Summary
Ubiquitin-mediated targeting of intracellular bacteria to the autophagy pathway is a key innate defense mechanism against invading microbes, including the important human pathogen Mycobacterium tuberculosis. However, the ubiquitin ligases responsible for catalyzing ubiquitin chains that surround intracellular bacteria are poorly understood. PARKIN is a ubiquitin ligase with a well-established role in mitophagy, and mutations in the PARKIN gene (Park2) lead to increased susceptibility to Parkinson’s disease. Surprisingly, genetic polymorphisms in the Park2 regulatory region are also associated with increased susceptibility to intracellular bacterial pathogens in humans, including Mycobacterium leprae and Salmonella typhi, but the function of PARKIN in immunity remains unexplored. Here we show that PARKIN plays a role in ubiquitin-mediated autophagy of M. tuberculosis. Both PARKIN-deficient mice and flies are sensitive to various intracellular bacterial infections, suggesting PARKIN plays a conserved role in metazoan innate defense. Moreover, our work reveals an unexpected functional link between mitophagy and infectious disease.
Eukaryotic cells target invading microbes to autophagosomes via a process termed xenophagy, which plays a key role in innate immune defense. Various intracellular bacterial pathogens, including Mycobacterium tuberculosis, are targeted for xenophagy via a ubiquitin-mediated pathway that surrounds bacteria with conjugated ubiquitin chains1–3. Marking with polyubiquitin presumably recruits ubiquitin-binding autophagy adaptors such as p62, which in turn engage the autophagic machinery for autophagosome formation and delivery of bacteria to the lysosome1,3,4. While ubiquitin binding adaptors are required for xenophagy, whether ubiquitin itself directly mediates bacterial autophagy is not clear as the identities of the ubiquitinated substrate(s) and ligase(s) responsible for coating cytosol-exposed bacteria are poorly understood.
In a fashion similar to xenophagy, the process of mitophagy eliminates damaged mitochondria through ubiquitin-mediated targeting to autophagosomes. A key step in mitophagy is marking of damaged mitochondria by the ubiquitin ligase PARKIN, which localizes to the organelle and directly ubiquitinates proteins on its surface5. Ubiquitin-tagged mitochondria are directed to the autophagosome pathway via p626–8 and several other factors10, ultimately delivering the organelle to the lysosome5.
Park2 mutations in humans are well-known risk factors for the development of Parkinson’s disease, but polymorphisms in the regulatory region of Park2, some of which result in decreased PARKIN expression9, have been associated with increased susceptibility to the intracellular pathogens Mycobacterium leprae and Salmonella typhi10,11. Although a genetic link to increased infection risk has been identified, the function of PARKIN in immunity remains obscure. We have identified that PARKIN, similar to its role in mitophagy, is also important for innate defense against M. tuberculosis and other intracellular pathogens by promoting xenophagy. This work provides a possible mechanism underlying the human genetic studies linking PARKIN to increased susceptibility to bacterial infection and reveals a surprising connection between mitochondrial homeostasis and pathogen defense.
PARKIN in TB-ubiquitin colocalization
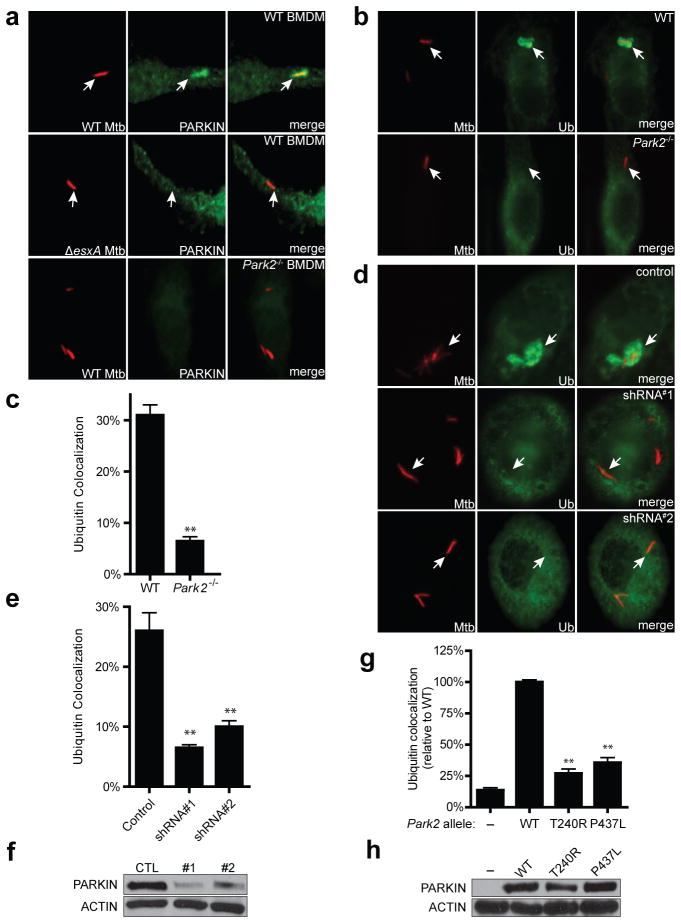
We have shown previously that upon infection of macrophages, M. tuberculosis bacilli that puncture phagosomal membranes via their ESX-1 secretion system gain access to the host cytosol but become enveloped by conjugated ubiquitin chains and are targeted to autophagosomes via p62 and NDP523. Although the role of ESX-1 in autophagy induction is likely complicated12, it is clear that approximately one-third of wild-type intracellular bacteria are targeted to autophagy during macrophage infection and that this plays a major role in host resistance to infection2,3. Because of the commonalities between mitophagy and autophagy of intracellular mycobacteria, and the links between Park2 polymorphisms and increased susceptibility to bacterial infection in humans, we hypothesized that PARKIN may also be recruited to M. tuberculosis-containing phagosomes and target them for ubiquitin-mediated autophagy. Indeed, after infection of murine bone marrow-derived macrophages (BMDMs) with M. tuberculosis expressing mCherry, we found that PARKIN localized to approximately 12% of wild-type M. tuberculosis phagosomes but not to ESX-1 mutants (Fig. 1a, Extended Data Fig. 1). Next, we infected BMDMs isolated from wild-type and Park2−/− mice and performed immunofluorescence co-localization experiments using antibodies that recognize polyubiquitin. As shown in Fig. 1b–c, Park2−/− BMDMs were severely defective for M. tuberculosis ubiquitin colocalization as compared to control macrophages, resulting in a significant reduction in ubiquitin-positive mycobacteria. Likewise, shRNA knock-down of PARKIN expression in human macrophage cell lines also resulted in a drastic reduction in ubiquitin localization with M. tuberculosis cells (Fig. 1d–f), indicating that PARKIN plays a conserved role in mycobacterium ubiquitination in mice and humans. Knock-down of LRSAM1, a ubiquitin ligase recently implicated in antibacterial defense and ubiquitination of Salmonella1,3,4,13, had no effect on ubiquitin or GFP-LC3 colocalization with M. tuberculosis (Extended Data Fig. 1b, c). Expression of wild-type Park2 in Park2−/− cells restored ubiquitin localization around M. tuberculosis cells (Fig. 1g, h). In contrast, Park2−/− BMDMs expressing either of two pathogenic RING domain mutant alleles that inactivate PARKIN’s E3 ligase activity, T240R or P437L3,4,14–16, failed to restore ubiquitin colocalization with M. tuberculosis (Fig. 1g, h). Taken together, these data demonstrate that Parkin and its E3 ligase activity are critical for the colocalization of ubiquitin with M. tuberculosis during infection.
Figure 1. PARKIN activity is required for M. tuberculosis-ubiquitin colocalization.
a, Wild-type (WT) and Park2−/− BMDMs were infected with mCherry-expressing M. tuberculosis for 4 h and immunostained using anti-PARKIN antibodies. b, Wild-type BMDMs were infected with mCherry-M. tuberculosis for 4 h and immunostained for polyubiquitin. c, Quantification of ubiquitin-positive M. tuberculosis from (b). Results are means ± SEM of three independent experiments (**P<0.001, paired Student’s t-test). d, U937 human macrophages expressing a scrambled shRNA (Control) or one of two different shRNAs targeting Park2 (shRNA#1, shRNA#2) were infected with mCherry-M. tuberculosis for 12 h and immunostained for polyubiquitin. e, Quantification of ubiquitin positive M. tuberculosis from (d), results are means ± SEM of three independent experiments (**P<0.005, Student’s t-test). f, PARKIN and actin expression in cells from (e) was determined by western blotting. g, Park2−/− BMDMs were transduced with lentivirus expressing BFP (−), wild-type PARKIN, or two separate mutant PARKIN alleles (T240R, P437L). Cells were infected with M. tuberculosis for 4 h and ubiquitin-M. tuberculosis colocalization was quantified and expressed relative to control BMDMs. Results are means ± SEM of three independent experiments (**P<0.005, paired Student’s t-test). h, PARKIN and actin expression in cells from (g) was determined by western blotting.
PARKIN mediates K63-linked polyubiquitin
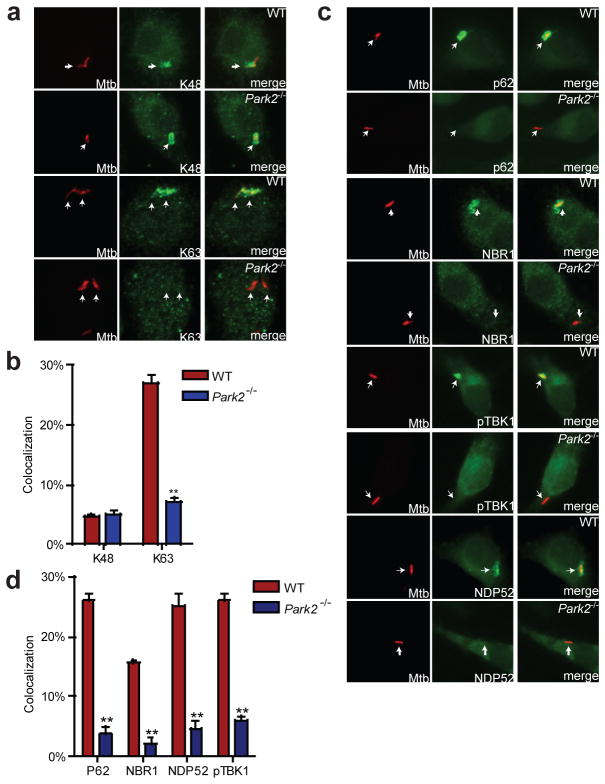
We showed previously that both K63- and K48-linked polyubiquitin chains accumulate around M. tuberculosis3. Because PARKIN is known to catalyze K63-linked ubiquitin chains5,17, we sought to determine the nature of the residual ubiquitin surrounding M. tuberculosis in Park2−/− BMDMs. Using ubiquitin linkage-specific antibodies5,18, we found that in wild-type BMDMs, approximately 26–29% of all intracellular bacteria (~90–95% of all ubiquitin-positive bacilli) co-localized with K63 ubiquitin, whereas only 5–7% bacilli stained for K48 (Fig. 2a–b). Additionally, expression of HA-epitope-tagged forms of K48 and K63 ubiquitin within BMDMs supported the notion that K63-linked polyubiquitin is more abundant surrounding M. tuberculosis than the K48-linked form (Extended Data Fig. 2). In Park2−/− BMDMs, however, there was a specific decrease in the number of K63-positive mycobacteria, while the K48-positive population remained unaffected (Fig. 2a–b, Extended Data Fig. 2). Previous electron microscopy studies indicated that though ubiquitin can localize directly with M. tuberculosis, the majority of ubiquitin is found on membranous structures surrounding M. tuberculosis-containing phagosomes15,19–21. To address whether bacterial or host proteins become ubiquitinated, we used a digitonin-based method that specifically permeabilizes the plasma membrane and leaves phagosomes intact with luminal contents inaccessible to antibodies10,22 (Extended Data Fig. 3a). As shown in Extended Data Fig. 3b, antibodies against poly-ubiquitin and K63-ubiquitin stained digitonin-permeabilized cells and resulted in colocalization with mCherry-expressing M. tuberculosis. Importantly, anti- M. tuberculosis antibodies failed to stain M. tuberculosis within digitonin-permeabilized cells and only stained cells after addition of Triton-X100 detergent, demonstrating that digitonin permeabilized cells contained intact phagosomes (Extended Data Fig. 3b, c). Taken together, these data suggest that PARKIN facilitates the linkage of K63-linked ubiquitin chains surrounding M. tuberculosis containing phagosomes, although the exact protein target(s) remain to be explored. Furthermore, this data also suggests at least one other ubiquitin ligase works independently of PARKIN to catalyze the K48-linked ubiquitination that surrounds a minor population of M. tuberculosis cells.
Figure 2. PARKIN mediates K63-ubiquitin colocalization of M. tuberculosis and recruitment of ubiquitin-autophagy receptors.
a, BMDMs were infected with mCherry-M. tuberculosis for 4 h and immunostained using linkage-specific ubiquitin antibodies. b, Quantification of K63 and K48 ubiquitin positive M. tuberculosis from (a). (**P<0.001, paired Student’s t-test). c, Same as (a) except immunostained for NDP52, p62, phospho-TBK1, or NBR1. d, Quantification of colocalization from (c), (N=3 per group, **P < 0.001).
PARKIN required for TB autophagy
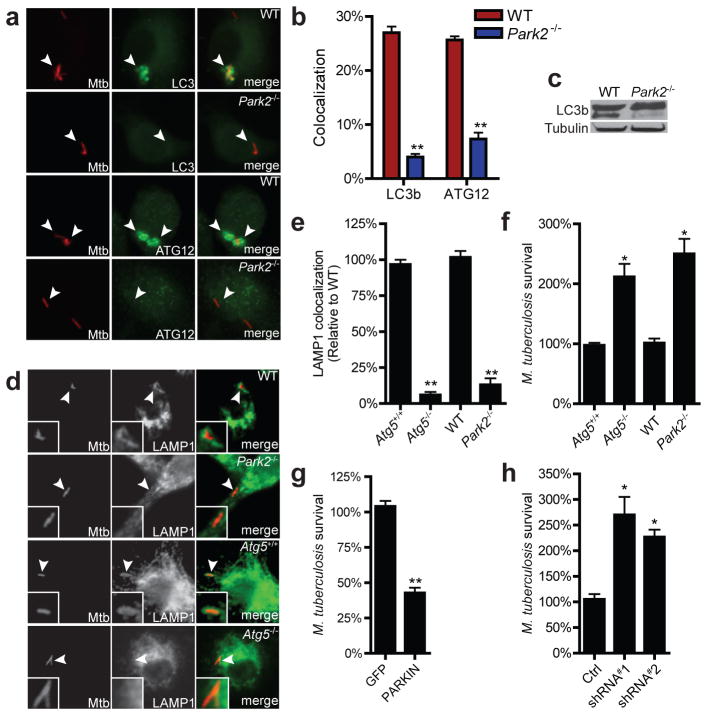
Ubiquitination coincides with autophagic targeting of M. tuberculosis, but a causal relationship has not been demonstrated. To determine whether Parkin-mediated ubiquitination directs autophagic targeting of M. tuberculosis, we infected wild-type and Park2−/− macrophages with M. tuberculosis and measured colocalization of bacilli with multiple markers of autophagy. Microscopy analysis of proteins involved in ubiquitin recognition (NBR1, NDP52, p62, phospho-TBK1) revealed reduced colocalization with M. tuberculosis in Park2−/− macrophages (Fig. 2c–d), suggesting that PARKIN-mediated ubiquitination directly leads to the recruitment of the proximal ubiquitin-adaptors that facilitate autophagic targeting of mycobacteria. Likewise, mycobacterial cells within infected Park2−/− BMDMs had reduced colocalization with autophagic proteins LC3 and ATG12 relative to infection of wild-type BMDMs (Fig. 3a–b), suggesting that the K63-linked polyubiquitin catalyzed by PARKIN is required for delivery of M. tuberculosis to autophagosomes. Consistent with this notion, Park2−/− cells were defective in conversion of LC3 to its activated, lipidated form, LC3-II, during M. tuberculosis infection, further demonstrating that Parkin is required for autophagy of mycobacteria (Fig. 3c).
Figure 3. PARKIN mediates autophagic targeting of M. tuberculosis and limits replication.
a, BMDMs were infected with mCherry-M. tuberculosis for 4 h and immunostained for LC3 or ATG12. b, Quantification of results from (a) (N=3 per group, **P < 0.001). c, Western blot analysis of LC3b from cell lysates from (a). d, BMDMs were infected with mCherry-M. tuberculosis for 6 h and immunostained for Lamp1. e, Quantification of results from (d) expressed relative to control BMDMs (N=3 per group, **P < 0.001). f, BMDMs were infected with M. tuberculosis and colony forming units (CFU) at t=0 and t=16 h were determined by plating. Results were normalized to t=0 (N=3 per group, *P < 0.02). g, 33 RAW264.7 macrophages transduced with lentivirus expressing GFP or PARKIN were infected with M. tuberculosis and CFU were determined after 16 h, results were normalized to GFP-expressing cells (N=3 per group, **P < 0.0076). h, U937 human macrophages expressing either scrambled or Park2 shRNAs were infected with M. tuberculosis for 36 h and CFU were determined. Results were normalized to t=0 (N=3 per group, *P < 0.02).
PARKIN limits TB replication
The autophagy pathway serves to limit M. tuberculosis replication in macrophages by delivering bacilli to the lysosome3,11. To determine if PARKIN mediated ubiquitination is required for autophagic targeting of M. tuberculosis to lysosomes, we infected BMDMs with M. tuberculosis and monitored co-localization with the lysosomal marker, LAMP1. During M. tuberculosis infection of wild-type BMDMs, approximately 30% of bacilli stained positive for LAMP1 at 6 hrs post-infection (Fig. 3d–e). In contrast, only 2–5% of bacilli colocalized with LAMP1 during M. tuberculosis infection of Park2−/− macrophages. This was similar to macrophages deficient for the essential autophagy protein, ATG5 (Fig. 3d–e)3,5. To test whether these differences led to changes in bacterial survival, we infected Park2−/− and Atg5−/− BMDMs with wild-type M. tuberculosis and determined bacterial viability by enumerating colony-forming units (CFUs). Infection of BMDMs deficient for either ATG5 or PARKIN resulted in a 2- and 2.5-fold increase in bacterial numbers, respectively, relative to control BMDMs by 12 hours post-infection (Fig. 3f). Conversely, overexpression of PARKIN in RAW 264.7 macrophages led to decreased bacterial replication (Fig. 3g). Importantly, knock-down of PARKIN expression in human U937 cells also led to an increase in bacterial replication during infection (Fig. 3h). Taken together, our data demonstrates that PARKIN-mediated ubiquitination leads to the autophagic targeting of M. tuberculosis and is essential for inhibition of mycobacterial replication in macrophages.
PARKIN mediates M. tuberculosis immunity
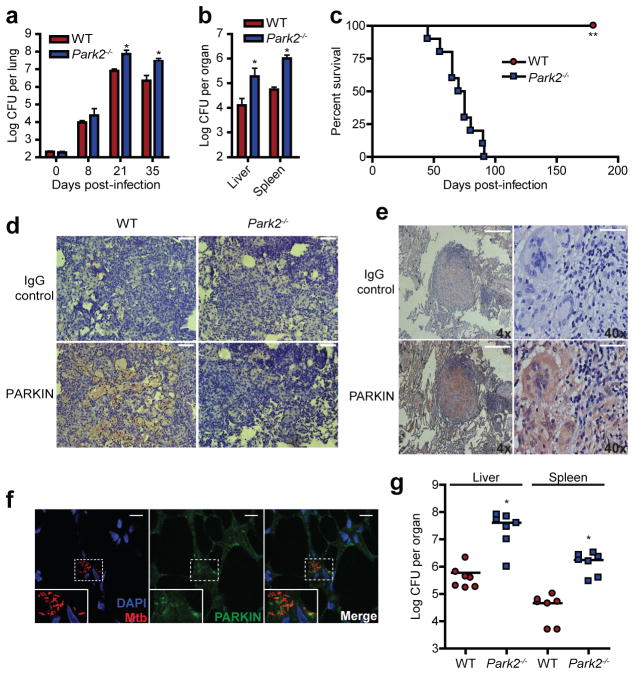
Polymorphisms within the regulatory region of Park2 in human populations have been identified as a common risk factor for increased susceptibility to Mycobacterium leprae and salmonella infection6,7,10,11, suggesting that PARKIN plays an important role in vivo against a broad range of intracellular bacterial infections. We began to test this by first determining whether PARKIN was required in vivo during M. tuberculosis infection of mice. We performed a low-dose aerosol infection of wild-type and Park2−/− knockout mice and determined mouse survival and bacterial burden within infected tissues. In comparison to infected wild-type mice, Park2−/− knockout mice had a 10-fold increase in bacterial CFUs within infected lungs, spleens and liver by 21 days post-infection (Fig. 4a, b). Furthermore, survival studies revealed that Park2−/− mice were extremely susceptible to M. tuberculosis as all infected mice succumbed to overwhelming infection by 85 days post-infection, while all infected wild-type mice remained alive and displayed no-overt signs of weight-loss or stress (Fig. 4c). Immunohistochemistry staining of infected mouse lungs revealed robust PARKIN expression in mouse granulomas within the central macrophage containing zone (Fig. 4d). In agreement with our mouse experiments, we also observed high expression of PARKIN within human lung granuloma tissue samples from M. tuberculosis infected patients (Fig. 4e, Extended Data Fig. 4). Further analysis of human lung specimens via confocal microscopy revealed the presence of PARKIN puncta within M. tuberculosis infected cells as well as in vivo colocalization of PARKIN with M. tuberculosis (Fig. 4f). Lastly, Park2−/− mice were also highly susceptible to another intracellular pathogen, L. monocytogenes, resulting in 10–20 fold higher bacterial burdens relative to wild-type mice within infected spleens and liver (Fig. 4g). Taken together, this data demonstrates that PARKIN is essential in vivo for controlling intracellular bacterial pathogens within mice and suggests an in vivo role for Parkin in human tuberculosis disease.
Figure 4. PARKIN is required for control of bacterial infection in vivo.
a, WT and Park2−/−mice were infected with wild-type M. tuberculosis via aerosol and lung bacterial burdens were determined by plating (means ± SD, N=5 per group, *P<0.02 by Student’s t-test). b, Enumeration of liver and spleen CFU from mice infected in (a) 21 days post-infection. (means ± SD, N=5 per group, *P<0.03 by Student’s t-test). c, Survival of M. tuberculosis-infected WT and Park2−/− mice (N=10, **P<0.001 by log-rank test). d, Immunohistochemistry staining of lung sections from infected mice 21 days post-infection, scale bar = 100μm. e, Immunohistochemistry staining of lung sections from a human patient with active tuberculosis. f, Confocal microscopy images of human lung sections from (e) immunostained for M. tuberculosis and PARKIN; DNA was visualized using DAPI. g, Mice were infected with WT L. monocytogenes via IP injection and bacterial burdens in livers and spleens were determined by plating (means ± SD, N=7 per group, *P<0.04 by Student’s t-test).
Conserved role of PARKIN in immunity
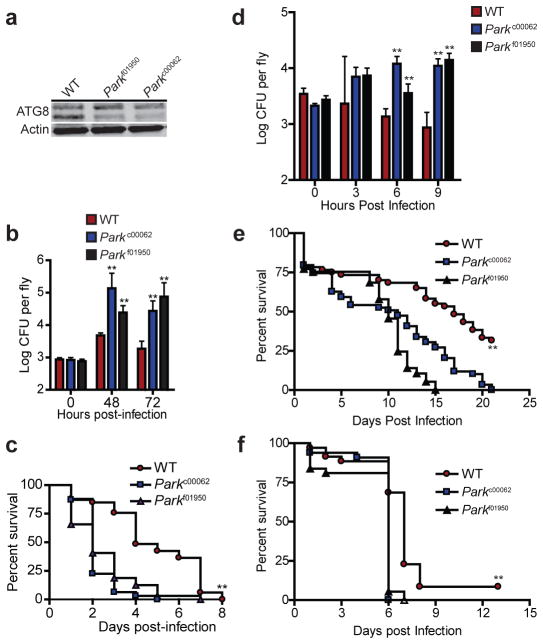
Park2 is present in all metazoans8,23, including Drosophila melanogaster and Caenorhabditis elegans, with well-characterized functions in mitochondrial maintenance and in models of Parkinson’s disease. Because xenophagy of intracellular pathogens is a highly evolutionarily conserved innate immune defense mechanism24, we sought to determine whether Parkin also plays an evolutionarily conserved role in immunity within non-mammalian organisms. We began by first analyzing PARKIN-deficient D. melanogaster strains using models of bacterial systemic infection. We obtained two mutant fly lines with independent disruptions of the Parkin gene (Parkf01950, Parkc00062) and infected them with L. monocytogenes, which has previously shown to induce autophagy within flies25,26. In contrast to wild-type infected flies, Parkin mutants were severely defective in ATG8/LC3 processing during infection (Fig. 5a), suggesting that Parkin plays a role in autophagic immunity within flies. Consistent with our results in mice, Parkin mutant flies were also highly susceptible to L. monocytogenes infection and led to 10–50 fold increases in bacterial burdens relative to wild-type infected flies (Fig. 5b). This was accompanied with decreased survival, with a median lifespan of two days following infection (Fig. 5c). In addition, Parkin mutant flies were also susceptible to other autophagy inducing intracellular pathogens such as S. Typhimurium and Mycobacterium marinum. By 9 hours post infection, Parkin deficient flies had a 10-fold increase in S. Typhimurium burden relative to wild-type flies (Fig. 5d). Moreover, PARKIN mutant flies had significantly decreased life spans upon infection with either S. Typhimurium (Fig. 5e) or M. marinum (Fig. 5f). Lastly, C. elegans strains deficient in the Parkin homolog (PDR-1) were also highly susceptible to S. Typhimurium infection (data not shown). All together, our data shows that Parkin homologs within lower metazoans are required for proper immune response to infection, suggesting an evolutionarily conserved role in innate immunity.
Figure 5. PARKIN is required for control of S. Typhimurium and M. marinum infection within flies.
a, WT and two independent Parkin-deficient D. melanogaster lines (Parkc00062, Parkf01950) were infected with L. monocytogenes via anterior abdomen injection. ATG8 processing was monitored in whole-fly protein lysates by western blotting. b, L. monocytogenes CFU from (a) were determined by plating (means ± SD, N=3–5, **P<0.001 by Student’s t-test) and c, survival of D. melanogaster was determined (N=33, **P<0.001 by log-rank test). d, e, Flies were infected with S. Typhimurium and d, bacterial CFU (means ± SD, N=3–5 per group, **P<0.009 by Student’s t-test) and e, fly survival were determined (N=59, **P<0.001 by log-rank test). f, Survival of flies infected with M. marinum (N=37, **P<0.0045 by log-rank test).
Discussion
Our findings reveal that PARKIN regulates a common cellular program by which metazoans mediate quality control of endogenous mitochondria (self) and eradicate harmful bacterial pathogens (non-self). Although these two activities are seemingly disparate, the evolutionary origin of mitochondria from a bacterial endosymbiont suggests that perhaps mitochondrial dysfunction triggers the recognition of the organelle as non-self. For example, mitochondria (and bacterial endosymbionts5,27) may actively evade PARKIN surveillance, but these inhibitory processes are overridden upon organelle damage. Alternatively, several studies have shown that damaged mitochondria may serve as a “danger-associated molecular pattern”, resulting in the activation of several innate immune receptors such as toll-like receptors and the NLRP3-inflammasome complex28,29. In view of recent studies showing that intracellular infection with several pathogens such as L. monocytogenes result in altered host mitochondria dynamics30, it is tempting to speculate that damaged mitochondria may serve as a signal for intracellular infection and activation of xenophagy.
Our results also provide a molecular explanation for increased bacterial susceptibility of humans with polymorphisms in the Park2 regulatory region3,10,11, broadening the role of PARKIN beyond mitochondrial homeostasis. Indeed, several recent studies have shown PARKIN to participate in a wide array of cellular processes such as apoptosis31, regulation of lipid metabolism32, and cytokine production upon infection33, all of which may contribute to the in vivo importance of PARKIN in immunity. Lastly, this work highlights the unexpected connection between mitochondrial-based neuronal disorders and susceptibility to bacterial infection in humans. Recent genome wide association studies on inflammatory bowel disease, which is linked to altered host-gut microbe interactions, have identified susceptibility SNPs within LRRK2 and Park7, two genes canonically associated with Parkinson’s disease34,35. Thus, we surmise that genes typically associated with neuronal maintenance or mitophagy may play broad roles in cellular homeostasis within various cell types.
Full Methods
Mice and macrophages
Park2−/− mice on the C57L/B6 background were a gift from K. Nakamura (Gladstone Institute)12,36. GFP-LC3 transgenic mice on the C57L/B6 were a gift from N. Mizushima and used were as previously described3. Wild-type C57BL/6 mice were purchased from Jackson laboratories. BMDMs were obtained from mouse femurs as previously described3 and cultured in DMEM H-21 supplemented with 20% FBS and 10% MCSF derived from 3T3-MCSF cells. U937 and Raw264.7 cells were obtained from ATCC and tested for mycoplasma prior to purchase. U937 monocytes were stimulated overnight with 20ng/ml of PMA (Sigma) prior to infections.
Antibodies
The following antibodies were used: LC3B (L10382 Invitrogen), NDP52 (Abnova D01), p62/SQSTM1 (Abnova 2C11), rabbit anti-drosophila ATG-8 (Gift from S. Cherry, U Penn), anti-Mtb (BEI Resources NR-13818), poly-ubiquitin (Enzo FK1), phospho-TBK1 (Cell Signaling #5483), Tubulin (Cell Signaling #2128), mouse-monoclonal Parkin (Cell Signaling #4211) and ATG12 (Cell Signaling #2011), rabbit polyclonal anti-Parkin (Abcam 15954), anti-NBR1 (Abcam 55474), humanized rabbit monoloncal antibodies specific for K63 or K48 (Gift from E. Brown lab at Genentech).
Bacterial strains
The following bacterial strains were used: M. tuberculosis (Erdman), L. monocytogenes (10403s), M. marinum (M), and S. Typhimurium (SL1344). Wild-type and ΔesxA mycobacteria expressing mCherry was previously described3.
Macrophage infection
For infections with M. tuberculosis, macrophages were infected as previously described3. Briefly, M. tuberculosis cultures were washed twice with PBS, gently sonicated to disperse clumps, and resuspended in DMEM supplemented with 10% horse serum. Media was removed from cells, monolayers overlaid with the bacterial suspension, and centrifuged for 10 min at 1,000 RPM. Cells were washed twice in PBS and returned to macrophage media. For determination of bacterial viability following infection, cells were lysed in 1% Triton-X 100 and plated on 7H10 solid media.
Western blotting
Protein lysates from cells and flies were obtained by lysis in RIPA buffer (Sigma) at the indicated time points. Micro BCA protein kit (Pierce) was used to measure protein levels and equal amounts of protein were electrophoresed on 4–20% Tris-HCL Criterion gels (Biorad), and transferred onto nitrocellulose membranes. Western blots were analyzed using an Odyssey Imager (Licor) according to manufacturer’s instructions. Western blot figures are a representative of at least two independent experiments.
Immunofluorescence
Infected cells were immunostained and visualized as previously described3. Briefly, macrophages were seeded onto poly-lysine coated coverslips and infected with M. tuberculosis as described above. Cells were infected at an MOI of 1, and fixed in 4%PFA for 20 min at the indicated time points. Cells were incubated with indicated primary antibodies for 2 h at room temperature in 5% milk, 0.05% saponin and visualized using secondary Alexa-fluor488 antibodies. For Parkin immunofluorescence, cells were stained using rabbit polyclonal anti-Parkin (Abcam15954) and an HRP conjugated donkey anti-rabbit secondary (1:100, Jackson Immunochemicals) followed by amplification with alexafluor-488 tyramide (Invitrogen). Colocalization studies were performed as blinded experiments, with a minimum count of 200 cells per coverslip and performed in triplicate. Data shown is the mean ± SEM of at least 3 experiments.
Mouse infection
Non-randomized mice (female 5–8 weeks old) were infected with M. tuberculosis via low-dose aerosol infection (200 CFU) as previously described37. Sample size (N=5 per group) was based on empirical evidence from previously published reports37 as the size necessary for adequate statistical analysis. Lungs, liver and spleens were harvested, homogenized, and plated on 7H10 agar plates. For survival experiments, infected mice were euthanized when they had lost 15% of their maximal body weight. For L. monocytogenes infections, mice were infected via intraperitoneal injection with 4 × 105 bacteria. 96 hours post infection, liver and spleen from infected mice were homogenized and serial dilutions were plated onto BHI agar plates. No blinding was done for these animal studies and all mice were housed and treated humanely using procedures described in an animal care protocol approved by University of California, San Francisco, Institutional Animal Care and Use Committee.
Fly strains and infections
The white1118 strain (Bloomington stock center, stock 6326) was used as the wild-type parental strain for all experiments. The Parkc00062 and Parkf01950 alleles are from the Exelexis piggyBac transposon collection38. Parkc00062 was obtained from Bloomington stock center and Parkf01950 was obtained from the Exelexis collection at Harvard. Infections were done as previously described39. Male 5 to 7 day-old files were anesthetized with CO2 and injected L. monocytogenes (1000 CFU), S. Typhimurium (2500 CFU), or M. marinum (1000 CFU) in 50 nl of culture into the anterior abdomen. Infected flies were homogenized in PBS supplemented with 1% Triton X-100 and serial dilutions were plated onto solid media. For survival analysis, the number of dead flies was counted daily and analyzed via log-rank test.
Lentiviral virus knockdown and complementation
Lentivirus expressing shRNAs targeting Park2 and LRSAM1 transcripts were generated using the Mission PLKO.1 lentivirus system from Sigma. A lentivirus expressing a non-targeting scrambled shRNA was used as a control. U937 cells were transduced with lentivirus per manufacturer’s instructions and stable cell lines were generated via selection with puromycin. For transgene expression of Park2 and Park2 mutants, full length Park2 was cloned into pBluescript vector and RING domain mutants were generated using Quick-Change site directed mutagenesis kit (Stratagene). HA-tagged ubiquitin constructs were obtained from Addgene31. Lentivirus expressing protein constructs were generated using the pLentiX system3. RAW264.7 cells were transduced and selected on puromycin for 1 week. BMDM cells expressing lentiviral constructs were generated by transducing marrow cells from knockout mice with lentivirus followed by differentiation into macrophages as described above. During day 3 of differentiation, cells were selected with 5ug/ml of puromycin.
Statistics
Statistical analysis of data was performed using GraphPad Prism software (Graphpad; San Diego, CA). Two-tailed unpaired Student’s t tests were used for analysis of microscopy images and mycobacterium CFU assays. The Kaplan-Meir method was used to analyze mouse survival.
RNA isolation and qPCR
RNA was isolated and purified from cells using the Trizol micro-midi RNA isolation kit (Invitrogen) per manufacturer’s instructions. For qPCR analysis 2μg of RNA was reverse transcribed using the VILO cDNA synthesis kit (Invitrogen) and qPCR analysis was performed in triplicate using a MJ Research Opticon 3200 machine. Data graphed in figures are representative of at least three independent experiments.
Digitonin permeabilization
Differential digitonin permeabilization of infected macrophages was performed as previously described.30
Immunohistochemistry of mouse specimens
Paraffin embedded specimens were deparaffinized in xylene, subjected to heat mediated antigen-retrieval in 10mM sodium citrate (pH 6.0), permeabilized in 0.2% Triton-100 (Sigma), treated with mouse on mouse blocking reagent (Vector Laboratories), and blocked in 5% donkey sera. Parkin was detected using a mouse monoclonal anti-Parkin antibody (Cell signal #4211) (1:50) and an HRP-conjugated donkey anti-rabbit secondary (1:250, Jackson immunochemicals), amplified with AB reagent (Vectastain) and detected using DAB reagent (Thermo Scientific). Images were acquired using a Zeiss Axioplan 2 microscope.
Immunohistochemistry and immunofluorescence of human specimens
Human lung biopsy specimens were obtained from patients with active tuberculosis. Paraffin embedded specimens were deparaffinized in xylene, subjected to heat mediated antigen-retrieval in 10mM sodium citrate (pH 6.0), permeabilized in 0.2% Triton-100 (Sigma) and blocked in 5% donkey sera. For immunohistochemistry, Parkin was detected using a rabbit polyclonal anti-Parkin antibody (abcam) (1:25) and an HRP-conjugated donkey anti-rabbit secondary (1:250, Jackson immunochemicals), amplified with AB reagent (Vectastain) and detected using DAB reagent (Thermo Scientific). Images were acquired using a Zeiss Axioplan 2 microscope. For immunofluorescence, Parkin was identified using rabbit polyclonal anti-Parkin (Abcam15954) at 1:25 and an HRP conjugated donkey anti-rabbit secondary (1:100, Jackson Immunochemicals) followed by amplification with tyramide (1:50, Perkin Elmer). TB was identified using guinea pig anti-TB (1:25,) and an Alexa 488 conjugated donkey anti-guinea pig secondary (1:100, Jackson Immunochemicals). Images were acquired using a Leica TCS SP5 confocal microscope. All human tissue specimens were obtained with consent. This study of human tissue specimens has been exempted under 45 CFR 46.101(b) and was approved by the Institutional Review Board at the University of Texas Southwestern Medical Center.
Supplementary Material
a, Quantification of PARKIN-positive M. tuberculosis in BMDMs from wild-type and Park2−/− mice, from Figure 1a. b, BMDMs from LC3-GFP transgenic mice were transduced with lentivirus expressing either a scrambled shRNA control (Ctrl) or shRNAs targeting either LRSAM1 or PARKIN. Lentiviral transduced cells were than infected with mCherry-expressing M. tuberculosis and the colocalization of GFP-LC3 and ubiquitin was quantified via immunofluorescence. *P<0.014, **P<0.008 by Student’s t-test b, RT-qPCR expression of LRSAM1 and PARKIN transcripts in lentiviral transduced cells from (a). Data shown is expressed relative to actin expression. *P<0.033, **P<0.0035 by Student’s t-test.
a, Wild-type BMDMs were transduced with lentivirus expressing HA-tagged constructs of wild-type ubiquitin (WT), ubiquitin with all lysine residues mutated to arginine except for lysine 63 (K63), or ubiquitin with all lysine residues mutated to arginine except for lysine 48 (K48). Transduced cells were then infected with mCherry-expressing M. tuberculosis and immunostained using anti-HA antibodies 4 hours post-infection. b, Quantification of HA-ubiquitin colocalization with M. tuberculosis from (a). **P<0.001 by Student’s t-test.
a, Cartoon model explaining digitonin differential permeabilization of macrophages and antibody accessibility to phagosomes. b, Microscopy images of Wild-type BMDMs were infected with mCherry expressing M. tuberculosis. Cells were immunostained via digitonin permeabilization alone or digitonin permeabilization with triton X-100 treatment. c, Quantification of ubiquitin colocalization with M. tuberculosis from (b). N.D., not determined.
Lung biopsy samples were obtained from three different human patients with active tuberculosis. Immunohistochemistry was performed on specimens using either anti-PARKIN, anti-M. tuberculosis or an IgG control antibody. Positive cells were visualized via DAB staining. Scale bar = 1003m.
Acknowledgments
We thank Noboru Mizushima, Sara Cherry, and Kassidy Huynh for mice and reagents. We are grateful to Sandy Johnson for use of his microscope, members of the Schneider lab for assistance with fly work and Dan Portnoy, Russell Vance, and Skip Virgin for helpful discussions. This work was supported by NIH grants R01 AI081727, P01 AI063302and R01 AI099439.
Footnotes
Author Contributions
A.C.C. and M.U.S. performed immunohistochemistry staining of tissues and confocal microscopy of human lungs. P.S.M., C.S.R., and G.S. performed listeria infections. J.S.A. performed all experiments involving Drosophila melanogaster. R.O.W. assisted in microscopy. P.S.M. performed all experiments involving M. tuberculosis. K.N. and D.S.S. provided reagents and resources. P.S.M. and J.S.C. conceived the study, designed the experiments, and wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Zhao Z, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host & Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host & Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild P, et al. Phosphorylation of the Autophagy Receptor Optineurin Restricts Salmonella Growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature cell biology. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 8.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 9.Chopra R, et al. Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups. PLoS Genet. 2013;9:e1003578. doi: 10.1371/journal.pgen.1003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mira MT, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 11.Ali S, et al. PARK2/PACRG polymorphisms and susceptibility to typhoid and paratyphoid fever. Clin Exp Immunol. 2006;144:425–431. doi: 10.1111/j.1365-2249.2006.03087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romagnoli A, et al. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells. Autophagy. 2012;8:1357–1370. doi: 10.4161/auto.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huett A, et al. The LRR and RING Domain Protein LRSAM1 Is an E3 Ligase Crucial for Ubiquitin-Dependent Autophagy of Intracellular Salmonella Typhimurium. Cell Host & Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponpuak M, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson’s disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D, et al. Parkin mono-ubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–38223. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim KL, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 19.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 20.Houben D, et al. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 2012;14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 21.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins CA, et al. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marín I, Ferrús A. Comparative genomics of the RBR family, including the Parkinson’s disease-related gene parkin and the genes of the ariadne subfamily. Mol Biol Evol. 2002;19:2039–2050. doi: 10.1093/oxfordjournals.molbev.a004029. [DOI] [PubMed] [Google Scholar]
- 24.Moy RH, Cherry S. Antimicrobial autophagy: a conserved innate immune response in Drosophila. J Innate Immun. 2013;5:444–455. doi: 10.1159/000350326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yano T, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voronin D, Cook DAN, Steven A, Taylor MJ. Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proceedings of the National Academy of Sciences. 2012;109:E1638–46. doi: 10.1073/pnas.1203519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 29.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stavru F, Bouillaud F, Sartori A, Ricquier D, Cossart P. Listeria monocytogenes transiently alters mitochondrial dynamics during infection. Proceedings of the National Academy of Sciences. 2011;108:3612–3617. doi: 10.1073/pnas.1100126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson BN, Berger AK, Cortese GP, LaVoie MJ. The ubiquitin E3 ligase parkin regulates the proapoptotic function of Bax. Proceedings of the National Academy of Sciences. 2012;109:6283–6288. doi: 10.1073/pnas.1113248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim KY, et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. The Journal of clinical investigation. 2011;121:3701–3712. doi: 10.1172/JCI44736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Leseleuc L, et al. PARK2 mediates interleukin 6 and monocyte chemoattractant protein 1 production by human macrophages. PLoS Negl Trop Dis. 2013;7:e2015. doi: 10.1371/journal.pntd.0002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg MS, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 37.Ohol YM, et al. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host & Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 39.Ayres JS, Schneider DS. A Signaling Protease Required for Melanization in Drosophila Affects Resistance and Tolerance of Infections. PLoS Biol. 2008;6:e305. doi: 10.1371/journal.pbio.0060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a, Quantification of PARKIN-positive M. tuberculosis in BMDMs from wild-type and Park2−/− mice, from Figure 1a. b, BMDMs from LC3-GFP transgenic mice were transduced with lentivirus expressing either a scrambled shRNA control (Ctrl) or shRNAs targeting either LRSAM1 or PARKIN. Lentiviral transduced cells were than infected with mCherry-expressing M. tuberculosis and the colocalization of GFP-LC3 and ubiquitin was quantified via immunofluorescence. *P<0.014, **P<0.008 by Student’s t-test b, RT-qPCR expression of LRSAM1 and PARKIN transcripts in lentiviral transduced cells from (a). Data shown is expressed relative to actin expression. *P<0.033, **P<0.0035 by Student’s t-test.
a, Wild-type BMDMs were transduced with lentivirus expressing HA-tagged constructs of wild-type ubiquitin (WT), ubiquitin with all lysine residues mutated to arginine except for lysine 63 (K63), or ubiquitin with all lysine residues mutated to arginine except for lysine 48 (K48). Transduced cells were then infected with mCherry-expressing M. tuberculosis and immunostained using anti-HA antibodies 4 hours post-infection. b, Quantification of HA-ubiquitin colocalization with M. tuberculosis from (a). **P<0.001 by Student’s t-test.
a, Cartoon model explaining digitonin differential permeabilization of macrophages and antibody accessibility to phagosomes. b, Microscopy images of Wild-type BMDMs were infected with mCherry expressing M. tuberculosis. Cells were immunostained via digitonin permeabilization alone or digitonin permeabilization with triton X-100 treatment. c, Quantification of ubiquitin colocalization with M. tuberculosis from (b). N.D., not determined.
Lung biopsy samples were obtained from three different human patients with active tuberculosis. Immunohistochemistry was performed on specimens using either anti-PARKIN, anti-M. tuberculosis or an IgG control antibody. Positive cells were visualized via DAB staining. Scale bar = 1003m.