Abstract
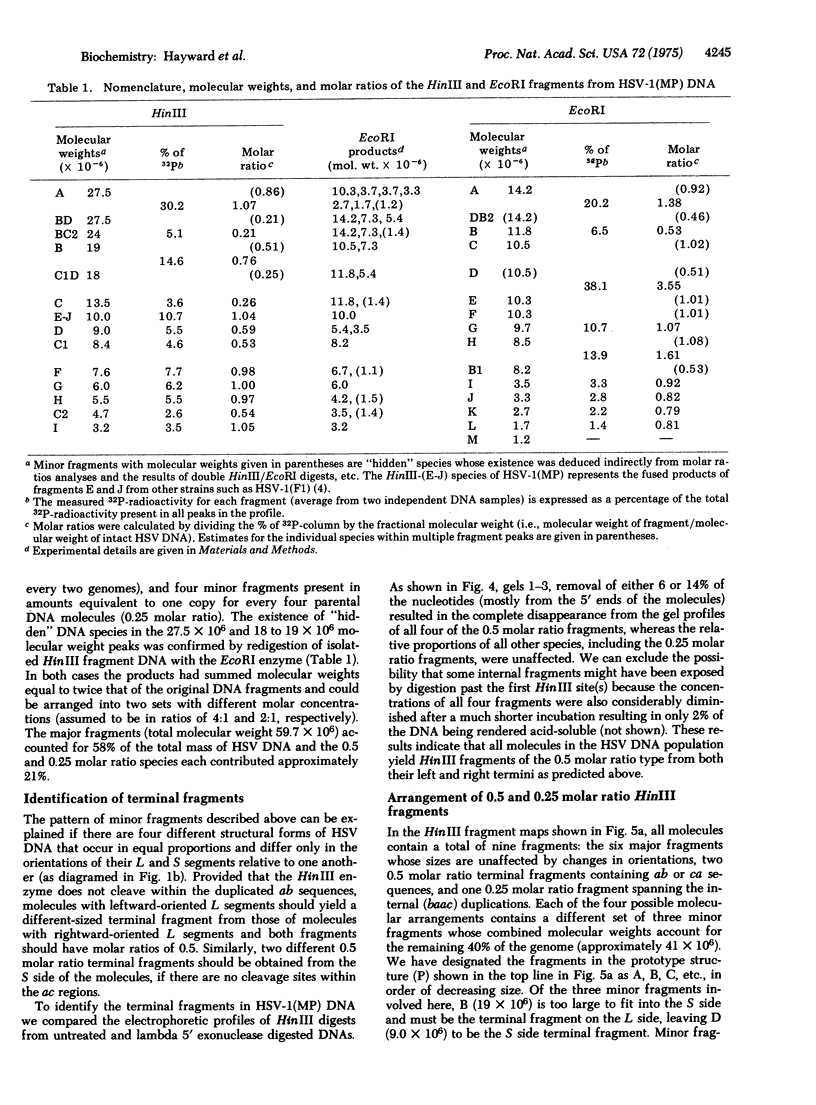
Intact DNA molecules extracted from HSV-1 (herpes simplex virus 1, human herpes virus 1) strain MP virions have a molecular weight of approximately 97 X 10(6), but cleavage with the HinIII restriction enzyme yields fourteen fragments with summed molecular weights of 160 X 10(6). Six "major" fragments occur once in every molecule in the population and account for 60% of the genetic information. Four "minor" fragments are present in amounts equivalent to one copy for every two genomes (0.5 molar ratio) and the other four occur only once in every four molecules (0.25 molar ratio). The minor fragments can be arranged into four equimolar sets, each with summed molecular weights that account for the remaining 40% of the genome. Treatment with lambda 5' exonuclease revealed that all molecules contain 0.5 molar ratio fragments at both termini. These observations and the results of similar analyses of the EcoRI and double HinIII/EcoRI digests indicate that there are four distinct structural forms of HSV DNA which differ only in the relative orientations of two subregions, designated L and S. The L and S segments consist of 82 and 18% of the sequences, respectively, and each has inverted terminally redundant regions that correspond to the internal duplications observed by electron microscopy. The DNA from other strains of HSV-1 and 2 also consists of equal proportions of all four possible permutations of the L and S segments. These unusual features of HSV DNA molecules have novel implications with regard to the genetic map and the mode of replication and evolution of herpes simples viruses.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Jeppesen P. G., Katagiri K. J., Delius H. Mapping the DNA fragments produced by cleavage by lambda DNA with endonuclease RI. Nature. 1973 Jan 12;241(5385):120–123. doi: 10.1038/241120a0. [DOI] [PubMed] [Google Scholar]
- Bronson D. L., Dreesman G. R., Biswal N., Benyesh-Melnick M. Defective virions of herpes simplex viruses. Intervirology. 1973;1(3):141–153. doi: 10.1159/000148841. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Jacob R. J., Honess R. W., Hayward G. S., Locker H., Roizman B. Anatomy of herpes simplex virus DNA. III. Characterization of defective DNA molecules and biological properties of virus populations containing them. J Virol. 1975 Jul;16(1):153–167. doi: 10.1128/jvi.16.1.153-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Herpes vimplex virus: genome size and redundancy studied by renaturation kinetics. J Virol. 1971 Oct;8(4):591–593. doi: 10.1128/jvi.8.4.591-593.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom R. H., Alwine J. C., Steinhart W. L., Hill C. W. Terminal repetitions in herpes simplex virus type 1 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):679–681. doi: 10.1101/sqb.1974.039.01.081. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Frenkel N., Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci U S A. 1975 May;72(5):1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Electron microscope heteroduplex study of the heterogeneity of Mu phage and prophage DNA. Virology. 1974 Mar;58(1):229–239. doi: 10.1016/0042-6822(74)90157-3. [DOI] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Regulation of lambda exonuclease. I. Properties of lambda exonuclease purified from lysogens of lambda T11 and wild type. J Mol Biol. 1966 Jul;18(2):235–250. doi: 10.1016/s0022-2836(66)80243-7. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. P., Summers W. C. Structure and function of herpesvirus genomes. I. comparison of five HSV-1 and two HSV-2 strains by cleavage their DNA with eco R I restriction endonuclease. J Virol. 1975 Apr;15(4):726–732. doi: 10.1128/jvi.15.4.726-732.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear P. G., Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972 Jan;9(1):143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]





