Abstract
Objective
MEIS1 is a Hox cofactor known for its role in development and strongly linked to normal and leukemic hematopoiesis. Although previous studies have focused on identifying protein partners of MEIS1 and its transcriptionaly regulated targets, little is known about the upstream transcriptional regulators of this tightly regulated gene. Understanding the regulation of MEIS1 is important toward the understanding of normal hematopoiesis and leukemogenesis.
Materials and Methods
Here we describe our studies focusing on the evolutionary conserved putative MEIS1 promoter region. Phylogenetic sequence analysis followed and reporter assays in MEIS1 expressing (K562) and non-expressing (HL60) leukemic cell line models were used to identify key regulatory regions and potential transcription factor binding sites within the candidate promoter region followed by functional and expression studies of one identified regulator in both cell lines and primary human cord blood and leukemia samples.
Results
The chromatin status of MEIS1 promoter region is associated with MEIS1 expression. Truncation and mutation studies coupled with reporter assays revealed that a conserved ETS family member binding site located 289bp upstream of the annotated human MEIS1 transcription start site (AHTSS) is required for promoter activity. Of the three ETS family members tested, only ELF1 was enriched on the MEIS1 promoter as assessed by both electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP) experiments in K562. This finding was confirmed in MEIS1 expressing primary human samples. Moreover, siRNA -mediated knockdown of ELF1 in K562 cells was associated with a decreased MEIS1 expression.
Conclusions
We conclude that the ETS transcription factor ELF1 is an important positive regulator of MEIS1 expression.
Keywords: MEIS1, Gene regulation, Leukemia, Homeobox, ETS family member
MEIS1 (myeloid ectropic viral integration site 1) belongs to the TALE (three amino acid loop extension) family of homeodomain-containing proteins and is a HOX cofactor. Key roles of this transcription factor are now documented both in normal development and disease formation (1–4). Decreased MEIS1 levels are associated with Restless leg syndrome (RLS) (5, 6). Overexpression of MEIS1, on the other hand, has been linked to leukemia (3, 4).
Meis1 was first identified as a common integration site of B-ecotropic provirus in a spontaneous mouse leukemia model, BXH-2 (7). The up regulation of Meis1 was often associated with the proviral insertional coactivation of Hoxa7 and Hoxa9 (8). Consistent with these initial observations, subsequent studies revealed that engineered overexpresssion of MEIS1 was a potent collaborating event with overexpresssed Hox genes or NUP98-HOX fusion genes that accelerates the onset of AML in murine bone marrow transplantation models (9–12). MEIS1 is frequently upregulated in human primary acute myelogenous leukemia (AML) and acute lymphocytic leukemia (ALL) samples (13–15). Recently Wong et al. have shown that MEIS1 is an essential and rate-limiting regulator of MLL-leukemia stem cell (LSC) potential in murine models (16).
In normal hematopoiesis, Meis1 expression appears to be tightly regulated with highest expression in the most primitive stem/progenitor cell enriched subpopulations and rapid downregulation in later stages of hematopoietic development (15, 17). Consistent with this, Meis1 knockout (Meis1−/−) mouse embryos die in utero between 12.5dpc - 14.5dpc with a dramatic reduction in myeloerythroid progenitors and extensive hemorrhaging due to the lack of megakaryocytes. Strikingly, Meis1−/− fetal liver cells fail to radio protect lethally irradiated recipient mice and compete poorly in hematopoietic competitive reconstitution assays (18). Taken together, these studies indicate that strict regulated expression of MEIS1 is essential for normal hematopoiesis and that deregulated overexpression of MEIS1 plays a major role in leukemogenesis.
While downstream transcriptional targets of MEIS1 are now emerging, little is known about how MEIS1 expression itself is regulated. Pan-chromatin modifiers, such as the trithorax group member, MLL, or the polycomb group member, EZH2 appear to be important regulators of MEIS1 (19, 20). Transcription factors such as CREB, GFI1 and recently HOXA9 also have been reported to be involved in MEIS1 regulation, but the molecular mechanisms of how they affect MEIS1 transcription are unclear (21–23). Detailed comparative sequence analysis of the 140 kb human MEIS1 locus reveals a high degree of evolutionary conservation not only in the exons, but also in the non-coding regions, namely introns and adjacent upstream and downstream regions. This suggests that multiple cis-regulatory elements and multiple transcription factors are involved in MEIS1 transcriptional regulation. As an initial step to elucidate the mechanisms of MEIS1 regulation, we have focused on the putative MEIS1 promoter region. We show that the chromatin status of this region is related to MEIS1 expression and identify key regions required for promoter activity. Truncation and mutation studies reveal that a conserved ETS family member binding site located 289bp upstream of the annotated human MEIS1 transcription start site (AHTSS) is required for promoter activity. Additional studies including ChIP analysis in MEIS1 expressing human primary cells or a cell line K562 and knockdown studies in K562 identify ELF1 as a strong candidate positive regulator of MEIS1.
Material and methods
Sequence analysis
The UCSC genome browser (http://genome.ucsc.edu/) was used to obtain the snap shot of the human MEIS1 locus (chr2:66,500,000–66,700,000) and the MEIS1 promoter region (chr2:66,515,181–66,516,635 in the Mar. 2006, hg 18 version). The settings were: Base position dense; Ref gene full; CpG island full; HAIB TFBS full with the selection of Pol II and TAF1 raw data in K562 cells; Conservation full with selected representative species: mouse, frog and zebrafish; AceView full for the promoter region. Candidate transcription factor binding sites were predicted through TRANSFAC (http://www.generegulation.com/pub/databases.html) (24); TFS search (http://www.cbrc.jp/research/db/TFSEARCH.html) (25) and JASPAR database (http://jaspar.genereg.net) (26).
Human primary samples and cell culture
Cord blood was obtained from consenting mothers undergoing cesarean delivery of healthy, full-term infants. Leukemic samples were obtained from chronic phase (CP) CML patients at the time of initial diagnosis with informed consent according to protocols approved by the Research Ethics Board of the University of British Columbia. To enrich for hematopoietic stem cell/progenitor cells, light density cells were first isolated by centrifugation on ficoll-hypaque and then cells expressing the following lineage (lin) markers: CD2, CD3, CD14, CD16, CD19, CD24, CD56, CD66b and glycophorin A were removed immunomagnetically using a column (StemSep, StemCell Technologies, Vancouver, BC) following manufacturer’s recommendation. Cells were then cryopreserved with DMSO and fetal bovine serum (FBS) until further use. Thawed cells were used for RNA isolation and ChIP assays. K562 cells and HL60 cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) FBS, 100U/mL penicillin, 100U/mL streptomycin. All cultures were maintained at 37 °C in 5% CO2.
Reverse transcription and real-time PCR
Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. cDNAs from different samples, were generated using the Superscript III system (Invitrogen) with random primers. Real-time PCR was performed with cDNA corresponding to 20ng RNA as template and gene- specific primers in a thermal cycler (7900 HT Fast Real-Time PCR System, Applied Biosystems, Foster City, CA, USA) at the following conditions: 95°C for 5 min followed by 40 cycles at 94°C for 45 s and 60°C for 1 min; the last cycle was followed by a 5 min extension at 60°C. Three quantitative PCR replicates were done for each sample. Gene-specific primers were designed using the Universal Probe Library Assay Design Center (https://www.roche-applied-science.com/, Roche diagnostics, Indianapolis, IN, USA). See supplementary Table 1 for primers’ sequence.
DNase I hypersensitivity assay
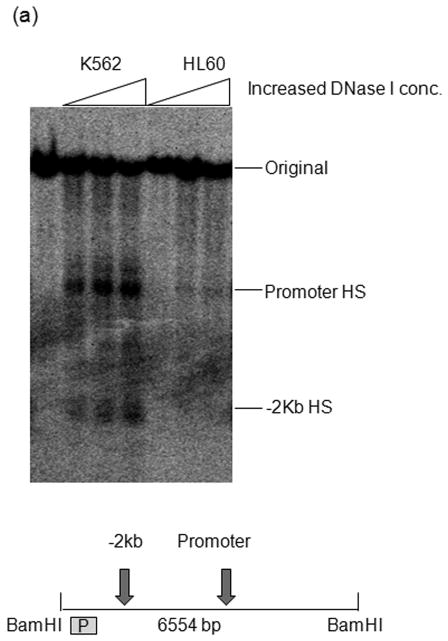
DNase I was purchased from Worthington Biochemical Corporation (Lakewood, NJ, USA). Nuclei isolation and DNase I digestion were done as described previously (27). Nuclei from 10 million K562 or HL60 cells were treated for each of the different DNase I concentrations ranging from 3 unit/ml to 24 unit/ml for 10min. The purified DNA was digested with BamH I (NEB, Ipswich, MA, USA), separated on a 0.8 % agarose gel and subjected to Southern blotting as described (27). The probe shown in Figure 6 was a 0.5 kb fragment (hg 18, Chr2:66,513,458–66,513,961), generated by PCR and sequencing confirmed.
Fig. 6.
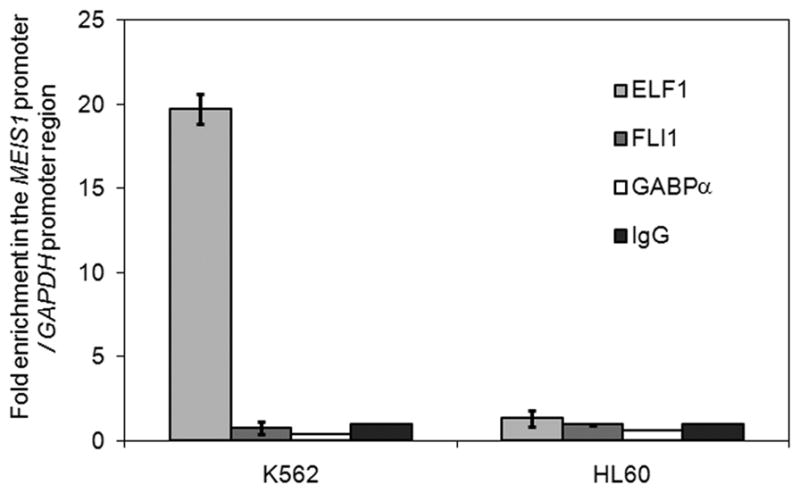
ELF1 is enriched in MEIS1 promoter in MEIS1 expressing K562 cell, where no enrichment is detected in no MEIS1 expressing HL60 cell lines through the ChIP analysis. ChIP analysis is done with the antibodies to three ETS family members, ELF1, FLI1 and GABPα and normal rabbit IgG. Relative enrichment of the MEIS1 promoter region by the specific antibody is shown as the ratio of the PCR readings of the MEIS1 promoter primer set over the internal control GAPDH promoter primer set adjusted by the ratio in the normal rabbit IgG control.
Chromatin immunoprecipitation (ChIP) assay
Rabbit polyclonal antibodies against histone H3 acetylated at lysines 9 and 14 (06-599) and normal rabbit IgG (12–370) were purchased from Upstate Biotechnology (Lake Placid, NY, USA). ELF1 antibody (C-20 X), FLI1 antibody (C-19 X) and GABPα antibody (H-2 X) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Syber green PCR kits were from Invitrogen. ChIP assays were performed as previously described with modifications (28). Human GAPDH was selected as a reference control. The primers used for real-time PCR were in Supplementary Table 1. Real time PCR was performed in the 7900 HT Fast Real-Time PCR System (Applied Biosystems) as described above. All data were expressed as the ratio of the quantitative PCR readings of a given primer set over the internal reference control at least in duplicate, from at least two independent experiments. Results were analyzed with RQ manager version 1.2 software (Applied Biosystem) relative to normal rabbit IgG control using the 2ΔΔCt method.
DNA methylation analysis of the MEIS1 promoter region
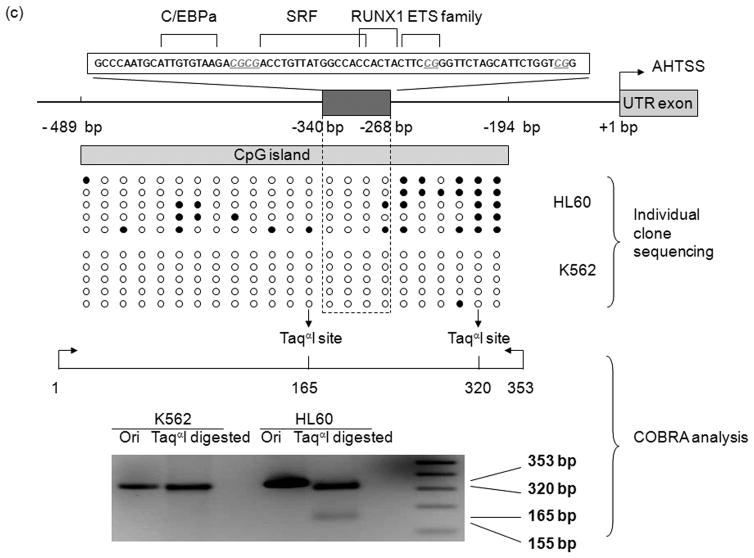
Genomic DNA was modified with sodium bisulfite as described (29). Converted DNA was used as a template in the following first round of PCR amplifying the MEIS1 promoter region with forward and reverse flanking bisulfite primers. After initial denaturation for 7 min at 95°C, 35–40 cycles were performed, each consisting of 1 min at 95°C, 1 min at 48°C + 0.1°C/cycle and 50 s at 72°C with a final extension of 7 min at 72°C. 2 μl of the first PCR was used for semi-nested amplification using MEIS1 promoter forward and reverse nested bisulfite primers (supplementary Table 1). The same amplification conditions were used as for the first-round PCR. The resulting products were fractionated on 1% agarose gels and the correct size bands were extracted using the MinElute gel extraction kit (Qiagen Inc., Mississauga, ON, Canada). The purified products were either subjected to combined bisulfite restriction analysis (COBRA) or directly cloned into the TOPO-TA cloning vector (Invitrogen). Sequencing was performed using the M13 reverse primer by McGill University and Genome Québec Innovation Centre sequencing facility. All clones (five representative clones for each cell line) included in the figures are unique as either they contain a different CpG pattern; unique unconverted cytosines in non-CpG position given the majority (>95%) of such cytosines are successfully converted; or unique non-cytosine associated mutations due to the Taq polymerase PCR related error. For COBRA, the gel-purified PCR fragments were digested with TaqαI restriction enzymes (NEB). TaqαI recognizes and cuts 5′-TCGA-3′, where the C (deoxycytidine) need to be originally methylated in the genomic DNA and therefore would not have been converted to T (thymidine) by sodium bisulfite treatment.
Generation of luciferase reporter constructs
Promoter fragments were generated by PCR using forward primers with a Kpn I (NEB) site inserted at the 5′ end and reverse primers with a Bgl II (NEB) site inserted at the 3′ end. PCR products were digested with Kpn I/Bgl II and cloned into pGL3 firefly luciferase basic vector (Promega, Madison, WI, USA). The primers used were as followings: 500 bp, 340 bp, 305 bp, 268 bp, 200bp upstream forward and 20 bp downstream reverse (supplementary Table 1). To generate the designed mutations, an A to G mutation was first introduced into the 267 bp position upstream of the MEIS1 initiation site through a PCR-based mutation approach to generate an EcoR I site in the 500 bp promoter construct. The promoter activities were tested in the luciferase assay and the EcoR I mutation did not result in significant change as compared to the wild type promoter (Supplementary figure 2). Annealed double stranded oligomers containing the region from 340 bp to 268 bp upstream of the MEIS1 initiation site with specific mutations or deletions were used to substitute the region between Pst I/EcoR I sites in the 500 bp MEIS1 construct with EcoR I mutation. The oligomers used to introduce mutations were in Supplementary Table 2.
Cell transfection and luciferase assays
K562 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were then incubated at 37°C for 36 h before assaying for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized relative to Renilla luciferase activity for each transfection and calculated as fold increase over pGL3 firefly luciferase basic vector (pGL3Basic). At least two independent transfection and two replicates each time were performed for each individual construct. In Figure 4, the mutation or deletion effects were normalized to the 500 bp MEIS1 promoter with the EcoR I mutation.
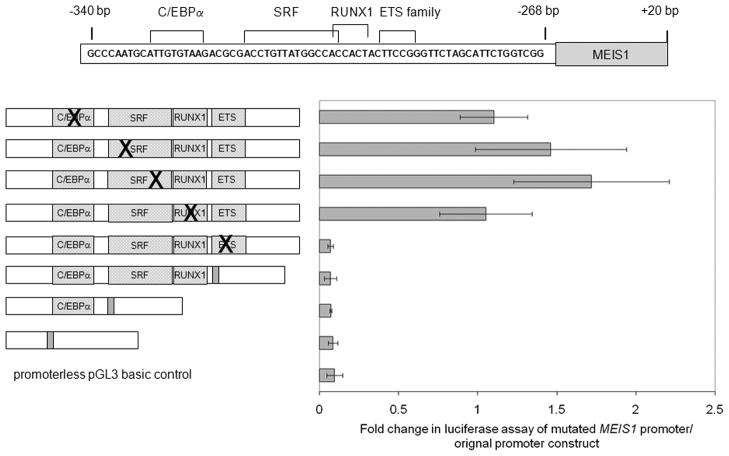
Fig. 4.
The predicted ETS family member binding site is important for the activity of MEIS1 promoter. Point mutations or deletion of individual predicted transcription factor binding sites were introduced into the original promoter construct as indicated on the left and assayed for promoter activities by luciferase assay as shown on the right in K562 cells. Promoter activity is shown relative to the original promoter construct (mean ±sd for 2 experiments each done in duplicate).
Electrophoretic mobility shift assay (EMSA)
Double stranded probes were generated by annealing the following oligomers to their complementary oligomers: wild type, 5′-GTTATGGCCACCACTACTTCCGGGTTCTAG-3′; and mutation, 5′-GTTATGGCCACCACTACAGATCTGTTCTAG-3′. Nuclear extraction, probe labeling and the gel shift assay were preformed as described (30). Protein concentration was determined using the Qubit Quantitation Platform (Invitrogen).
siRNA knockdown of ELF1
K562 cells (5 × 104 per well) were seeded in 96-well plates and transfected with 1 μM Accell SMARTpool human ELF1 siRNA or Accell Non-Targeting Pool siRNA using 100 μL Accell siRNA Delivery media (Dharmacon, Lafayette, CO, USA). An untreated sample was also included where the media was exchanged but no siRNA was added. Seventy-two hours later, total RNA collection and RT-PCR were carried out as described above. Three wells were pooled per treatment. Experiments were replicated three times. Real-time PCR was performed using the cDNA as template as described above.
Results
The MEIS1 promoter region is highly conserved
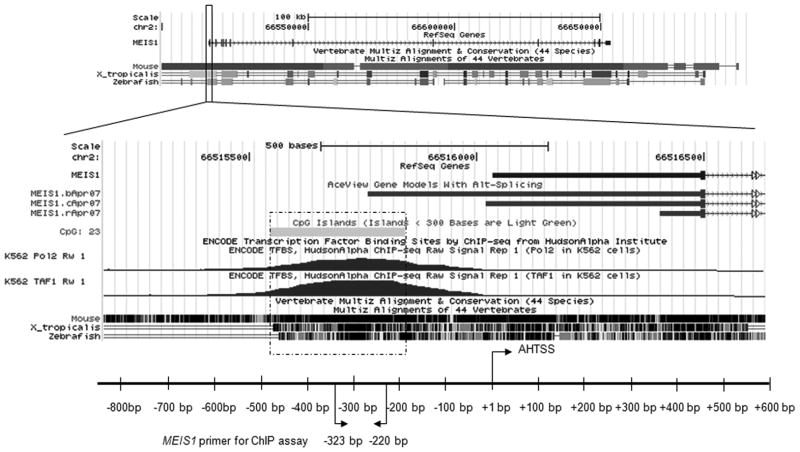
To understand the transcriptional regulation of MEIS1, we examined the human MEIS1 gene for regions of high evolutionary conservation using the UCSC genome browser (Figure 1). The MEIS1 gene spans a large chromatin region of ~140 kb in both the human and mouse genomes and its sequence is evolutionarily conserved not only in all of its exons, but also in many non-coding regions including several intronic regions and adjacent downstream and upstream genomic regions (Figure 1). The existence of multiple highly conserved regions outside of exonic sequence suggests the existence of multiple cis-regulatory elements and a high degree of complexity in the transcriptional control of MEIS1 expression. As an initial step toward understanding the transcriptional regulation of MEIS1, we focused on the putative promoter region between 267 bp to 403 bp upstream of the annotated human transcription start site (AHTSS). This region shows more than 70% identity between human and zebrafish sequence and is within a CpG island (between 194 bp to 489 bp upstream of the AHTSS). Interestingly, there is no TATA box before the AHTSS and results of genome wide ChIP analysis reveal peaks of RNA polymerase II or TAF1 binding at the end of this CpG island, around 184 bp to 283 bp upstream relative to the AHTSS in human erythroleukemia K562 cells which express MEIS1. The UCSC database AceView track, which provides a comprehensive representation of non-redundant sequences of all public mRNA sequences, suggests that an alternative initiation site exists within this region at 274 bp upstream of the AHTSS.
Fig. 1.
MEIS1 region is evolutionarily conserved. Top panel is the snap shot of the human MEIS1 locus between chr2: 66,500,000–66,700,000 (hg18) in the UCSC genome browser. The conservation tracks comparisons of human to mouse, frog (x_tropicalis) and zebrafish within this region are displayed. Bottom panel depicts an expanded detailed view of the MEIS1 promoter region. The annotated human transcription start site (AHTSS) of the reference gene MEIS1 is marked as position +1 bp (x axis).
The chromatin status of the MEIS1 promoter region is related to its expression
In order to assess the chromatin status of the MEIS1 promoter region, we performed DNase I hypersensitivity assay, ChIP with acetylated H3 antibody and DNA methylation analysis in two human leukemia cell lines that expresses or does not express MEIS1: K562 and HL60, respectively (Supplementary figure 1). The MEIS1 promoter region was DNase I hypersensitive in K562 cells but only minimally sensitive in HL60 cells (Figure 2a). Consistent with these findings, the level of acetylated histone H3 at the MEIS1 promoter region was low in HL60 but high in K562 cells which showed a level comparable to the highly expressed house-keeping gene, GAPDH (Figure 2b). Moreover, the CpG island region of the MEIS1 promoter was more methylated in HL60 than in K562 cells as assessed by bisulfite sequencing and COBRA analysis (Figure 2c). Together these data reveal a strong correlation of epigenetic modification of the MEIS1 promoter region and its expression.
Fig. 2.
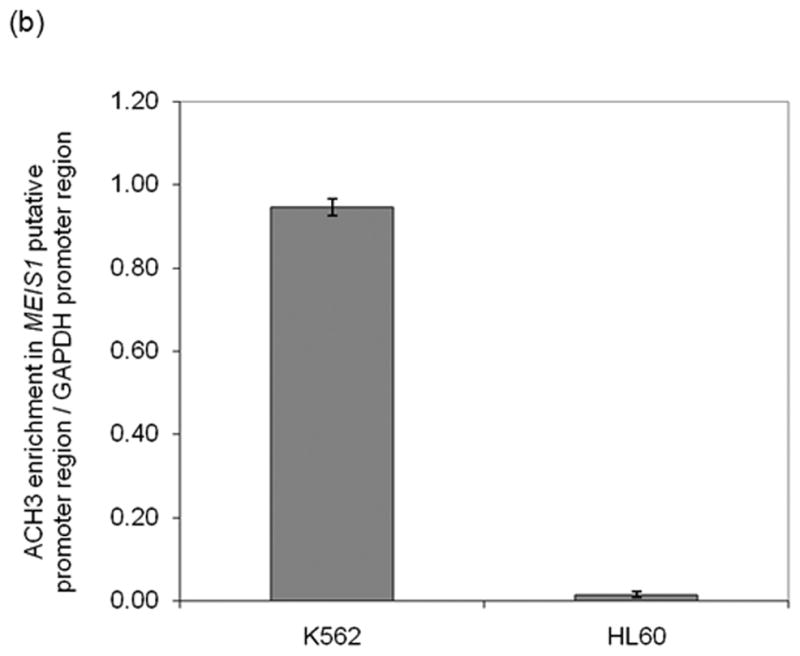
MEIS1 promoter region is accessible in MEIS1 expressing K562 cells, but not in non MEIS1 expressing HL60 cells. A) DNase I hypersensitivity assay. In K562 cells, a strong DNase I hypersensitive site (HS) was formed around MEIS1 promoter region, which was very weak in HL60 cells. B) Real time PCR results from ChIP assay with acetylated Histone H3 K9/27 antibody. MEIS1 promoter region had a similar level of acetylated Histone H3 K9/27 as the housekeeping gene GAPDH promoter region in K562, but was barely acetylated in HL60 cells. C) DNA methylation status in the CpG island of MEIS1 promoter. Ten individual clones were sent for sequencing and five unique representative clones for each cell line were shown. Open circles: unmethylated CpG; closed circles: methylated CpG. Putative transcription factor binding sites such as C/EBPα, SRF and ETS family member are shown in scale with the particular CpG sites highlighted. PCR products of the MEIS1 promoter region corresponding to 167 bp to 520 bp upstream to the AHTSS region contains two TaqαI restriction enzyme digestion sites at the 165 and 320 positions. There are more methylated CpGs in the MEIS1 promoter region in HL60 than in K562 cells as seen from both direct sequencing and COBRA analysis.
The region between 268 bp to 305 bp upstream of the MEIS1 AHTSS contributes strongly to promoter activity
The putative promoter region, which is between 500 bp upstream to 20 bp downstream of the MEIS1 AHTSS, was inserted into the pGL3 basic vector and this construct (referred to as the 500 bp promoter construct in the following text) was tested by luciferase reporter assay. As shown in Supplementary figure 2, this region conferred strong promoter activity that was comparable (~50%) to the activity of the positive control SV40 promoter in K562 cells. To delineate the functional sites within this region, we assessed the promoter activity of a truncation series of the promoter region as depicted in Figure 3. Removal of the region between 305 bp to 500 bp upstream of the AHTSS caused an ~66% reduction in promoter activity compared to the full length promoter construct. Further deletion of the region between 268 bp to 305 bp resulted in a further significant decrease of activity to essentially background levels. Additional removal of the region between 200 bp to 268 bp region slightly increased the activity but not significantly above that of background. This suggests that the 268 bp to 305 bp region upstream of AHTSS is crucial for MEIS1 promoter function and the region between 305 bp to 500 bp also positively contributes to its activity.
Fig. 3.
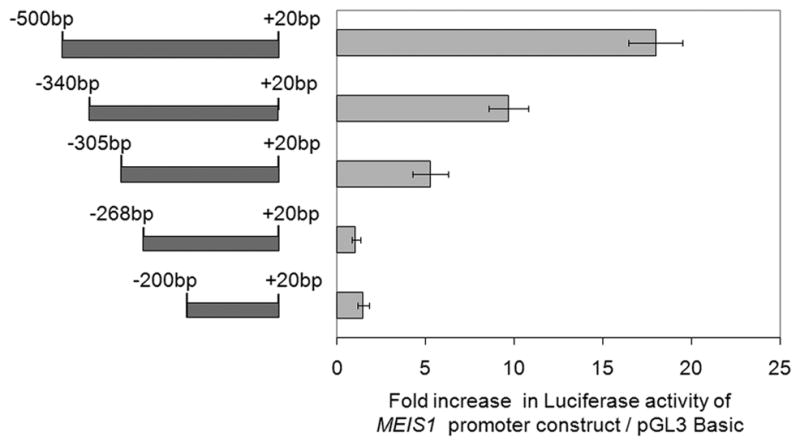
The conserved region between 268 bp to 305 bp upstream of the AHTSS contributes to the MEIS1 promoter activity. Different sized promoter regions of MEIS1 as shown on the left were fused to the pGL3 basic construct and assayed for their promoter activities by luciferase assay as shown on the right in K562 cells. Promoter activity is shown relative to the pGL3 basic vector (mean ±sd for 2 experiments each done in duplicate).
The predicted ETS family member binding site is required for the MEIS1 promoter activity
Sequence analysis of the 268 bp to 305 bp upstream promoter region revealed many predicted transcription factor binding sites, including for serum response factor (SRF), RUNX1 and ETS family members (Figure 4). Luciferase reporter assays were then performed using the 500 bp promoter construct with several of these binding sites individually mutated or deleted (Figure 4). A predicted C/EBPα binding site around site 330 bp was also included in this mutation study due to the importance of this transcription factor during hematopoietic differentiation (31). These transcription factor binding sites are well preserved during evolution (Supplementary figure 3). As shown in Figure 4, mutations of the predicted C/EBPα, SRF or RUNX1 binding sites did not significantly diminish the promoter activity. However, all the constructs with mutation or deletions affecting the predicted ETS family member binding site resulted in a dramatic decrease of the promoter activity to background levels.
ELF1 and GABPα bind to the predicted ETS family member binding site in vitro
To elucidate which factor bound to the predicted ETS family member binding site, we performed EMSA with K562 cell nuclear extracts. Using a labeled oligo probe corresponding to the putative ETS binding region between 277 bp to 294 bp upstream of the AHTSS we detected a specific band that was readily competed off with a cold wild type probe but not by a cold probe with a mutation in the ETS binding site (Figure 5). Moreover, this band was supershifted by antibodies to the ETS family members, ELF1 and GABPα, but not FLI1. These data suggest that ELF1 and GABPα may interact with each other and form a single complex with the putative ETS binding region. However, it could also be that this band includes multiple complexes, at least one involving ELF1, another involving GABPα, with identical mobility. From the ChIP results described below, the latter seems more likely, where ELF1 would bind independently from the GABPα. Together, these data reveal that ETS family members, ELF1 and GABPα have the ability to interact with the predicted ETS binding site at the MEIS1 promoter region in vitro.
Fig. 5.
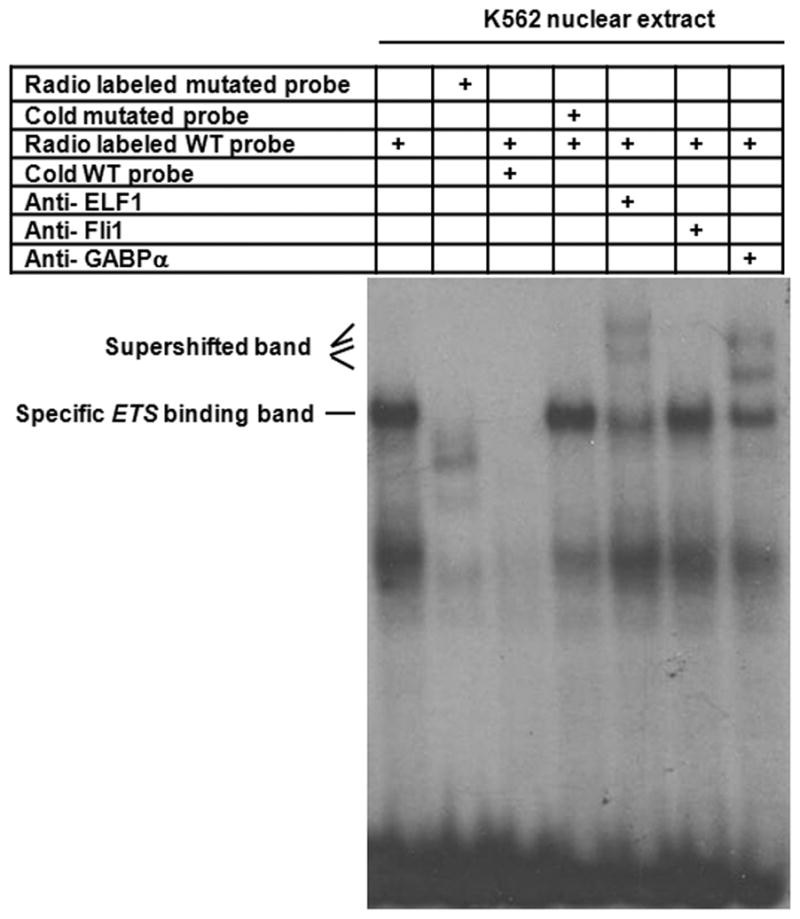
ELF1 binds to the predicted ETS family member binding site in EMSA. Nuclear extracts were prepared from K562 cells and incubated with radio-labeled probe containing the region of 277 bp to 294 bp upstream of the AHTSS prior to gel electrophoresis. The specific ETS family member binding band, which cannot be competed by the cold ETS site mutated probe, was super shifted by the antibodies to either ELF1 or GABPα, but not by the antibody of FLI1.
ELF1, but not FLI1 or GABPα is enriched at the MEIS1 promoter region in a MEIS1 expressing human cell line
To identify the factors that are able to bind the predicted ETS family member binding site under native chromatin conditions, we performed chromatin immunoprecipitation in MEIS1 expressing and non-expressing cells, K562 and HL60 respectively, using antibodies to three ETS family members. As shown in Figure 6, ELF1 strongly bound to the MEIS1 promoter region in K562 cells with an ~ 20 fold enrichment compared to GAPDH control; there was no significant enrichment using antibodies to FLI1 or GABPα in the same region. Interestingly, no binding of ELF1 was detected at the MEIS1 promoter region in HL60 cells (Figure 6) despite the abundant levels of ELF1 in these cells (Supplementary figure 1), suggesting that accessibility of this region is conferred by factors additional to ELF1.
The strong binding of ELF1 to MEIS1 promoter region is observed also in MEIS1 expressing human primary cells
We extended the ChIP experiments to human primary cells from cord blood and primary leukemic cells from patient samples. The hematopoietic stem cell/progenitor cells enriched fraction from cord blood or the leukemic patients express high levels of ELF1 and MEIS1, while for total, non-enriched cord blood only a high level of ELF1, but not MEIS1, was detected at high levels (supplementary Figure 4). As shown in Figure 7, ELF1 significantly bound to the MEIS1 promoter region with an ~5 fold and ~10 fold enrichment compared to GAPDH control in the hematopoietic stem cell/progenitor cells enriched fraction from a representative leukemic patient and cord blood sample respectively, while no enrichment was detected in the un-fractionated total cord blood sample.
Fig. 7.
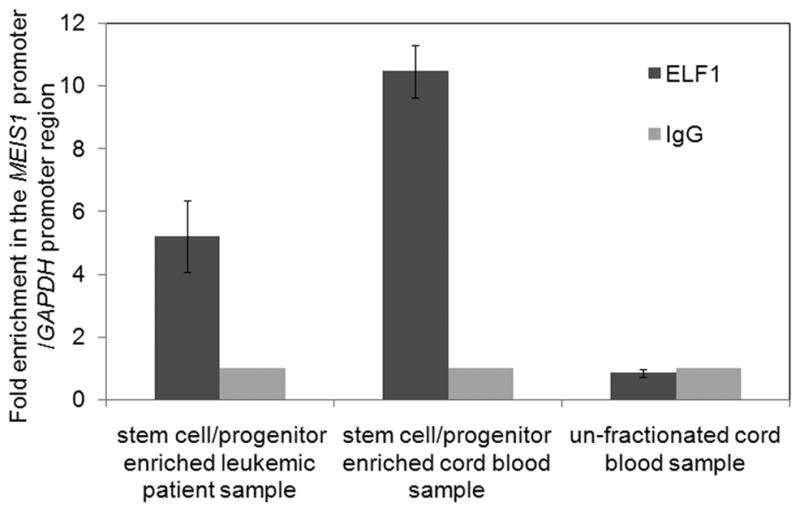
ELF1 is enriched at the MEIS1 promoter in MEIS1 expressing human primary cells though the ChIP analysis. ChIP analysis was performed with the antibodies to ELF1 and normal rabbit IgG as described in Figure 6. The samples used were the un-fractionated cord blood sample, the stem cell/progenitor enriched fraction from a representative leukemic patient sample or the cord blood sample through the lineage depletion methods.
Knockdown of ELF1 through siRNA decreases the expression of MEIS1
To further test the functional role of ELF1 in MEIS1 transcription, siRNA was used to knockdown ELF1. We carried out the experiments in the stem/progenitor enriched fraction from human primary cells. Due to the extremely high expression level of both ELF1 and MEIS1 in both samples (supplementary Figure 4) and the low ELF1 knockdown efficiency (< 20%) we achieved in these primary hematopoietic cells, we did not observe significant change of MEIS1 level. However, in K562 cells, as shown in Figure 8, siRNA knockdown was quite efficient with an ~80% reduction of ELF1 mRNA levels with a concordant reduction (~35%) in MEIS1 mRNA levels. In contrast, the scrambled control siRNA did not significantly affect expression of ELF1 or MEIS1 compared to the untreated K562 cells. These data suggest that ELF1 is positively involved in the transcriptional regulation of MEIS1.
Fig. 8.
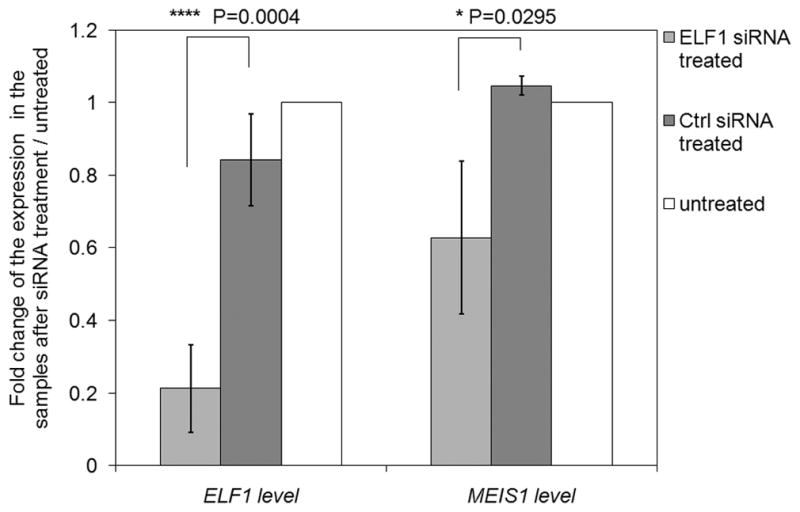
Decreasing ELF1 level through siRNA knockdown decreases the expression of MEIS1 in K562 cells. The change of the expression is shown as the ratio of the PCR readings of specific primer set (either ELF1 or MEIS1) over the internal GAPDH primer set in the treated samples adjusted by the ratio in the untreated samples.
Discussion
As an initial step to understand the transcriptional regulation of MEIS1 we focused on its highly conserved promoter region. Using several chromatin assays in MEIS1-expressing and non-expressing human leukemic cell lines, we demonstrated that MEIS1 expression correlates with an active epigenetic signature of this region. This region displayed a strong promoter activity as shown by the luciferase assays in the MEIS1 expressing human leukemic cell line: K562. Truncation and mutation studies have identified an ETS family binding site that was required for the promoter activity in luciferase assay. Detailed testing of three ETS family members, namely, ELF1, GABPα and FLI1 in our current work shows that ELF1 is a strong candidate as a positive regulator of MEIS1.
ELF1 is an important ETS family member and can act both as an activator and a repressor to regulate transcription of various genes (32). ELF1, as well as FLI1 have been included in the transcriptional network of hematopoietic stem cells (HSC) as they are the regulators of several important HSC factors, such as SCL (32). ELF1 is highly expressed in mouse HSC cells and slightly decreased as the cells differentiate into lineage committed cells (33). Previous studies revealed the presence of ELF1 in all the AML patient samples tested (34). This indicated the constant availability of this factor during MEIS1 expression in at least the hematopoietic system. Our data revealed that ELF1, but not GABPα or FLI1 displayed strong enrichment in the MEIS1 promoter region both from EMSA and ChIP experiments in K562 cells. This was consistent with a recent genome wide ChIP study performed in a human T leukemia cell line, Jurkat, where ELF1 but not the other two ETS family members tested, GABPα and ETS1, was enriched at the MEIS1 promoter region (35). This strong association of ELF1 to the MEIS1 promoter region was also confirmed in human primary samples enriched with the hematopoietic stem/progenitor cells where MEIS1 was highly expressed. We further showed through the siRNA knockdown experiments that the decreased ELF1 level was associated with a decreased MEIS1 expression in K562 cells. Intriguingly, there is a reported alternative MEIS1 transcript initiation site located about 17 bp downstream of the ETS family binding region and the peaks of Pol II and TAF1 binding are more adjacent to this initiation site compared to the AHTSS in the K562 cells (Figure 1). The presence of this ETS family member binding site in the MEIS1 promoter region, therefore, might serve as a recruiting dock for the transcription initiation complex. Consistent with this hypothesis, a systematic analysis of regulatory motifs showed the abundance of ETS family binding sites in human promoters. Moreover, the binding motifs of ETS family members such as ELK1, GABPα and ETS2 have a strong position bias of −23/24 relative to the transcription start site (36) and ETS family members such as Pu.1 have been shown to have the ability to interact with TBP and thus to be directly linked to transcription initiation (37).
While MEIS1 knockout mice display a severe phenotype, ELF1 knockout mice show mild defects (38). This may be explained by functional redundancy of the ETS gene family which consists of 27 members (39). Indeed, the binding sites of ETS family members are highly similar to each other with an internal conserved DNA binding core motif, GGAA/T. In addition, many factors coexist in a given cell. It is not uncommon that several ETS family members co-occupy the same binding site and function as either co-activators or competitors depending on different biological conditions. Over expression of ETS family members is frequently associated with malignant conditions. For example, ELF4, a member of ELF1 subfamily was also expressed in all the AML samples tested in the same study as ELF1 and might also contribute to leukemia formation (34). ETS2 and ERG were often unregulated due to their increased gene copies in several leukemic studies (40, 41). However, a recent study showed that ectopically over expressed ETS2, ERG or FLI1 in GATA1 knockout mice fetal liver cells actually decreased endogenous MEIS1 level and therefore these three EST family members might be the negative regulators of MEIS1 under the condition studied (42).
In HL60 cells where MEIS1 expression is negligible, ELF1 was not enriched at the MEIS1 promoter region. However, ELF1 was expressed at levels comparable to that of K562 cells and had binding activity detectable by EMSA (data not shown). Our data thus indicate that the presence of ELF1 is not sufficient for MEIS1 activation, likely due to requirements of additional factors to provide the requisite accessible chromatin. Candidates include pan-chromatin epigenetic modifiers such as modifiers of histone acetylation and DNA methylation as suggested by the upregulation of MEIS1 by the combination of TSA and 5 aza-2′ deoxycytidine in an AML cell line (19, 20, 43). Additionally, specific transcription factors, such as SRF or RUNX1, for which there are predicted binding sites in the MEIS1 promoter, may be also required under specific biological conditions which could not be revealed through the transient luciferase assay system in K562 cells. Interestingly, SRF has been shown to be able to interact with the ELK1 sub group of ETS family members and RUNX1 is able to interact with multiple ETS family members including ELF1, ELF4, NERF2, ETS1, PU1 and FLI1(44).
In summary, our current study identifies a key region in the MEIS1 promoter and shows that the ETS family member, especially ELF1 as a positive regulator of MEIS1. The MEIS1 locus spans about 140 kb in both human and mouse genome. There are multiple highly conserved motifs in addition to the promoter region studies here. This suggests additional cis-regulatory regions, which remain to be elucidated and are currently under the investigation in our lab.
Supplementary Material
Acknowledgments
We sincerely thank other lab members in Dr. Keith Humphries laboratory and Drs. Yan Xing, Juan Hou, Wei wei and Pamela Hoodless for helpful discussions. This work was supported by a Terry Fox Foundation Program Project award and additional funding from the Canadian Institutes of Health Research, the Children’s Leukemia Research Association and the British Columbia Cancer Foundation.
Footnotes
Conflict of Interest
No financial interest/relationships with financial relating to the topic of this article have been declared.
References
- 1.Spieker N, van Sluis P, Beitsma M, et al. The MEIS1 oncogene is highly expressed in neuroblastoma and amplified in cell line IMR32. Genomics. 2001;71:214–221. doi: 10.1006/geno.2000.6408. [DOI] [PubMed] [Google Scholar]
- 2.Crijns AP, de Graeff P, Geerts D, et al. MEIS and PBX homeobox proteins in ovarian cancer. Eur J Cancer. 2007;43:2495–2505. doi: 10.1016/j.ejca.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 3.Mercader N, Leonardo E, Azpiazu N, et al. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- 4.Argiropoulos B, Yung E, Humphries RK. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev. 2007;21:2845–2849. doi: 10.1101/gad.1619407. [DOI] [PubMed] [Google Scholar]
- 5.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 6.Xiong L, Catoire H, Dion P, et al. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. 2009;18:1065–10747. doi: 10.1093/hmg/ddn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura T, Largaespada DA, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 9.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001;20:350–361. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pineault N, Buske C, Feuring-Buske M, et al. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 2003;101:4529–4538. doi: 10.1182/blood-2002-08-2484. [DOI] [PubMed] [Google Scholar]
- 12.Pineault N, Abramovich C, Ohta H, Humphries RK. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell Biol. 2004;24:1907–1917. doi: 10.1128/MCB.24.5.1907-1917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 14.Rozovskaia T, Feinstein E, Mor O, et al. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4: 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 15.Imamura T, Morimoto A, Takanashi M, et al. Frequent co-expression of HoxA9 and Meis1 genes in infant acute lymphoblastic leukaemia with MLL rearrangement. Br J Haematol. 2002;119:119–121. doi: 10.1046/j.1365-2141.2002.03803.x. [DOI] [PubMed] [Google Scholar]
- 16.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 18.Hisa T, Spence SE, Rachel RA, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- 20.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 21.Esparza SD, Chang J, Shankar DB, Zhang B, Nelson SF, Sakamoto KM. CREB regulates Meis1 expression in normal and malignant hematopoietic cells. Leukemia. 2008;22:665–667. doi: 10.1038/sj.leu.2404933. [DOI] [PubMed] [Google Scholar]
- 22.Horman SR, Velu CS, Chaubey A, et al. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113:5466–5475. doi: 10.1182/blood-2008-09-179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu YL, Fong S, Ferrell C, Largman C, Shen WF. HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol Cell Biol. 2009;29:5181–5192. doi: 10.1128/MCB.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matys V, Fricke E, Geffers R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinemeyer T, Wingender E, Reuter I, et al. Databases on Transcriptional Regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:364–370. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryne JC, Valen E, Tang MH, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36(Database issue):D102–106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Zhang M, Duan Z, Stamatoyannopoulos G. Structural analysis and mapping of DNase I hypersensitivity of HS5 of the beta-globin locus control region. Genomics. 1999;61:183–193. doi: 10.1006/geno.1999.5954. [DOI] [PubMed] [Google Scholar]
- 28.Duan ZJ, Fang X, Rohde A, Han H, Stamatoyannopoulos G, Li Q. Developmental specificity of recruitment of TBP to the TATA box of the human gamma-globin gene. Proc Natl Acad Sci USA. 2002;99:5509–5514. doi: 10.1073/pnas.072084499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouhi A, Gagnier L, Takei F, Mager DL. Evidence for epigenetic maintenance of Ly49A monoallelic gene expression. J Immunol. 2006;176:2991–2999. doi: 10.4049/jimmunol.176.5.2991. [DOI] [PubMed] [Google Scholar]
- 30.Maksakova IA, Mager DL. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J Virol. 2005;79:13865–13874. doi: 10.1128/JVI.79.22.13865-13874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller BU, Pabst T. C/EBPalpha and the pathophysiology of acute myeloid leukemia. Curr Opin Hematol. 2006;13:7–14. doi: 10.1097/01.moh.0000190110.08156.96. [DOI] [PubMed] [Google Scholar]
- 32.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 33.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 34.Fukushima T, Miyazaki Y, Tsushima H, et al. The level of MEF but not ELF-1 correlates with FAB subtype of acute myeloid leukemia and is low in good prognosis cases. Leukemia Res. 2003;27:387–392. doi: 10.1016/s0145-2126(02)00214-x. [DOI] [PubMed] [Google Scholar]
- 35.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie X, Lu J, Kulbokas EJ, Golub TR, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aittomäki S, Yang J, Scott EW, Simon MC, Silvennoinen O. Molecular basis of Stat1 and PU.1 cooperation in cytokine-induced Fcγ receptor I promoter activation. Int Immunol. 2004;16:265–274. doi: 10.1093/intimm/dxh037. [DOI] [PubMed] [Google Scholar]
- 38.Garrett-Sinha LA, Dahl R, Rao S, Barton KP, Simon MC. PU.1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood. 2001;97:2908–2912. doi: 10.1182/blood.v97.9.2908. [DOI] [PubMed] [Google Scholar]
- 39.Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;21:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taub JW, Huang X, Matherly LH, et al. Expression of chromosome 21-localized genes in acute myeloid leukemia: differences between Down syndrome and non-Down syndrome blast cells and relationship to in vitro sensitivity to cytosine arabinoside and daunorubicin. Blood. 1999;94:1393–1400. [PubMed] [Google Scholar]
- 41.Marcucci G, Baldus CD, Ruppert AS, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 42.Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood. 2009;113:3337–3347. doi: 10.1182/blood-2008-08-174813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasa A, Carnicer MJ, Aventín A, et al. MEIS 1 expression is downregulated through promoter hypermethylation in AML1-ETO acute myeloid leukemias. Leukemia. 2004;18:1231–1237. doi: 10.1038/sj.leu.2403377. [DOI] [PubMed] [Google Scholar]
- 44.Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.