19). The geodesic distance is computed by the algorithm of Dijkstra on a weighted graph, whose vertices correspond to the data points, the edges link data points whose euclidean distance is less than , and the weights wG(i, j) are the euclidean distances, that is,
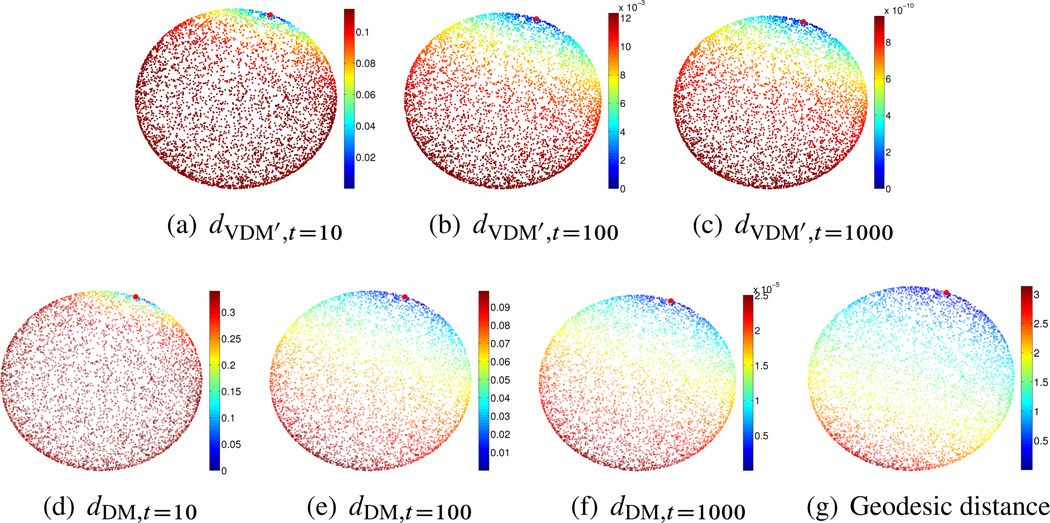
𝕊2 case: We sampled n = 5000 points uniformly from 𝕊2 = {x ∈ ℝ3 : ∥x∥ = 1} ⊂ ℝ3 and set ∊PCA = 0.1 and . For the truncated vector diffusion distance, when t = 10, we find that the number of eigenvectors whose eigenvalue is larger (in magnitude) than is mVDM = mVDM(t = 10, δ = 0.2) = 16 (recall the definition of m(t, δ) that appears after (3.19)). The corresponding embedded dimension is mVDM(mVDM + 1)/2, which in this case is 16 · 17/2 = 136. Similarly, for t = 100, mVDM = 6 (embedded dimension is 6 · 7/2 = 21), and when t = 1000, mVDM = 6 (embedded dimension is again 21). Although the first eigenspace (corresponding to the largest eigenvalue) of the connection Laplacian over 𝕊2 is of dimension 6, there are small discrepancies between the top 6 numerically computed eigenvalues, due to the finite sampling. This numerical discrepancy is amplified upon raising the eigenvalues to the tth power, when t is large, e.g., t = 1000. For demonstration purposes, we remedy this numerical effect by artificially setting λl = λl for l = 2, 3, …, 6. For the truncated diffusion distance, when t = 10, mDM = 36 (embedded dimension is 36 − 1 = 35), when t = 100, mDM = 4 (embedded dimension is 3), and when t = 1000, mDM = 4 (embedded dimension is 3). Similarly, we have the same numerical effect when t = 1000, that is, µ2, µ3, and µ4 are close but not exactly the same, so we again set µl = µ2 for l = 3, 4. The results are shown in Figure 6.2.
Figure 6.2.
𝕊2 case. Top: truncated vector diffusion distances for t = 10, t = 100, and t = 1000. Bottom: truncated diffusion distances for t = 10, t = 100, and t = 1000, and the geodesic distance. The reference point from which distances are computed is marked in red.
𝕋2 case: We sampled n = 5000 points (u, v) uniformly over the [0, 2π) × [0, 2π) square and then mapped them to ℝ3 using the following transformation, which defines the surface 𝕋2 as
Notice that the resulting sample points are not uniformly distributed over 𝕋2. Therefore, the usage of S1 and D1 instead of S and D is important if we want the eigenvectors to approximate the eigenvector fields of the connection Laplacian over 𝕋2. We used ∊PCA = 0:2 and , and find that for the truncated vector diffusion distance, when t = 10, the embedded dimension is 2628, when t = 100, the embedded dimension is 36, and when t = 1000, the embedded dimension is 3. For the truncated diffusion distance, when t = 10, the embedded dimension is 130; when t = 100, the embedded dimension is 14; and when t = 1000, the embedded dimension is 2. The results are shown in Figure 6.3.
Figure 6.3.
𝕋2 case. Top: truncated vector diffusion distances for t = 10, t = 100, and t = 1000. Bottom: truncated diffusion distances for t = 10, t = 100, and t = 1000, and the geodesic distance. The reference point from which distances are computed is marked in red.
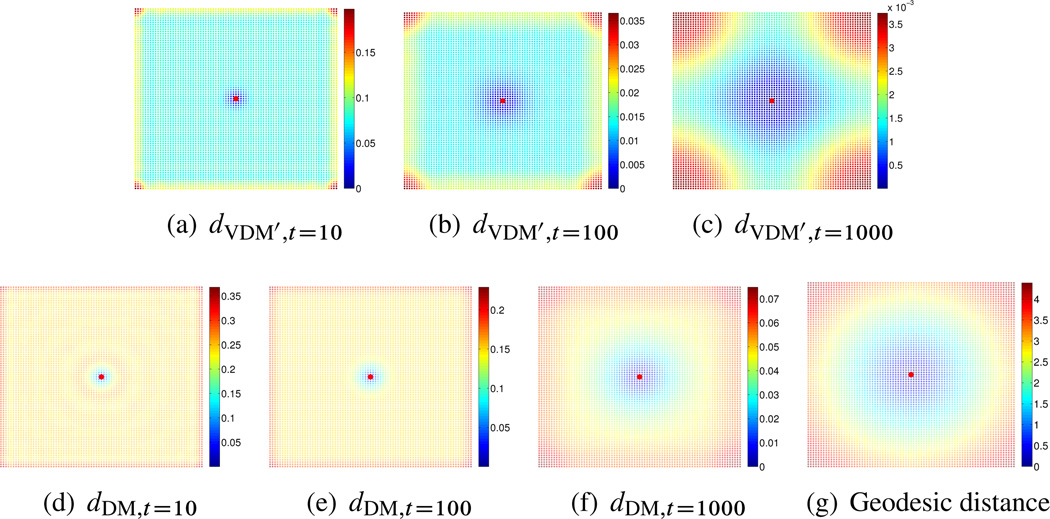
One-dimensional interval case: We sampled n = 5000 equally spaced grid points from the interval [−π, π] ⊂ ℝ1 and set ∊PCA = 0.01 and . For the truncated vector diffusion distance, when t = 10, the embedded dimension is 120, when t = 100, the embedded dimension is 15, and when t = 1000, the embedded dimension is 3. For the truncated diffusion distance, when t = 10, the embedded dimension is 36, when t = 100, the embedded dimension is 11, and when t = 1000, the embedded dimension is 3. The results are shown in Figure 6.4.
Figure 6.4.
One-dimensional interval case. Top: truncated vector diffusion distances for t = 10, t = 100, and t = 1000. Bottom: truncated diffusion distances for t = 10, t = 100, and t = 1000, and the geodesic distance. The reference point from which distances are computed is marked in red.
Square case: We sampled n = 6561 = 812 equally spaced grid points from the square [0, 2π] × [0, 2π] and fixed ∊PCA = 0:01 and . For the truncated vector diffusion distance, when t = 10, the embedded dimension is 20;100 (we only calculate the first 200 eigenvalues); when t = 100, the embedded dimension is 1596; and when t = 1000, the embedded dimension is 36. For the truncated diffusion distance, when t = 10, the embedded dimension is 200 (we only calculate the first 200 eigenvalues); when t = 100, the embedded dimension is 200; and when t = 1000, the embedded dimension is 28. The results are shown in Figure 6.5.
Figure 6.5.
Square case. Top: truncated vector diffusion distances for t = 10, t = 100, and t = 1000. Bottom: truncated diffusion distances for t = 10, t = 100, and t = 1000, and the geodesic distance. The reference point from which distances are computed is marked in red.
7 Out-of-Sample Extension of Vector Fields
Let so that 𝒳, 𝒴 ⊂ ℳd where ℳ is embedded in ℝp by ι. Suppose X is a smooth vector field that we observe only on 𝒳 and want to extend to 𝒴. That is, we observe the vectors ι*X(x1), ι*X(x2), …, ι*X(xn) ∈ ℝp and want to estimate ι*X(y1), ι*X(y2), …, ι*X(ym). The set 𝒳 is assumed to be fixed, while the points in 𝒴 may arrive on the fly and need to be processed in real time. We propose the following Nyström scheme for extending X from 𝒳 to 𝒴.
In the preprocessing step we use the points x1, x2; …, xn for local PCA, alignment, and vector diffusion mapping as described in Sections 2 and 3. That is, using local PCA, we find the p × d matrices Oi (i = 1, 2, …, n) such that the columns of Oi are an orthonormal basis for a subspace that approximates the embedded tangent plane ι*Txiℳ; using alignment, we find the orthonormal d × d matrices Oij that approximate the parallel transport operator from Txjℳ to Txiℳ; and using wij and Oij we construct the matrices S and D and compute (a subset of) the eigenvectors v1, v2, …, vnd and eigenvalues λ1, λ2, …, λnd of D−1S.
We project the embedded vector field ι*X(xi) ∈ ℝp into the d-dimensional subspace spanned by the columns of Oi and define Xi ∈ ℝd as
| (7.1) |
We represent the vector field X on 1D4B3 by the vector x of length nd, organized as n vectors of length d, with
We use the orthonormal basis of eigenvector fields v1; v2, …, vnd to decompose x as
| (7.2) |
where al = xΤvl. This concludes the preprocessing computations.
Suppose y ∈ 𝒴 is a “new” out-of-sample point. First, we perform the local PCA step to find a p × d matrix, denoted Oy whose columns form an orthonormal basis to a d-dimensional subspace of ℝp that approximates the embedded tangent plane ι*Tyℳ. The local PCA step uses only the neighbors of y among the points in 𝒳 (but not in 𝒴) inside a ball of radius centered at y.
Next, we use the alignment process to compute the d × d orthonormal matrix Oy,i between xi and y by setting
Notice that the eigenvector fields satisfy
We denote the extension of vl to the point y by ṽl(y) and define it as
| (7.3) |
To finish the extrapolation problem, we denote the extension of x to y by x̃(y) and define it as
| (7.4) |
where m(δ) = maxl|λl| > and > 0 is some fixed parameter to ensure the numerical stability of the extension procedure (due to the division by λl in (7.3); 1/δ can be regarded as the condition number of the extension procedure).
8 The Continuous Case: Heat Kernels
As discussed earlier, in the limit n → ∞ and ∊ ∞ 0 considered in (5.2), the normalized graph Laplacian converges to the Laplace-Beltrami operator, which is the generator of the heat kernel for functions (0-forms). Similarly, in the limit n → ∞ considered in (5.3), we get the connection Laplacian operator, which is the generator of a heat kernel for vector fields (or 1-forms). The connection Laplacian ▽2 is a self-adjoint, second-order elliptic operator defined over the tangent bundle Tℳ. It is well-known [16] that the spectrum of ▽2 is discrete inside ℝ−, and the only possible accumulation point is −∞. We will denote the spectrum as , where 0 ≤ λ0 ≤ λ1 ≤ …. From classical elliptic theory (see, for example, [16]), we know that et▽2 has the kernel
where ▽2Xn = −λnXn. Also, the eigenvector fields Xn of ▽2 form an orthonormal basis of L2(Tℳ). In the continuous setup, we define the vector diffusion distance between x; y ∈ ℳ using . An explicit calculation gives
| (8.1) |
It is well-known that the heat kernel kt(x, y) is smooth in x and y and analytic in t [16], so for t > 0 we can define a family of vector diffusion mappings Vt that map any x ∈ ℳ into the Hilbert space ℓ2 by
| (8.2) |
which satisfies
| (8.3) |
The vector diffusion distance dVDM,t (x, y) between x ∈ M and y ∈ M is defined as
| (8.4) |
which is clearly a distance function over ℳ. In practice, due to the decay of e−(λn+λm)t only pairs (n, m) for which λn + λm is not too large are needed to get a good approximation of this vector diffusion distance. As in the discrete case, the dot products 〈Xn (x), Xm(x)〉 are invariant to the choice of basis for the tangent space at x.
We now study some properties of the vector diffusion map Vt (8.2). First, we claim that for all t > 0, the vector diffusion mapping Vt is an embedding of the compact Riemannian manifold ℳ into ℓ2.
THEOREM 8.1. Given a d-dimensional closed Riemannian manifold (ℳ, g) and an orthonormal basis composed of the eigenvector fields of the connection Laplacian ▽2, then for any t > 0, the vector diffusion map Vt is a diffeomorphic embedding of ℳ in ℓ2.
PROOF. We show that Vt: ℳ → ℓ2 is continuous in x by noting that
| (8.5) |
From the continuity of the kernel kt(x, y) it is clear that as y → x. Since ℳ is compact, it follows that Vt(ℳ) is compact in ℓ2. Finally, we show that Vt is one-to-one. Fix x ≠ y and a smooth vector field X that satisfies 〈X(x),X(x)〉 ≠ 〈X(y), X(y)〉. Since the eigenvector fields form a basis for L2 (Tℳ), we have
where cn = ∫ℳ〈X,Xn〉dV. As a result,
Since 〈X(x), X(x)〉 ≠ 〈X(y), X(y)〉, there exist n, m ∈ ℕ such that
which shows that Vt (x) ≠ Vt (y); i.e., Vt is one-to-one. From the fact that the map Vt is continuous and one-to-one from ℳ, which is compact, onto Vt (ℳ), we conclude that Vt is an embedding.
Next, we demonstrate the asymptotic behavior of the vector diffusion distance dVDM,t(x, y) and the diffusion distance dDM,t (x, y) when t is small and x is close to y. The following theorem shows that in this asymptotic limit both the vector diffusion distance and the diffusion distance behave like the geodesic distance.
THEOREM 8.2. Let (ℳ, g) be a smooth d-dimensional closed Riemannian manifold. Suppose x, y ∈ ℳ so that x = expy(v), where v ∈ Ty ℳ. For any t > 0, when ∥v∥2 ≪ t ≪ 1 we have the following asymptotic expansion of the vector diffusion distance:
Similarly, when ∥v∥2 ≪ t ≪ 1, we have the following asymptotic expansion of the diffusion distance:
PROOF. Fixing y and a normal coordinate around y, we define j(x, y) = |det(dv expy)|, where x = expy(v), v ∈ Txℳ. Suppose ∥v∥ is small enough so that x = expy(v) is away from the cut locus of y. It is well-known that the heat kernel kt(x, y) for the connection Laplacian ▽2 over the vector bundle ℰ possesses the following asymptotic expansion when x and y are close [5, p. 84] or [11]:
| (8.6) |
, where ∥·∥l is the Cl norm,
| (8.7) |
N > d/2, and Φi is a smooth section of the vector bundle ℰ ⊗ ℰ over ℳ × ℳ. Moreover, Φ0(x, y) = Px, y is the parallel transport from ℰy to ℰx. In the VDM setup, we take ℰ = Tℳ, the tangent bundle of ℳ. Also, by [5, prop. 1.28], we have the following expansion:
| (8.8) |
Equations (8.7) and (8.8) lead to the following expansion under the assumption ∥v∥2 ≪ t:
In particular, for ∥v∥ = 0 we have
Thus, for ∥v∥2 ≪ t ≪ 1, we have
| (8.9) |
By the same argument we can carry out the asymptotic expansion of the diffusion distance dDM,t (x, y). Denote the eigenfunctions and eigenvalues of the Laplace-Beltrami operator Δ by φn and µn. We can rewrite the diffusion distance as follows:
| (8.10) |
where k̃t is the heat kernel of the Laplace-Beltrami operator. Note that the Laplace- Beltrami operator is equal to the connection Laplacian operator defined over the trivial line bundle over ℳ. As a result, equation (8.7) also describes the asymptotic expansion of the heat kernel for the Laplace-Beltrami operator as
Putting these facts together, we obtain
| (8.11) |
, when ∥v∥2 ≪ t ≪ 1.
9 Application of VDM to Cryo-Electron Microscopy
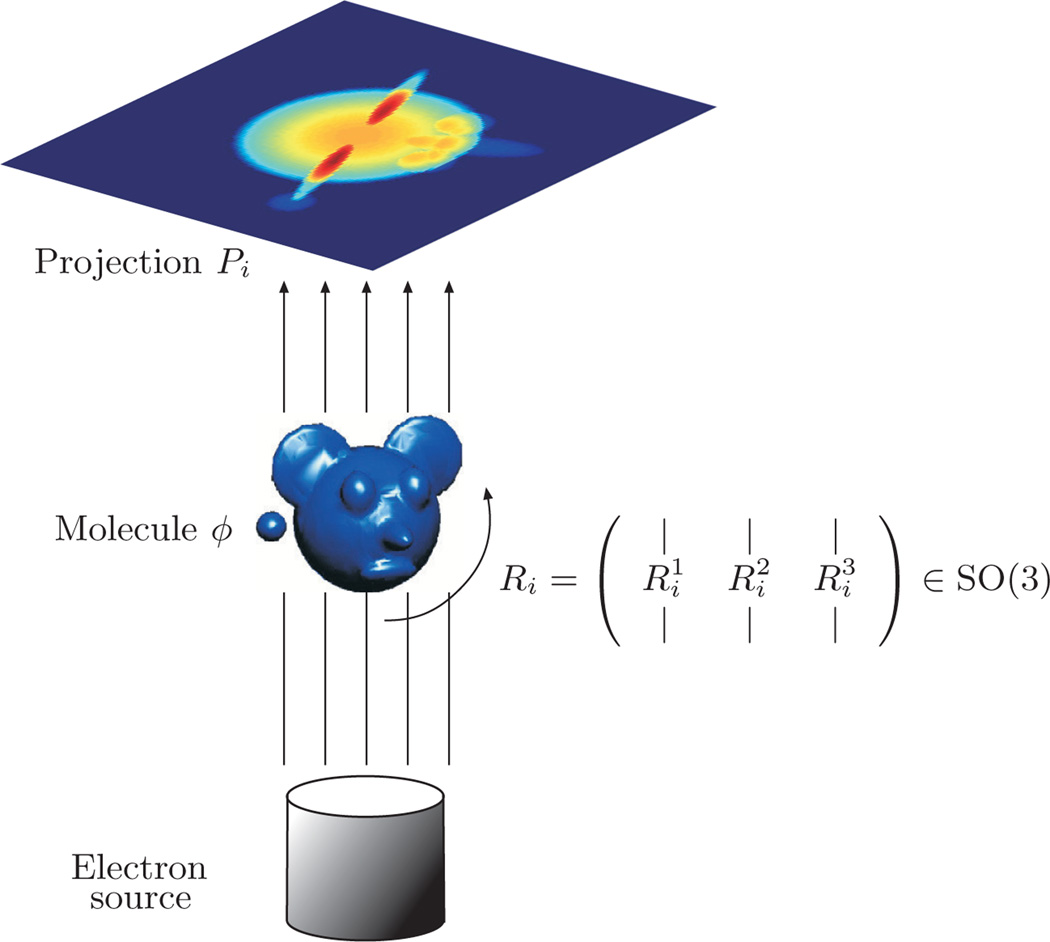
In addition to being a general framework for data analysis and manifold learning, VDM is useful for performing robust multireference rotational alignment of objects, such as one-dimensional periodic signals, two-dimensional images, and three-dimensional shapes. In this section, we briefly describe the application of VDM to a particular multireference rotational alignment problem of two-dimensional images that arise in the field of cryo-electron microscopy (cryo-EM). A more comprehensive study of this problem can be found in [19, 37]. It can be regarded as a prototypical multireference alignment problem, and we expect many other multireference alignment problems that arise in areas such as computer vision and computer graphics to benefit from the proposed approach.
The goal in cryo-EM [14] is to determine three-dimensional macromolecular structures from noisy projection images taken at unknown random orientations by an electron microscope, i.e., a random computational tomography (CT). Determining three-dimensional macromolecular structures for large biological molecules remains vitally important, as witnessed, for example, by the 2003 Chemistry Nobel Prize, co-awarded to R. MacKinnon for resolving the three-dimensional structure of the Shaker K+ channel protein, and by the 2009 Chemistry Nobel Prize, awarded to V. Ramakrishnan, T. Steitz, and A. Yonath for studies of the structure and function of the ribosome. The standard procedure for structure determination of large molecules is X-ray crystallography. The challenge in this method is often more in the crystallization itself than in the interpretation of the X-ray results, since many large proteins have so far withstood all attempts to crystallize them.
In cryo-EM, an alternative to X-ray crystallography, the sample of macromolecules is rapidly frozen in an ice layer so thin that their tomographic projections are typically disjoint; this seems the most promising alternative for large molecules that defy crystallization. The cryo-EM imaging process produces a large collection of tomographic projections of the same molecule, corresponding to different and unknown projection orientations. The goal is to reconstruct the three-dimensional structure of the molecule from such unlabeled projection images, where data sets typically range from 104 to 105 projection images whose size is roughly 100 × 100 pixels. The intensity of the pixels in a given projection image is proportional to the line integrals of the electric potential induced by the molecule along the path of the imaging electrons (see Figure 9.1). The highly intense electron beam destroys the frozen molecule, and it is therefore impractical to take projection images of the same molecule at known different directions as in the case of classical CT. In other words, a single molecule can be imaged only once, rendering an extremely low signal-to-noise ratio (SNR) for the images (see Figure 9.2 for a sample of real microscope images), mostly due to shot noise induced by the maximal allowed electron dose (other sources of noise include the varying width of the ice layer and partial knowledge of the contrast function of the microscope). In the basic homogeneity setting considered hereafter, all imaged molecules are assumed to have the exact same structure; they differ only by their spatial rotation. Every image is a projection of the same molecule but at an unknown random three-dimensional rotation, and the cryo-EM problem is to find the three-dimensional structure of the molecule from a collection of noisy projection images.
Figure 9.1.
Schematic drawing of the imaging process: every projection image corresponds to some unknown three-dimensional rotation of the unknown molecule.
Figure 9.2.

A collection of four real electron microscope images of the E. coli 50S ribosomal subunit; courtesy of Dr. Fred Sigworth (Yale Medical School).
The rotation group SO(3) is the group of all orientation-preserving orthogonal transformations about the origin of the three-dimensional euclidean space ℝ3 under the operation of composition. Any three-dimensional rotation can be expressed using a 3 × 3 orthogonal matrix
satisfying RRΤ = RΤR = I3×3 and det R = 1. The column vectors R1, R2, R3 of R form an orthonormal basis to ℝ3. To each projection image P there corresponds a 3 × 3 unknown rotation matrix R describing its orientation (see Figure 9.1). Excluding the contribution of noise, the intensity P(x, y) of the pixel located at (x, y) in the image plane corresponds to the line integral of the electric potential induced by the molecule along the path of the imaging electrons, that is,
| (9.1) |
where φ : ℝ3 ↦ ℝ is the electric potential of the molecule in some fixed “laboratory” coordinate system. The projection operator (9.1) is also known as the X-ray transform [29].
We therefore identify the third column R3 of R as the imaging direction, also known as the viewing angle of the molecule. The first two columns R1 and R2 form an orthonormal basis for the plane in ℝ3 perpendicular to the viewing angle R3. All clean projection images of the molecule that share the same viewing angle look the same up to some in-plane rotation. That is, if Ri and Rj are two rotations with the same viewing angle are two orthonormal bases for the same plane. On the other hand, two rotations with opposite viewing angles give rise to two projection images that are the same after reflection (mirroring) and some in-plane rotation.
As projection images in cryo-EM have extremely low SNR, a crucial initial step in all reconstruction methods is “class averaging” [14, 41]. Class averaging is the grouping of a large data set of n noisy raw projection images P1, P2; …, Pn into clusters such that images within a single cluster have similar viewing angles (it is possible to artificially double the number of projection images by including all mirrored images). Averaging rotationally aligned noisy images within each cluster results in “class averages”; these are images that enjoy a higher SNR and are used in later cryo-EM procedures such as the angular reconstitution procedure [40] that requires better-quality images. Finding consistent class averages is challenging due to the high level of noise in the raw images as well as the large size of the image data set. A sketch of the class-averaging procedure is shown in Figure 9.3.
Figure 9.3.
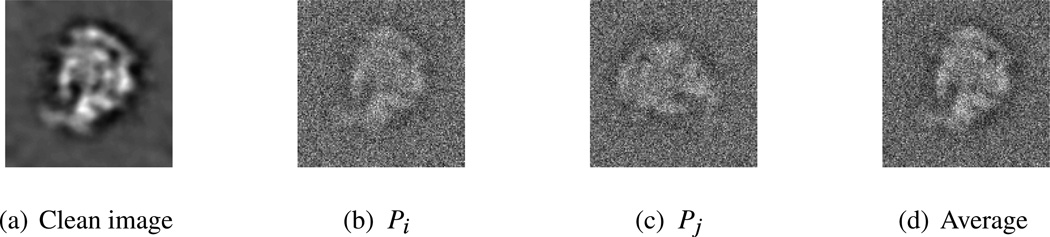
(a) A clean simulated projection image of the ribosomal subunit generated from its known volume. (b) Noisy instance of (a), denoted Pi, obtained by the addition of white Gaussian noise. For the simulated images we chose the SNR to be higher than that of experimental images in order for image features to be clearly visible. (c) Noisy projection, denoted Pj, taken at the same viewing angle but with a different in-plane rotation. (d) Averaging the noisy images (b) and (c) after in-plane rotational alignment. The class average of the two images has a higher SNR than that of the noisy images (b) and (c), and it has better similarity with the clean image (a).
Penczek, Zhu, and Frank [31] introduced the rotationally invariant K-means clustering procedure to identify images that have similar viewing angles. Their rotationally invariant distance dRID(i, j) between image Pi and image Pj is defined as the euclidean distance between the images when they are optimally aligned with respect to in-plane rotations (assuming the images are centered)
| (9.2) |
, where R(θ) is the rotation operator of an image by an angle θ in the counterclockwise direction. Prior to computing the invariant distances of (9.2), a common practice is to center all images by correlating them with their total average , which is approximately radial (i.e., has little angular variation) due to the randomness in the rotation. The resulting centers usually miss the true centers by only a few pixels (as can be validated in simulations during the refinement procedure). Therefore, like [31], we also choose to focus on the more challenging problem of rotational alignment by assuming that the images are properly centered, while the problem of translational alignment can be solved later by solving an overdetermined linear system.
It is worth noting that the specific choice of metric to measure proximity between images can make a big difference in class averaging. The cross-correlation and euclidean distance (9.2) are by no means optimal measures of proximity. In practice, it is common to denoise the images prior to computing their pairwise distances. Although the discussion that follows is independent of the particular choice of filter or distance metric, we emphasize that filtering can have a dramatic effect on finding meaningful class averages.
The invariant distance between noisy images that share the same viewing angle (with perhaps a different in-plane rotation) is expected to be small. Ideally, all neighboring images of some reference image Pi in a small invariant distance ball centered at Pi should have similar viewing angles, and averaging such neighboring images (after proper rotational alignment) would amplify the signal and diminish the noise.
Unfortunately, due to the low SNR, it often happens that two images of completely different viewing angles have a small invariant distance. This can happen when the realizations of the noise in the two images match well for some random in-plane rotational angle, leading to spurious neighbor identification. Therefore, averaging the nearest-neighbor images can sometimes yield a poor estimate of the true signal in the reference image.
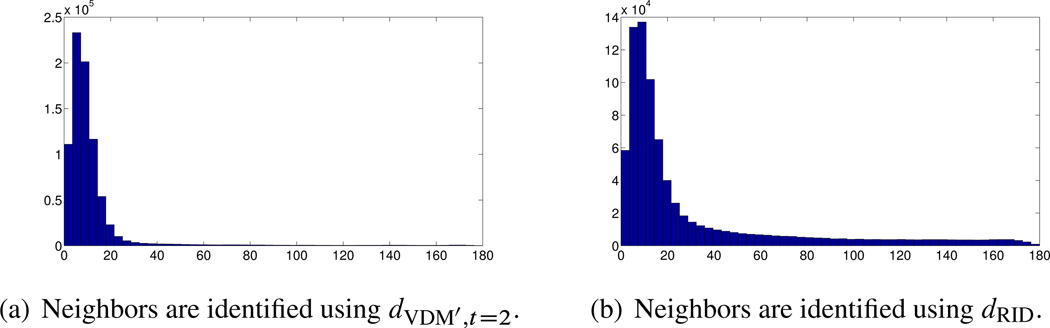
The histograms of Figure 9.5 demonstrate the ability of small rotationally invariant distances to identify images with similar viewing directions. For each image we use the rotationally invariant distances to find its 40 nearest neighbors among the entire set of n = 40;000 images. In our simulation we know the original viewing directions, so for each image we compute the angles (in degrees) between the viewing direction of the image and the viewing directions of its 40 neighbors. Small angles indicate successful identification of “true” neighbors that belong to a small spherical cap, while large angles correspond to outliers. We see that for there are no outliers, and all the viewing directions of the neighbors belong to a spherical cap whose opening angle is about 8°. However, for lower values of the SNR, there are outliers, indicated by arbitrarily large angles (all the way to 180°).
Figure 9.5.
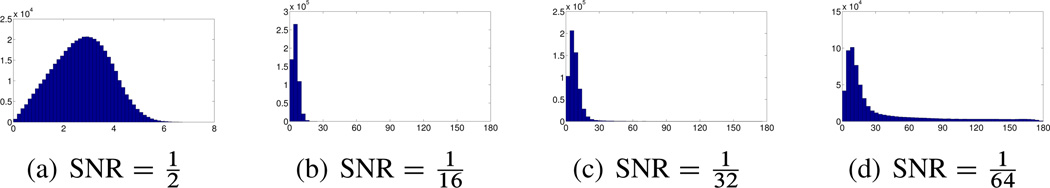
Histograms of the angle (in degrees, x-axis) between the viewing directions of 40,000 images and the viewing directions of their 40 nearest neighboring images as found by computing the rotationally invariant distances. (Courtesy of Zhizhen Zhao, Princeton University)
Clustering algorithms, such as the K-means algorithm, perform much better than naive nearest-neighbors averaging, because they take into account all pairwise distances, not just distances to the reference image. Such clustering procedures are based on the philosophy that images that share a similar viewing angle with the reference image are expected to have a small invariant distance not only to the reference image but also to all other images with similar viewing angles. This observation was utilized in the rotationally invariant K-means clustering algorithm [31]. Still, due to noise, the rotationally invariant K-means clustering algorithm may suffer from misidentifications at the low SNR values present in experimental data.
VDM is a natural algorithmic framework for the class-averaging problem, as it can further improve the detection of neighboring images even at lower SNR values. The rotationally invariant distance neglects an important piece of information, namely, the optimal angle that realizes the best rotational alignment in (9.2):
| (9.3) |
In VDM, we use the optimal in-plane rotation angles θij to define the orthogonal transformations Oij and to construct the matrix S in (3.1). The eigenvectors and eigenvalues of D−1S (other normalizations of S are also possible) are then used to define the vector diffusion distances between images.
This VDM based classification method has proven to be quite powerful in practice. We applied it to a set of n = 40;000 noisy images with . For every image we found the 40 nearest neighbors using the vector diffusion metric. In the simulation we knew the viewing directions of the images, and we computed for each pair of neighbors the angle (in degrees) between their viewing directions. The histogram of these angles is shown in Figure 9.6 (left panel). About 92% of the identified images belong to a small spherical cap of opening angle 20° whereas this percentage is only about 65% when neighbors are identified by the rotationally invariant distances (right panel). We remark that for , the percentage of correctly identified images by the VDM method goes up to about 98%.
Figure 9.6.
: Histogram of the angles (x-axis, in degrees) between the viewing directions of each image (out of 40;000) and its 40 neighboring images. Left: neighbors are post-identified using vector diffusion distances. Right: neighbors are identified using the original rotationally invariant distances dRID.
The main advantage of the algorithm presented here is that it successfully identifies images with similar viewing angles even in the presence of a large number of spurious neighbors, that is, even when many pairs of images with viewing angles that are far apart have relatively small rotationally invariant distances. In other words, the VDM-based algorithm is shown to be robust to outliers.
10 Summary and Discussion
This paper introduced vector diffusion maps, an algorithmic and mathematical framework for analyzing data sets where scalar affinities between data points are accompanied by orthogonal transformations. The consistency among the orthogonal transformations along different paths that connect any fixed pair of data points is used to define an affinity between them. We showed that this affinity is equivalent to an inner product, giving rise to the embedding of the data points in a Hilbert space and to the definition of distances between data points, to which we referred as vector diffusion distances.
For data sets of images, the orthogonal transformations and the scalar affinities are naturally obtained via the procedure of optimal registration. The registration process seeks to find the optimal alignment of two images over some class of transformations (also known as deformations), such as rotations, reflections, translations, and dilations. For the purpose of vector diffusion mapping, we extract from the optimal deformation only the corresponding orthogonal transformation (rotation and reflection). We demonstrated the usefulness of the vector diffusion map framework in the organization of noisy cryo-electron microscopy images, an important step towards resolving three-dimensional structures of macromolecules. Optimal registration is often used in various mainstream problems in computer vision and computer graphics, for example, in optimal matching of three-dimensional shapes. We therefore expect the vector diffusion map framework to become a useful tool in such applications.
In the case of manifold learning, where the data set is a collection of points in a high-dimensional euclidean space, but with a low-dimensional Riemannian manifold structure, we detailed the construction of the orthogonal transformations via the optimal alignment of the orthonormal bases of the tangent spaces. These bases are found using the classical procedure of PCA. Under certain mild conditions about the sampling process of the manifold, we proved that the orthogonal transformation obtained by the alignment procedure approximates the parallel transport operator between the tangent spaces. The proof required careful analysis of the local PCA step, which we believe is interesting in its own right. Furthermore, we proved that if the manifold is sampled uniformly, then the matrix that lies at the heart of the vector diffusion map framework approximates the connection Laplacian operator. Following spectral graph theory terminology, we call that matrix the connection Laplacian of the graph. Using different normalizations of the matrix we proved convergence to the connection Laplacian operator also for the case of nonuniform sampling. We showed that the vector diffusion mapping is an embedding and proved its relation with the geodesic distance using the asymptotic expansion of the heat kernel for vector fields. These results provide the mathematical foundation for the algorithmic framework that underlies the vector diffusion mapping.
We expect many possible extensions and generalizations of the vector diffusion mapping framework. We conclude by mentioning a few of them.
Topology of the data. In [36] we showed how the vector diffusion mapping can determine if a manifold is orientable or nonorientable, and in the latter case to embed its double covering in a euclidean space. To that end we used the information in the determinant of the optimal orthogonal transformation between bases of nearby tangent spaces. In other words, we used just the optimal reflection between two orthonormal bases. This simple example shows that vector diffusion mapping can be used to extract topological information from the point cloud. We expect more topological information can be extracted using appropriate modifications of the vector diffusion mapping.
Hodge and higher-order Laplacians. Using tensor products of the optimal orthogonal transformations it is possible to construct higher-order connection Laplacians that act on p-forms (p ≥ 1). The index theorem [16] relates topological structure with geometrical structure. For example, the so-called Betti numbers are related to the multiplicities of the harmonic p-forms of the Hodge Laplacian. For the extraction of topological information it would therefore be useful to modify our construction in order to approximate the Hodge Laplacian instead of the connection Laplacian.
Multiscale, sparse, and robust PCA. In the manifold learning case, an important step of our algorithm is local PCA for estimating the bases for tangent spaces at different data points. In the description of the algorithm, a single-scale parameter ∊PCA is used for all data points. It is conceivable that a better estimation can be obtained by choosing a different, location-dependent scale parameter. A better estimation of the tangent space Txiℳ may be obtained by using a location-dependent scale parameter ∊PCA,i due to several reasons: nonuniform sampling of the manifold, varying curvature of the manifold, and global effects such as different pieces of the manifold that are almost touching at some points (i.e., varying the “condition number” of the manifold). Choosing the correct scale was recently considered in [28], where a multiscale approach was taken to resolve the optimal scale. We recommend the incorporation of such multiscale PCA approaches into the vector diffusion mapping framework. Another difficulty that we may face when dealing with real-life data sets is that the underlying assumption about the data points being located exactly on a low-dimensional manifold does not necessarily hold. In practice, the data points are expected to reside off the manifold, either due to measurement noise or due to the imperfection of the low-dimensional manifold model assumption. It is therefore necessary to estimate the tangent spaces in the presence of noise. Noise is a limiting factor for successful estimation of the tangent space, especially when the data set is embedded in a high-dimensional space and noise affects all coordinates [23]. We expect recent methods for robust PCA [7] and sparse PCA [6, 24] to improve the estimation of the tangent spaces and as a result to become useful in the vector diffusion map framework.
Random matrix theory and noise sensitivity. The matrix S that lies at the heart of the vector diffusion map is a block matrix whose blocks are either d × d orthogonal matrices Oij or the zero blocks. We anticipate that for some applications the measurement of Oij would be imprecise and noisy. In such cases, the matrix S can be viewed as a random matrix, and we expect tools from random matrix theory to be useful in analyzing the noise sensitivity of its eigenvectors and eigenvalues. The noise model may also allow for outliers, for example, orthogonal matrices that are uniformly distributed over the orthogonal group O(d) (according to the Haar measure). Notice that the expected value of such random orthogonal matrices is 0, which leads to robustness of the eigenvectors and eigenvalues even in the presence of a large number of outliers (see, for example, the random matrix theory analysis in [35]).
Compact and noncompact groups and their matrix representation. As mentioned earlier, the vector diffusion mapping is a natural framework to organize data sets for which the affinities and transformations are obtained from an optimal alignment process over some class of transformations (deformations). In this paper we focused on utilizing orthogonal transformations. At this point the reader has probably asked herself the following question: Is the method limited to orthogonal transformations, or is it possible to utilize other groups of transformations such as translations, dilations, and more? We note that the orthogonal group O(d) is a compact group that has a matrix representation and remark that the vector diffusion mapping framework can be extended to such groups of transformations without much difficulty. However, the extension to noncompact groups, such as the euclidean group of rigid transformation, the general linear group of invertible matrices, and the special linear group is less obvious. Such groups arise naturally in various applications, rendering the importance of extending the vector diffusion mapping to the case of noncompact groups.
Figure 2.1.
The orthonormal basis of the tangent plane Txiℳ is determined by local PCA using data points inside a euclidean ball of radius centered at xi. The bases for Txiℳ and Txjℳ are optimally aligned by an orthogonal transformation Oij that can be viewed as a mapping from Txjℳ to Txiℳ.
Figure 9.4.
Simulated projection with various levels of additive Gaussian white noise.
Acknowledgment
A. Singer was partially supported by grant DMS-0914892 from the National Science Foundation, by grant FA9550-09-1-0551 from the Air Force Office of Scientific Research, by grant R01GM090200 from the National Institute of General Medical Sciences, and by the Alfred P. Sloan Foundation. H.-T. Wu acknowledges support by Federal Highway Administration Grant DTFH61-08-C-00028. The authors would like to thank Charles Fefferman for various discussions regarding this work and the anonymous referee for his useful comments and suggestions. They also express gratitude to the audiences of the seminars at Tel Aviv University, the Weizmann Institute of Science, Princeton University, Yale University, Stanford University, University of Pennsylvania, Duke University, Johns Hopkins University, and the Air Force, where parts of this work were presented in 2010 and 2011.
Appendix A
Some Differential Geometry Background
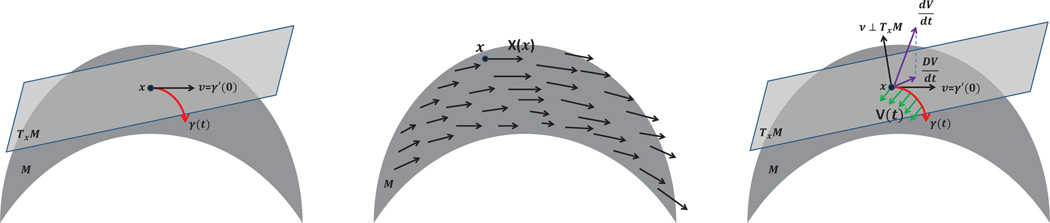
The purpose of this appendix is to provide the required mathematical background for readers who are not familiar with concepts such as the parallel transport operator, connection, and the connection Laplacian. We illustrate these concepts by considering a surface ℳ embedded in ℝ3.
Given a function f (x) : ℝ3 → ℝ, its gradient vector field is given by
Through the gradient, we can find the rate of change of f at x ∈ ℝ3 in a given unit vector v ∈ ℝ3, using the directional derivative:
Define ▽vf(x) := ▽f(x)(v).
Let X be a vector field on ℝ3,
It is natural to extend the derivative notion to a given vector field X at x ∈ ℝ3 by mimicking the derivative definition for functions in the following way:
| (A.1) |
where v ∈ ℝ3. Following the same notation for the directional derivative of a function, we denote this limit by ▽vX(x). This quantity tells us that at x, following the direction v, we compare the vector field at two points x and x + tv, and see how the vector field changes. While this definition looks good at first sight, we now explain that it has certain shortcomings that need to be fixed in order to generalize it to the case of a surface embedded in ℝ3.
Consider a two-dimensional smooth surface ℳ embedded in ℝ3 by ι. Fix a point x ∈ ℳ and a smooth curve γ(t) : (−∊, ∊); → ℳ ⊂ ℝ3 where ∊ ≪ 1 and γ(0) = x. We call γʹ(0) ∈ ℝ3 a tangent vector to ℳ at x. The two-dimensional subspace in ℝ3 spanned by the collection of all tangent vectors to ℳ at x is defined to be the tangent plane at x and denoted by Txℳ;7 see Figure A.1 (left panel). Having defined the tangent plane at each point x ∈ ℳ, we define a vector field X over ℳ to be a differentiable map that maps x to a tangent vector in Txℳ.8
Figure A.1.
Left: a tangent plane and a curve γ. Middle: a vector field. Right: the covariant derivative.
We now generalize the definition of the derivative of a vector field over ℝ3 (A.1) to define the derivative of a vector field over ℳ. The first difficulty we face is how to make sense of “X(x +tv),” since x + tv does not belong to ℳ. This difficulty can be tackled easily by changing the definition (A.1) a bit by considering the curve γ: (−∊, ∊) → ℳ ⊂ ℝ3 so that γ(0) = x and γʹ(0) = v. Thus, (A.1) becomes
| (A.2) |
where v ∈ ℝ3. In ℳ, the existence of the curve γ: (−∊, ∊) → ℝ3 and γ(0) = x and γʹ(0) = v is guaranteed by the classical ordinary differential equation theory. However, (A.2) still cannot be generalized to ℳ directly even though X(γ(t)) is well-defined. The difficulty we face here is how to compare X(γ(t)) and X(x), that is, how to make sense of the subtraction X(γ(t)) − X(γ(0)). It is not obvious since a priori we do not know how Tγ(t)ℳ and Tγ(0)ℳ are related. The way we proceed is by defining an important notion in differential geometry called “parallel transport,” which plays an essential role in our VDM framework.
Fix a point x ∈ ℳ and a vector field X on ℳ, and consider a parametrized curve γ: (−∊, ∊) → ℳ so that γ(0) = x. Define a vector-valued function V : (−∊, ∊) → ℝ3 by restricting X to γ, that is, V(t) = X(γ(t)). The derivative of V is well-defined as usual:
where h ∈ (−∊; ∊). The covariant derivative is defined as the projection of onto Tγ(h)ℳ. Then, using the definition of , we consider the following equation:
where w ∈ Tγ(0)ℳ. The solution W(t) exists by the classical ordinary differential equation theory. The solution W(t) along γ(t) is called the parallel vector field along the curve γ(t), and we also call W(t) the parallel transport of w along the curve γ(t) and denote W(t) = Pγ(t),γ (0)w.
We come back to address the initial problem: how to define the “derivative” of a given vector field over a surface ℳ. We define the covariant derivative of a given vector field X over ℳ as follows:
| (A.3) |
where γ: (−∊, ∊) → ℳ with γ(0) = x ∈ ℳ, γʹ (0) = v ∈ Tγ(0)ℳ. This definition says that if we want to analyze how a given vector field at x ∈ ℳ changes along the direction v, we choose a curve γ so that γ(0) = x and γʹ(0) = v, and then “transport” the vector field value at point γ(t) to γ(0) = x so that the comparison of the two tangent planes makes sense. The key point of the whole story is that without applying parallel transport to transport the vector at point γ(t) to Tγ(0)ℳ, then the subtraction X(γ(t)) – X(γ(0)) ∈ ℝ3 in general does not live on Txℳ, which distorts the notion of derivative. For comparison, let us reconsider definition (A.1). Since at each point x ∈ ℝ3, the tangent plane at x is Txℝ3 = ℝ3, the subtraction X(x + tv) – X(x) always makes sense. To be more precise, the true meaning of X(x+tv) is Pγ(0),γ(t)X(γ(t)), where Pγ(0),γ(t) = id, and γ(t) = x + tv.
With the above definition, when X and Y are two vector fields on ℳ, we define ▽XY to be a new vector field on ℳ so that
Note that X(x) ∈ Txℳ. We call ▽ a connection on ℳ. (The notion of connection can be quite general. For our purposes, this definition is sufficient.)
Once we know how to differentiate a vector field over ℳ, it is natural to consider the second-order differentiation of a vector field. The second-order differentiation of a vector field is a natural notion in ℝ3. For example, we can define a second-order differentiation of a vector field X over ℝ3 as follows:
| (A.4) |
where x, y, z are standard unit vectors corresponding to the three axes. This definition can be generalized to a vector field over ℳ as follows:
| (A.5) |
where X is a vector field over ℳ, x ∈ ℳ, and E1, E2 are two vector fields on ℳ that satisfy ▽EiEj = 0 for i, j = 1, 2. The condition ▽EiEj = 0 (for i, j = 1, 2) is needed for technical reasons. (Please see [32] for details.) Note that in the ℝ3 case (A.4), if we set E1 = x, E2 = y, and E3 = z, then ▽EiEj = 0 for i, j = 1, 2, 3. The operator ▽2 is called the connection Laplacian operator, which lies at the heart of the VDM framework. The notion of an eigenvector field over ℳ is defined to be the solution of the following equation:
for some λ ∈ ℝ. The existence and other properties of the eigenvector fields can be found in [16]. Finally, we comment that all the above definitions can be extended to the general manifold setup without much difficulty, where, roughly speaking, a “manifold” is the higher-dimensional generalization of a surface. (We will not provide details in the manifold setting, and refer readers to standard differential geometry textbooks, such as [32].)
Appendix B
Proofs of Theorems 5.3, 5.5, and 5.6
Throughout this appendix, we adapt Assumption 5.1 and Definition 5.2 (see also Table 1.1). We divide the proof of Theorem 5.3 into four theorems, each of which has its own interest. The first theorem, Theorem B.1, states that the columns of the matrix Oi that are found by local PCA (see (2.4)) form an orthonormal basis to a d-dimensional subspace of ℝp that approximates the embedded tangent plane ι*Txiℳ. The proven order of approximation is crucial for proving Theorem 5.3. The proof of Theorem B.1 involves geometry and probability theory.
THEOREM B.1. In addition to Assumption 5.1, suppose KPCA ∈ C2([0, 1]). If ∊PCA = O(n−2/(d+2)) and then, with high probability (w.h.p.), the columns of the p × d matrix Oi, which is determined by local PCA, form an orthonormal basis to a d-dimensional subspace of ℝp that deviates from in the following sense:
| (B.1) |
where Θi is a p × d matrix whose columns form an orthonormal basis to ι*Txiℳ. Let the minimizer in (B.1) be
| (B.2) |
and denote by Qi the p × d matrix
| (B.3) |
and el(xi) the lth column of Qi. The columns of Qi form an orthonormal basis to ι*Txiℳ, and
| (B.4) |
If then, w.h.p.
Better convergence near the boundary is obtained for ∊PCA = O(n−2/(d+1)), which gives
for , and
for .
Theorem B.1 may seem a bit counterintuitive at first glance. When considering data points in a ball of radius , it is expected that the order of approximation would be O(∊PCA), while equation (B.1) indicates that the order of approximation is higher . The true order of approximation for the tangent space, as observed in (B.4), is still O(∊PCA). The improvement observed in (B.1) is of relevance to Theorem B.2, and we relate it to the probabilistic nature of the PCA procedure, more specifically, to a large-deviation result for the error in the law of large numbers for the covariance matrix that underlies PCA. Since the convergence of PCA is slower near the boundary, then for manifolds with boundary we need a smaller ∊PCA. Specifically, for manifolds without boundary we choose ∊PCA = O(n−2/(d+2)), and for manifolds with boundary we choose ∊PCA = O(n−2/(d+1)). We remark that the first choice also works for manifolds with boundary at the expense of a slower convergence rate.
The second theorem, Theorem B.2, states that the d × d orthonormal matrix Oij which is the output of the alignment procedure (2.5), approximates the parallel transport operator Pxi,xj from xj to xi along the geodesic connecting them. Assuming that (here, ∊ is different than ∊PCA), the order of this approximation is whenever xi, xj are away from the boundary. This result is crucial for proving Theorem 5.3. The proof of Theorem B.2 uses Theorem B.1 and is purely geometric.
THEOREM B.2. Consider satisfying that the geodesic distance between xi and xj is . For ∊PCA = O(n−2/(d+2)) w.h.p., Oij approximates Pxi, xj in the following sense:
| (B.6) |
where is an orthonormal set determined by local PCA. For
| (B.7) |
For∊PCA = O(n−2/(d+1)), the orders of ∊PCA in the error terms change according to Theorem B.1.
The third theorem, Theorem B.3, states that the n × n block matrix is a discrete approximation of an integral operator over smooth sections of the tangent bundle. The integral operator involves the parallel transport operator. The proof of Theorem B.3 mainly uses probability theory.
THEOREM B.3. In addition to Assumption 5.1 suppose ∊PCA = O(n−2/(d+2)). For we have w.h.p.
| (B.8) |
where T∊, α is defined in (5.2), is the orthonormal set determined by local PCA, and Oij is the optimal orthogonal transformation determined by the alignment procedure.
For we have w.h.p.
| (B.9) |
For ∊PCA = O(n−2/(d+1)) the orders of ∊PCA in the error terms change according to Theorem B.1.
The fourth theorem, Theorem B.4, states that the operator T∊, α can be expanded in powers of , where the leading-order term is the identity operator, the second-order term is the connection Laplacian operator plus some possible potential terms, and the first- and third-order terms vanish for vector fields that are sufficiently smooth. For α = 1, the potential terms vanish, and as a result, the second-order term is the connection Laplacian. The proof is based on geometry.
THEOREM B.4. For we have
| (B.10) |
where T∊, α is defined in (5.2).
COROLLARY B.5. Under the same conditions and notation as in Theorem B.4 if X ∈ C3(Tℳ), then for all we have
| (B.11) |
Putting Theorems B.1, B.3, and B.4 together, we now prove Theorem 5.3.
PROOF OF THEOREM 5.3. Suppose . By Theorem B.3, w.h.p.
where ∊PCA = O(n−2/(d+2)), and Theorem B.1 was used to replace ul(xi) by el(xi). Using Theorem B.4 for the right-hand side of (B), we get
For ∊ = O(n−2/(d+4)), upon dividing by ∊, the three error terms are
Clearly the three error terms vanish as n → ∞. Specifically, the dominant error is O(n−1/(d+4)), which is the same as . As a result, in the limit n → ∞, almost surely,
as required.
B.1 Preliminary Lemmas
For the proofs of Theorems B.1 through B.4, we need the following lemmas: LEMMA B.6. In polar coordinates around x ∈ ℳ, the Riemannian measure is given by
, where θ ∈ Txℳ, ∥θ∥ = 1, t > 0 and
PROOF. Please see [32].
The following lemma is needed in Theorems B.1 and B.2.
LEMMA B.7. Fix x ∈ ℳ and denote by expx the exponential map at x and by the exponential map of ℝp at ι(x). With the identification of Tι(x) ℝp with ℝp, for v ∈ Txℳ with ∥v∥ ≪ 1 we have
| (B.12) |
Furthermore, for w ∈ Txℳ ≅ ℝd, we have
| (B.13) |
PROOF. Define , that is,
| (B.14) |
Since φ can be viewed as a function from Txℳ ≅ ℝd to Tι,(x)ℝp≅ ℝp we can Taylor-expand it to get
where the equality holds since φ(0) = 0 and for all w ∈ Tι(x)ℝp if we identify Tι(x)ℝp with ℝp. To conclude (B.12), we claim that
which comes from the chain rule and the fact that
| (B.15) |
Indeed, we have that d from the definition of the exponential map, where is the pullback tangent bundle, and
where v ∈ Txℳ, v(t) = tv ∈ Txℳ, and vʹ(t) = v ∈ TtvTxℳ. The claim (B.15) for k = 1 follows when t = 0. The result for k ≥ 2 follows from a similar argument. Hence
which gives us (B.12) since ▽dι =Π and ▽2dι = ▽Π.
Next we show (B.13). When w ∈ TvTxℳ, we view d[ι○expx].(w) as a function from Txℳ≅ ℝd to ℝp so that when ∥v∥ ≪ 1, Taylor expansion gives us
| (B.16) |
here d and ▽ are understood as the ordinary differentiation over ℝd. To simplify the calculation of ▽(d[ι ○ expx].(w))|0 (v) and ▽2(d[ι ○ expx].(w))|0 (v,v), we denote
and
where u, v, w ∈ ℝd. By (B.12), when ∥v∥ is small enough, we have
where R(v) is the remainder term in the Taylor expansion:
Thus
| (B.17) |
since
Similarly, from (B.17), when ∥u∥ is small enough we have
| (B.18) |
and
| (B.19) |
As a result, from (B.17), (B.18), and (B.19) we have that
Plugging them into (B.16) we get (B.13) as required.
LEMMA B.8. Suppose x, y ∈ ℳ such that y = expx(tθ)where θ ∈ Txℳ and ∥θ∥ = 1. If t ≪ 1, then h = ∥ι(x) – ι(y)∥ ≪ 1 satisfies
| (B.20) |
PROOF. Please see [9] or apply (B.12) directly.
LEMMA B.9. Fixx ∈ ℳ and y = expx(tθ)where θ ∈ Txℳ and ∥θ∥ = 1. Let be the normal coordinate on a neighborhood U of x; then for a sufficiently small t we have
| (B.21) |
for all l = 1, 2, …, d.
PROOF. Choose an open subset U ⊂ ℳ small enough and find an open neighborhood B of 0 ∈ Txℳ so that expx : B → U is diffeomorphic. It is well-known that
where Jl(t) is the Jacobi field with Jl(0) = 0 and ▽tJl (0) = ∂l(x). By applying Taylor’s expansion in a neighborhood of t = 0, we have
Since , the following relationship holds:
| (B.22) |
Thus we obtain
| (B.23) |
On the other hand, from (B.13) in Lemma B.7 we have
| (B.24) |
Putting (B.23) and (B.24) together, it follows that, for l = 1, 2, … d,
| (B.25) |
B.2 Proof of Theorem B.1
Fix . Denote by the standard orthonormal basis of ℝpthat is, vk has 1 in the kth entry and 0 elsewhere. We can properly translate and rotate the embedding ι so that ι(xi) = 0 and the first d components {v1, v2; …, vd} ⊂ ℝp form the orthonormal basis of ι*Txiℳ, and we find a normal coordinate around xi so that ι*∂k(xi) = vk. Instead of directly analyzing the matrix Bi that appears in the local PCA procedure given in (2.3), we analyze the covariance matrix , whose eigenvectors coincide with the left singular vectors of Bi. Throughout this proof, we let K = KPCA to simply the notation. We rewrite Ξi as
| (B.26) |
where
| (B.27) |
and
| (B.28) |
Let denote the geodesic ball of radius around xi. We apply the same variance error analysis as in [34, sec. 3] to approximate Ξi. Since the points xi are independent identically distributed (i.i.d.), Fj, j ≠ i , are also i.i.d.; by the law of large numbers one expects
| (B.29) |
where F = F1,
| (B.30) |
and
| (B.31) |
In order to evaluate the first moment 𝔼F (k, l) of (B.31), we note that for y = expxi v, where v ∈ Txiℳ, by (B.12) in Lemma B.7 we have
| (B.32) |
By substituting (B.32) into (B.31), applying Taylor’s expansion, and combining Lemma B.8 and Lemma B.6, we have
where the last equality holds since integrals involving odd powers of θ must vanish due to the symmetry of the sphere 𝕊 d−1. Note that 〈ι*θ, vk〉 = 0 when k = d + 1, d + 2, … p. Therefore,
| (B.33) |
where is a positive constant. Similar considerations give the second moment of F(k, l) as
| (B.34) |
Hence, the variance of F(k, l) becomes
| (B.35) |
We now move on to establish a large-deviation bound on the estimation of by its mean 𝔼Fj(k, l). For that purpose, we measure the deviation from the mean value by α and define its probability by
| (B.36) |
To establish an upper bound for the probability pk,l(n, α), we use Bernstein’s inequality; see, e.g., [22]. Define
Clearly Yj (k, l) are zero mean i.i.d. random variables. From the definition of Fj (k, l) (see (B.27) and (B.28)) and from the calculation of its first moment (B.33), it follows that Yj (k, l) are bounded random variables. More specifically,
| (B.37) |
.
Consider first the case k, l = 1, 2, …, d, for which Bernstein’s inequality gives
| (B.38) |
From (B.38) it follows that for
and
| (B.39) |
the probability pk, l(n, α) is exponentially small.
Similarly, for k, l = d + 1, d + 2, …, p, we have
, which means that for
and (B.39), the probability pk, l(n, α) is exponentially small.
Finally, for k = d+1, d+2, …, p, l = 1, 2, …, d or l = d+1, d+2; …, p, and k = 1, 2, …, d, we have
, which means that for
and (B.39), the probability pk,l(n, α) is exponentially small. The condition (B.39) is quite intuitive as it is equivalent to n∊PCAd/2 ≫ 1, which says that the expected number of points inside should be large.
As a result, when (B.39) holds, w.h.p., the covariance matrix Ξi is given by
| (B.40) |
| (B.41) |
| (B.42) |
where Id×d is the identity matrix of size d×d, and 0m×mʹ is the zero matrix of size m × mʹ. The error term in (B.41) is the bias term due to the curvature of the manifold, while the error term in (B.42) is the variance term due to finite sampling (i.e., finite n). In particular, under the condition in the statement of the theorem for the sampling rate, namely, ∊PCA = O(n−2/(d+2)), we have w.h.p.
| (B.43) |
Note that by definition Ξi is symmetric, so we rewrite (B.43) as
| (B.44) |
where I is the d × d identity matrix, A is a d × d symmetric matrix, C is a d × (p – d) matrix, and B is a (p – d) × (p – d) symmetric matrix. All entries of A, B, and C are O(1).
Denote by uk and λk, k = 1, 2, …, p, the eigenvectors and eigenvalues of Ξi, where the eigenvectors are orthonormal and the eigenvalues are listed in decreasing order. Using regular perturbation theory, we find that λk = D∊PCAd/2+1 (1 + O (∊PCA1/2)) (for k = 1, 2, …, d), and that the expansion of the first d eigenvectors is given by
| (B.45) |
where are orthonormal eigenvectors of A satisfying . Indeed, a direct calculation gives us
| (B.46) |
where v3/2, v2 ∈ ℝd and z1, z3/2 ∈ ℝp–d. On the other hand,
| (B.47) |
where γ2 ∈ R. Matching orders of ∊PCA between (B.46) and (B.47), we conclude that
| (B.48) |
Note that the matrix appearing in (B.48) is singular and its null space is spanned by the vector wk so the solvability condition is . We mention that A is a generic symmetric matrix generated due to random finite sampling, so almost surely the eigenvalue is simple.
Denote by Oi the p × d matrix whose kth column is the vector uk. We measure the deviation of the d-dimensional subspace of ℝp spanned by uk, k = 1, 2, …, d, from ιTxiM by
| (B.49) |
where Θi is a p×d matrix whose kth column is vk (recall that vk is the kth standard unit vector in ℝp). Let Ô be the d × d orthonormal matrix
Then,
| (B.50) |
which completes the proof for points away from the boundary.
Next, we consider . The proof is almost the same as the above, so we just point out the main differences without giving the full details. The notations Ξi, Fj (k, l), pk,l(n, α), and Yj (k, l) refer to the same quantities. Here the expectation of Fj (k, l) is
| (B.51) |
Due to the asymmetry of the integration domain when xi is near the boundary, we do not expect 𝔼Fj (k, l) to be the same as (B.33) and (B.34), since integrals involving odd powers of θ do not vanish. In particular, when l = d + 1, …, p, k = 1, 2, …, d or k = d + 1, d + 2, …, p, l = 1, 2, …, d, 𝔼F(k, l) becomes
which is O(∊PCAd/2+3/2). Note that for the bias term in the expansion of the covariance matrix differs from (B.41) when l = d + 1, d = 2, …, p, k = 1, 2, …, d or k = d +1, d +2, …, p, l = 1, 2, …, d. Similar calculations show that
| (B.52) |
| (B.53) |
| (B.54) |
Similarly, Yj (k, l) are also bounded random variables satisfying
| (B.55) |
Consider first the case k, l = 1, 2, …, d, for which Bernstein’s inequality gives
| (B.56) |
From (B.56) it follows that w.h.p.
provided (B.39). Similarly, for k, l = d + 1, d + 2, …, p, we have
which means that w.h.p.
provided (B.39). Finally, for k = d + 1, d + 2, …, p, l = 1, 2, …, d, or l = d + 1, d + 2, …, p, k = 1, 2, …, d, we have
which means that w.h.p.
provided (B.39). As a result, under the condition in the statement of the theorem for the sampling rate, namely, ∊PCA = O(n−2/(d+2)), we have w.h.p.
Then, by the same argument as in the case when , we conclude that
Similar calculations show that for ∊PCA = O(n−2/(d+1)) we get
B.3 Proof of Theorem B.2
Denote by Oi the p × d matrix whose columns ul(xi), l = 1, 2, …, d, are orthonormal inside ℝp as determined by local PCA around xi. As in (B.3), we denote by el(xi) the lth column of Qi where Qi is a p × d matrix whose columns form an orthonormal basis of ι*Txiℳ so by Theorem B.1 , which is the case of focus here (if ∊PCA = O(n−2/(d+1)), then ).
Fix xi and the normal coordinate around xi so that ι*∂l(xi) = el(xi). Let xj = expxi tθ, where θ ∈ Txiℳ, . Then, by the definition of the parallel transport, we have
| (B.57) |
and since the parallel transport and the embedding ι are isometric, we have
| (B.58) |
Local PCA provides an estimation of an orthonormal basis spanning ι*Txiℳ, which is free up to O(d). Thus, there exists R ∈ O(p) so that ι*Txjℳ is invariant under R and el(xj) = Rι*Pxj,xi ∂l (xi) for all l = 1, 2, …, d. Hence we have the following relationship:
| (B.59) |
where
On the other hand, since , Lemma B.9 gives us
We now analyze the right-hand side term by term. Note that since ι*Txjℳ is invariant under R, we have . For the O(t) term, we have
| (B.60) |
where the second equality is due to Lemma B.9 and the third equality holds since Π(θ, ∂l(xi)) is perpendicular to ι*∂r(xi) for all l, r = 1, 2, …, d. Moreover, the Gauss equation gives us
which means the matrix
is symmetric.
Fix a vector field X on a neighborhood around xi so that X(xi) = θ. By definition we have
| (B.61) |
Viewing Tℳ as a subbundle of Tℝpwe have the equation of Weingarten:
| (B.62) |
where AΠ(X, X)∂l and are the tangential and normal components of ▽∂l, respectively. Moreover, the following equation holds:
| (B.63) |
By evaluating (B.61) and (B.62) at xi we have
| (B.64) |
where the third equality holds since are perpendicular to ι*∂l(xi) and the last equality holds by (B.63). Due to the symmetry of the second fundamental form, we know that the matrix
is symmetric.
Similarly we have
| (B.65) |
Since by (B.63), which we denoted earlier by S1 and used the Gauss equation to conclude that it is symmetric.
To estimate the last term, we work out the following calculation by using the isometry of the parallel transport:
| (B.66) |
Denote , which is symmetric by the definition of ℛ.
By (B.60), (B.64), (B.65), and (B.66) we have
| (B.67) |
where S:= −S1 − S2/3 + S1/6 − S3/6 is a symmetric matrix.
Suppose that both xi and xj are not in . To finish the proof, we have to understand the relationship between which is rewritten as
| (B.68) |
From (B.1) in Theorem B.1, we know
which is equivalent to
| (B.69) |
Due to (B.67) we have Qj = Qi R̄+t2QiSR̄+O(t3), which together with (B.69) gives
| (B.70) |
Together with the fact that derived from (B.67), we have
| (B.71) |
Recall that the following relationship between Oi and Qi holds (B.45)
| (B.72) |
when the embedding ι is translated and rotated so that it satisfies ι(xi) = 0, the first d standard unit vectors {v1, v2, … vd} ⊂ ℝp form the orthonormal basis of ι*Txiℳ, and the normal coordinates . Similarly, the following relationship between Oj and Qj holds (B.45):
| (B.73) |
when the embedding ι is translated and rotated so that it satisfies ι(xj) = 0, the first d standard unit vectors {v1, v2, … vd} ⊂ ℝp form the orthonormal basis of ι*Txjℳ, and the normal coordinates around xj satisfy ι*∂k(xj) = R̄ι*Pxj,xi∂k(xi)=vk. Also, recall that ι*Txjℳis invariant under the rotation R and from Lemma B.9, ek(xi) and ek(xj) are related by . Therefore,
| (B.74) |
when expressed in the standard basis of ℝp so that the first d standard unit vectors {v1, v2, … vd} ⊂ ℝp form the orthonormal basis of ι*Txiℳ. Hence, plugging (B.74) into (B.71) gives
| (B.75) |
Inserting (B.71) and (B.75) into (B.68), we conclude
| (B.76) |
Recall that Oij is defined as Oij = UVΤ, where U and V come from the singular value decomposition of . As a result,
Since R̄ΤSR̄ is symmetric, we rewrite R̄ΤSR̄ = UΣUΤ, where U is an orthonormal matrix and Σ is a diagonal matrix with the eigenvalues of R̄ΤSR̄ on its diagonal. Thus,
Since the Hilbert-Schmidt norm is invariant to orthogonal transformations, we have
Since UΤR̄ΤOU is orthogonal, the minimizer must satisfy , as otherwise the sum of squares of the matrix entries would be larger. Hence we conclude
Applying (B.59) and (B.45), we conclude
This concludes the proof for points away from the boundary.
When xi and xj are in , by the same reasoning as above we get
This concludes the proof. Similar results hold for ∊PCA = O(n−2/(d+1)) by using the results in Theorem B.1.
B.4 Proof of Theorem B.3
We demonstrate the proof for the case when the data is uniformly distributed over the manifold. The proof for the nonuniform sampling case is the same but more tedious. Note that when the data is uniformly distributed, T∊,α = T∊,0 for all 0 < α ≤ 1, so in the proof we focus on analyzing T∊ := T∊, 0. Denote K∊ := K∊, 0. Fix . We rewrite the left-hand side of (B.8) as
| (B.77) |
where
Since x1, x2; …, xn are i.i.d. random variables, then Gj for j ≠ i are also i.i.d. random variables. However, the random vectors Fj for j ≠ i are not independent, because the computation of Oij involves several data points, which leads to possible dependency between Oij1 and Oij2. Nonetheless, Theorem B.2 implies that the random vectors Fj are well approximated by the i.i.d. random vectors that are defined as
| (B.78) |
and the approximation is given by
| (B.79) |
where we use ∊PCA = O(n−2/(d+2)) (the following analysis can be easily modified to apply to the case ∊PCA = O(n−2/(d+1)).
Since Gj when j ≠ i are identical and independent random variables and when j ≠ i are identical and independent random vectors, we hereafter replace and Gj by Fʹ and G in order to ease notation. By the law of large numbers we should expect the following approximation to hold:
| (B.80) |
where
| (B.81) |
and
| (B.82) |
In order to analyze the error of this approximation, we make use of the result in [34, eq. (3.14), p. 132] to conclude a large-deviation bound on each of the d coordinates of the error. Together with a simple union bound, we obtain the following large-deviation bound:
| (B.83) |
where C1 and C2 are some constants (related to d). This large-deviation bound implies that w.h.p. the variance term is . As a result,
which completes the proof for points away from the boundary. The proof for points inside the boundary is similar.
B.5 Proof of Theorem B.4
We begin the proof by citing the following lemma from [9, lemma 8]: LEMMA B.10. Suppose f ∈ 𝒞3(ℳ) and then
where .
By Lemma B.10, we get
| (B.84) |
which leads to
| (B.85) |
Plug (B.85) into the numerator of T∊,αX(x):
where
We evaluate A and B by changing the integration variables to polar coordinates, and the odd monomials in the integral vanish because the kernel is symmetric. Thus, applying Taylor’s expansion to A leads to
which after rearrangement becomes
From the definition of z(x) it follows that
Suppose is an orthonormal basis of Txℳ and express . A direct calculation shows that
and similarly
Therefore, the first three terms of A become
and the last term is simplified to
Next, we consider B. Note that since there is an ∊ in front of B, we only need to consider the leading-order term. Let to simplify notation. Thus, applying Taylor’s expansion to each of the terms in the integrand of B leads to
In conclusion, the numerator of T∊,αX(x) becomes
Similar calculation of the denominator of the T∊,αX(x) gives
where
We apply Lemma B.10 to C and D and get
In conclusion, the denominator of T∊,αX(x) is
Putting all the above together, we have
In particular, when α = 1, we have
B.6 Proof of Theorem 5.5
Suppose miny∈∂ℳd(x, y) =∊̃. Choose a normal coordinate {∂1, ∂2; …, ∂d} on the geodesic ball B∊1/2(x) around x so that x0 = expx(∊̃∂d(x)). Due to the Gauss lemma, we know span {∂1(x0), ∂1(x1), …, ∂d–1(x0)}=Tx0∂ℳ and ∂d(x0) is outer normal at x0.
We focus first on the integral appearing in the numerator of T∊, 1X(x):
We divide the integral domain into slices Sη defined by
where η ∈ [−∊1/2,∊1/2] and u = (u1, u2, …, ud–1) ∈ ℝd–1. By Taylor’s expansion and (B.85), the numerator of T∊,1X becomes
| (B.86) |
Note that in general the integral domain Sη is not symmetric with relation to (0, …, 0, η), so we will try to symmetrize Sη by defining the symmetrized slices:
where Ri(u1; …, ui, …, η) = (u1, …, −ui, …, η). Note that from (B.22) in Lemma B.9, the orthonormal basis {Px0,x∂1(x), Px0,x∂2(x), …, Px0,x∂d–1(x)} of Tx0∂ℳ differs from (∂1(x0),∂2(x0), … ∂d–1(x0)} by O(∊). Also note that up to error of order ∊3/2, we can express ∂ℳ ∩ B∊1/2(x) by a homogeneous degree 2 polynomial with variables {Px0,x∂1(x), Px0,x∂2(x), …, Px0,x∂d–1(x)}. Thus the difference between S̃η and Sη is of order ∊ and (B.86) can be reduced to
| (B.87) |
Next, we apply Taylor’s expansion to X(x):
Since
the Taylor expansion of X(x) becomes
| (B.88) |
Similarly for all i = 1, 2, …, d we have
| (B.89) |
Plugging (B.88) and (B.89) into (B.87) further reduces (B.86) into
| (B.90) |
The symmetry of the kernel implies that for i = 1, 2, …, d – 1,
| (B.91) |
and hence the numerator of T1,∊X(x) becomes
| (B.92) |
where
| (B.93) |
and
| (B.94) |
Similarly, the denominator of T∊,1X can be expanded as
| (B.95) |
which together with (B.92) gives us the following asymptotic expansion:
| (B.96) |
Combining (B.96) with (B.9) in Theorem B.3, we conclude the theorem.
B.7 Proof of Theorem 5.6
We denote the spectrum of ▽2 by and the corresponding eigenspaces by El := {X ∈ L2(Tℳ) : ▽2X = −λlX}, l = 0, 1, …. The eigenvector fields are smooth and form a basis for L2(Tℳ), that is,
Thus we proceed by considering the approximation through eigenvector field subspaces. To simplify notation, we rescale the kernel K so that .
Fix Xl ∈ El. When from Corollary B.5 we have uniformly
When , from Theorem 5.5 and the Neumann condition, we have uniformly
| (B.97) |
Note that we have
thus again by the Neumann condition at x0, (B.97) becomes
In conclusion, when uniformly we have
Note that when the boundary of the manifold is smooth, the measure of . We conclude that in the L2 sense,
| (B.98) |
Next we show how converges to e−t▽2. We know I + ∊▽2 is invertible on El with norm . Next, note that if B is a bounded operator with norm ∥B∥ < 1, we have the following bound for any s > 0 by the binomial expansion:
| (B.99) |
On the other hand, note that on El
| (B.100) |
Indeed, for X ∈ El we have and
by the binomial expansion. Thus we have the claim.
Putting all the above together, over El for all l ≥ 0, when we have
where the first equality comes from (B.98) and (B.100), and the third inequality comes from (B.99). Thus on . By taking ∊ → 0, the proof is completed.
Appendix C
Multiplicities of Eigen 1-Forms of Connection Laplacian over 𝕊n
All results and proofs in this section can be found in [15, 38]. Consider the following setting:
Denote by Ω1(𝕊n) the complexified smooth 1-forms, which is a G-module by (g · s)(x) = g · s(g−1x) for g ∈ G, s ∈ Ω1(𝕊n), and x ∈ 𝕊n. Over ℳ we have the Haar measure dµ defined by the bi-invariant Hermitian metric 〈·, ·〉, which comes from the Killing form of G and the Hodge Laplacian operator Δ = dδ+ δd defined by 〈·, ·〉. Since Δ is a self-adjoint and uniform second-order elliptic operator on Ω1(𝕊n), the eigenvalues λi are discrete and nonnegative real numbers, with an accumulation point only at ∞, and their related eigenspaces Ei are of finite dimension. We also know is dense in Ω1(𝕊n) in the topology defined by the inner product (f, g)𝕊n := ∫𝕊n 〈f, g〉dµ. Note that ℊ/ 𝓉⊗ℝ ℂ ≅ ℂn when G = SO(n + 1) and K = SO(n). Denote by V = ℂn the standard representation of SO(n). We split the calculation of the multiplicity of eigenforms of Δ over 𝕊n into four steps. Step 1. Clearly Ω1(𝕊n) is a reducible G-module. Fix an irreducible representation λ of G acting on Γλ and construct a G-homomorphism
by φ ⊗ v ↦ φ (v). We call the image the Γλ-isotypical summand in Ω1(𝕊n) with multiplicity dimℂHomG(Γλ, Ω1(𝕊n)). Then we apply the Frobenius reciprocity law:
Thus if we can calculate dimℂ HomK, we know how many copies of the irreducible representation Γλ are inside Ω1(𝕊n).
Denote by L1, L2, …, Ln the basis for the dual space of Cartan subalgebra of so(2n) or so(2n+1). Then L1, L2; …, Ln together with generate the weight lattice. The Weyl chamber of SO(2n + 1) is
and the edges of the W are thus the rays generated by the vectors L1, L1 + L2; …, L1 + L2 + … + Ln; for SO(2n), the Weyl chamber is
and the edges are thus the rays generated by the vectors L1, L1 + L2, …, L1 + L2 + …, + Ln–3 + Ln–2, L1 + L2 + … + Ln–3 + Ln–2 + Ln–1 + Ln, and L1 + L2 + … + Ln–3 + Ln–2 + Ln–1 – Ln.
To keep the notation unified, we denote the fundamental weights ωi separately. When G = SO(2n), denote
| (C.1) |
when G = SO(2n + 1), denote
| (C.2) |
THEOREM C.1.
When m = 2n + 1, the exterior powers ΛpV of the standard representation V of so(2n + 1) are the irreducible representations with the highest weight ωp when p < n and 2ωn when p = n.
When m = 2n, the exterior powers ΛpV of the standard representation V of so(2n) are the irreducible representations with the highest weight ωp when p ≤ n–1; when p = n, ΛnV splits into two irreducible representations with the highest weights 2ωm–1 and 2ωm.
PROOF. Please see [15] for details.
THEOREM C.2 (Branching Theorem). When G = SO(m) and K = SO(m – 1) the restriction of the irreducible representations of G to K will be decomposed as the direct sum of the irreducible representations of K in the following way: Let Γλ be an irreducible G-module over ℂ with the highest weight .
- If m = 2n
where ⊕ runs over all such that
with and λi simultaneously all integers or all half-integers. Here is the irreducible K-module with the highest weight . - If m = 2n+1
where ⊕ runs over all such that
with and λi simultaneously all integers or all half-integers. Here is the irreducible K-module with the highest weight .
PROOF. Please see [15] for details.
From the above theorems, we know how to calculate dimℂ HomG(Γλ,Ω1(ℳ)). To be more precise, since the right-hand side of HomK is the irreducible representation of K with the highest weight ω1 (or splits in the low-dimension case), we know the dimℂ HomK can be 1 only if has the same highest weight by Schur’s lemma and the classification theorem.
Step 2. In this step we relate the irreducible representation Γλ ⊂ Ω1(𝕊n) to the eigenvalue of the Hodge-Laplace operator Δ.
THEOREM C.3. Suppose Γµ ⊂ Ω1(𝕊n) is an irreducible G-module with the highest weight µ; then we have
where f ∈ Γµ, ρ is the half sum of all positive roots, and 〈·, ·〉 is induced inner product on the dual Cartan subalgebra of ℊ from the Killing form B.
PROOF. Please see [38] for details.
Note that for since R+={Li–Lj, Li+Lj,Li:i < j} for since R+={Li–Lj, Li+Lj : i < j}.
Combining these theorems, we know if Γµ is an irreducible representation of G, then it is an eigenspace of Δ with eigenvalue λ = 〈µ +2ρ, µ〉. In particular, if we can decompose the eigen 1-form space Eλ ⊂ Ω1(𝕊n) into an irreducible G-module and calculate the dimension of the irreducible G-module, we can determine the multiplicity of Eλ.
Step 3. Now we apply the Weyl character formula to calculate the dimension of Γλ for G.
THEOREM C.4.
- When m = 2n + 1, consider where λ1 ≥ λ2 ≥ … ≥ λn ≥ 0, the highest weight of an irreducible representation Γλ. Then
- When m = 2n, consider where λ1 ≥ λ2 ≥ … ≥|λn|, the highest weight of an irreducible representation Γλ. Then
PROOF. Please see [15] for details.
Step 4. We need the following theorem about real representations of G to solve the original problem.
TABLE C.1.
Eigenvalues and their multiplicity of 𝕊m, where m ≥ 2. l0,i = λi + n – i, m0,i = n – i, l1/2,i = , and .
| λ | eigenvalues | multiplicity | ||
|---|---|---|---|---|
| 𝕊2n, n ≥ 2 |
kL1, k ≥ 1 kL1 + L2, k ≥ 1 |
k(k + 2n − 1) (k + 1)(k + 2n − 2) |
|
|
| 𝕊2n+1, n ≥ 2 |
kL1, k ≥ 1 kL1 + L2, k ≥ 1 |
k(k + 2n) (k + 1)(k + 2n − 1) |
|
|
| 𝕊3 |
kL1, k ≥ 1 kL1 + L2, k ≥ 1 kL1 − L2, k ≥ 1 |
k(k + 2) (k + 1)2 (k + 1)2 |
(k + 1)2 k(k + 2) k(k + 2) |
|
| 𝕊2 | kL1 k ≥ 1 | k(k + 1) | 2(2k + 1) |
THEOREM C.5.
When n is odd, for any weight λ = a1ω1 + a2ω2 +… + an–1ωn–1 + anωn/2 of so2n+1ℝ, the irreducible representation Γλ with the highest weight λ is real if an is even or if n ≅ 0 or 3 mod 4, if an is odd and n ≡ 1 or 2 mod 4 then Γλ is quaternionic.
When n is even, the representation Γλ of so2n ℝ with highest weight λ = a1ω1 + a2ω2 + … + an–2ωn–2 + an–1ωn–1 + anωn will be complex if n is odd and an–1 ≠ an; it will be quaternionic if n ≡ 2 mod 4 and an–1 +an is odd; and it will be real otherwise.
PROOF. Please see [15] for details.
Combining Table C.1 and this theorem, we know all the eigen 1-form spaces are real form.
Step 5. Now we put all the above together. All the eigen 1-form spaces of 핊m as an irreducible representation of SO(m+1), m ≥ 2, are listed in Table C.1. The highest weight is denoted by λ.
Consider SO(3) for example. In this case, n = 1 and ℳ = 𝕊2. From the analysis in cryo-EM [18], we know that the multiplicities of eigenvectors are 6, 10, …, which echoes the above analysis. Moreover, the first few multiplicities of 𝕊3, 𝕊4, and 𝕊5 are 4, 6, 9, 16, 16, …; 5, 10, 14, …; and 6, 15, 20, …, respectively.
Footnotes
One of the main considerations in the presentation of this paper is to make it as accessible as possible, including to readers who are not familiar with differential geometry. Although the connection Laplacian is essential to the understanding of the mathematical framework that underlies VDM, and differential geometry is extensively used in Appendix B for the proof of Theorem 5.3, we do not assume knowledge of differential geometry in Sections 2 through 10 (except for some parts of Section 8) that detail the algorithmic framework. The concepts of differential geometry that are required for achieving basic familiarity with the connection Laplacian are explained in Appendix A.
Since Ni depends on ∊PCA, it should be denoted as Ni,∊PCA, but since ∊PCA is kept fixed it is suppressed from the notation, a convention that we use except for cases in which confusion may arise.
The solution is unique whenever is nonsingular, a condition that is satisfied whenever the distance between xi and xj is sufficiently small, due to bounded curvature.
The definition of the parallel transport operator is provided in Appendix A and in textbooks on differential geometry; see, e.g., [32, chap. 2].
We do not align a basis with itself, so the edge set E does not contain self-loops of the form (i, i).
Notice that the weights are only a function of the euclidean distance between data points; another possibility, which we do not consider in this paper, is to include the Grassmannian distance in the definition of the weight.
Here we abuse notation slightly. Usually Txℳ defined here is understood as the embedded tangent plane by the embedding ι of the tangent plane at x. Please see [32] for a rigorous definition of the tangent plane.
Contributor Information
A. Singer, Princeton University, Dedicated to the memory of Partha Niyogi, Fine Hall, Washington Road, Princeton, N.J. 08544-1000, amits@math.princeton.edu
H.-T. Wu, Princeton University, Dedicated to the memory of Partha Niyogi, Fine Hall, Washington Road, Princeton, N.J. 08544-1000, hauwu@math.princeton.edu
Bibliography
- 1.Arun KS, Huang TS, Blostein SD. Least-squares fitting of two 3-D point sets. IEEE Trans. Patt. Anal. Mach. Intell. 1987;9(no. 5):698–700. doi: 10.1109/tpami.1987.4767965. [DOI] [PubMed] [Google Scholar]
- 2.Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Computation. 2003;15(no. 6):1373–1396. [Google Scholar]
- 3.Belkin M, Niyogi P. Lecture Notes in Computer Science. Vol. 3559. Berlin: Springer; 2005. Towards a theoretical foundation for Laplacian-based manifold methods. Learning Theory; pp. 486–500. [Google Scholar]
- 4.Belkin M, Niyogi P. Advances in neural information processing systems. Cambridge, Mass: MIT Press; 2008. Convergence of Laplacian eigenmaps; pp. 1–31. [Google Scholar]
- 5.Berline N, Getzler E, Vergne M. Heat kernels and Dirac operators. Berlin: Springer; 2004. [Google Scholar]
- 6.Bickel PJ, Levina E. Covariance regularization by thresholding. Ann. Statist. 2008;36(no. 6):2577–2604. [Google Scholar]
- 7.Candès EJ, Li X, Ma Y, Wright J. Robust principal component analysis? J. ACM. 2011;58(no. 3):37. Art. 11. [Google Scholar]
- 8.Carlsson G, Ishkhanov T, de Silva V, Zomorodian A. On the local behavior of spaces of natural images. Int. J. Comput. Vis. 2008;76(no. 1):1–12. [Google Scholar]
- 9.Coifman RR, Lafon S. Diffusion maps. Appl. Comput. Harmon. Anal. 2006;21(no. 1):5–30. [Google Scholar]
- 10.Cucuringu M, Lipman Y, Singer A. Sensor network localization by eigenvector synchronization over the Euclidean group. ACM Transactions on Sensor Networks. doi: 10.1145/2240092.2240093. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeWitt B. The global approach to quantum field theory. New York: Oxford University Press; 2003. [Google Scholar]
- 12.Donoho DL, Grimes C. Hessian eigenmaps: Locally linear embedding techniques for high-dimensional data. Proc. Natl. Acad. Sci. USA. 2003;100(no. 10):5591–5596. doi: 10.1073/pnas.1031596100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan K, Hoffman AJ. Some metric inequalities in the space of matrices. Proc. Amer. Math. Soc. 1955;6(no. 1):111–116. [Google Scholar]
- 14.Frank J. Three-dimensional electron microscopy of macromolecular assemblies: visualization of biological molecules in their native state. 2nd ed. New York: Oxford University Press; 2006. [Google Scholar]
- 15.Fulton W, Harris J. Representation theory: a first course. New York: Springer; 1991. [Google Scholar]
- 16.Gilkey P. Mathematics Lecture Series, 4. Publish or Perish. Boston: 1974. The index theorem and the heat equation. [Google Scholar]
- 17.Goldberg M, Kim S. Some remarks on diffusion distances. J. Appl. Math. 2010;2010:17. Art. ID 464815. [Google Scholar]
- 18.Hadani R, Singer A. Representation theoretic patterns in three dimensional cryo-electron microscopy I—the intrinsic reconstitution algorithm. Ann. of Math. (2) 2011;174(no. 2):1219–1241. doi: 10.4007/annals.2011.174.2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadani R, Singer A. Representation theoretic patterns in three dimensional cryo-electron microscopy II—the class averaging problem. Found. Comput. Math. 2011;11(no. 5):589–616. doi: 10.1007/s10208-011-9095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hein M, Audibert J-Y, von Luxburg U. Learning theory, Lecture Notes in Computer Science. 3559. Berlin: Springer; 2005. From graphs to manifolds - weak and strong pointwise consistency of graph Laplacians; pp. 470–485. [Google Scholar]
- 21.Higham NJ. Computing the polar decomposition—with applications. SIAM J. Sci. Statist. Comput. 1986;7(no. 4):1160–1174. [Google Scholar]
- 22.Hoeffding W. Probability inequalities for sums of bounded random variables. J. Amer. Statist. Assoc. 1963;58:13–30. [Google Scholar]
- 23.Johnstone IM. International Congress of Mathematicians. Vol. 1. Zürich: European Mathematical Society; 2007. High dimensional statistical inference and random matrices; pp. 307–333. [Google Scholar]
- 24.Johnstone IM, Lu AY. On consistency and sparsity for principal components analysis in high dimensions. J. Amer. Statist. Assoc. 2009;104(no. 486):682–693. doi: 10.1198/jasa.2009.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller JB. Closest unitary, orthogonal and Hermitian operators to a given operator. Math. Mag. 1975;48(no. 4):192–197. Available at: http://www.jstor.org/stable/2690338. [Google Scholar]
- 26.Lafon S. Thesis. New Haven, Conn.: Yale University; 2004. Diffusion maps and geometric harmonics. [Google Scholar]
- 27.Lee A, Pedersen K, Mumford D. The nonlinear statistics of high-contrast patches in natural images. Int. J. Comput. Vis. 2003;54(no. 1–3):83–103. [Google Scholar]
- 28.Little AV, Lee J, Jung Y, Maggioni M. Estimation of intrinsic dimensionality of samples from noisy low-dimensional manifolds in high dimensions with multiscale SVD. 2009 IEEE/SP 15th Workshop on Statistical Signal Processing. 2009:85–88. [Google Scholar]
- 29.Natterer F. Classics in Applied Mathematics. Vol. 32. Philadelphia: Society for Industrial and Applied Mathematics (SIAM); 2001. The mathematics of computerized tomography. [Google Scholar]
- 30.Niyogi P, Smale S, Weinberger S. Finding the homology of submanifolds with high confidence from random samples. Discrete Comput. Geom. 2008;39(no. 1–3):419–441. [Google Scholar]
- 31.Penczek P, Zhu J, Frank J. A common-lines based method for determining orientations for N ≥ 3 particle projections simultaneously. Ultramicroscopy. 1996;63(no. 3–4):205–218. doi: 10.1016/0304-3991(96)00037-x. [DOI] [PubMed] [Google Scholar]
- 32.Petersen P. Riemannian geometry. New York: Springer; 2006. [Google Scholar]
- 33.Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290(no. 5500):2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 34.Singer A. From graph to manifold Laplacian: the convergence rate. Appl. Comput. Harmon. Anal. 2006;21(no. 1):128–134. [Google Scholar]
- 35.Singer A. Angular synchronization by eigenvectors and semidefinite programming. Appl. Comput. Harmon. Anal. 2011;30(no. 1):20–36. doi: 10.1016/j.acha.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer A, Wu H-T. Orientability and diffusion map. Appl. Comput. Harmon. Anal. 2011;31(no. 1):44–58. doi: 10.1016/j.acha.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer A, Zhao Z, Shkolnisky Y, Hadani R. Viewing angle classification of cryo-electron microscopy images using eigenvectors. SIAM J. Imaging Sci. 2011;4(no. 2):723–759. doi: 10.1137/090778390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor ME. Mathematical Surveys and Monographs. Vol. 22. Providence, RI: American Mathematical Society; 1986. Noncommutative harmonic analysis. [Google Scholar]
- 39.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(no. 5500):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 40.van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21(no. 2):111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 41.van Heel M, Gowen B, Matadeen R, Orlova E, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. Single-particle electron cryo-microscopy: towards atomic resolution. Quarterly Reviews of Biophysics. 2000;33(no. 4):307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Zha H. Principal manifolds and nonlinear dimensionality reduction via tangent space alignment. SIAM J. Sci. Comput. 2004;26(no. 1):313–338. [Google Scholar]