Abstract
Infectious bursal disease virus (IBDV) infection causes immunodeficiency in chickens. To understand cell-mediated immunity during IBDV infection, this study perform a detailed analysis of chicken γc chain (chCD132) and γc family cytokines, including interleukins 2, 4, 7, 9, and 15. The mouse anti-chCD132 monoclonal antibody (mAb) was first generated by the E.coli-expressed γc protein. Immunofluorescence assay further showed that γc was a protein located with the anti-chCD132 mAb on the surface of chicken's splenic mononuclear cells. Real-time quantitative RT-PCR revealed that the chCD132 mRNA transcript was persistently downregulated in embryo fibroblasts, spleen and thymus of chickens infected with IBDV. Correspondingly during IBDV infection, the transcription of five γc family cytokines was downregulated in the thymus and presented an imbalance in the spleen. Fluorescence-activated cell sorting analyses also indicated that the percentage of CD132+CD8+ T cells linearly decreased in the bursa of IBDV-infected chickens. These results confirmed that IBDV infection disturbed the in vivo balance of CD132 and γc family cytokine expression and that IBDV-induced immunodeficiency involved cellular networks related to the γc family.
Introduction
Gamma c chain (γc or CD132) is identified as a cellular receptor responsible for the high affinity and signal transduction of IL-2 and is a shared cellular receptor of type I cytokines including IL-2, -4, -7, -9, -15, and -21 [1]. ILs sharing CD132 are named γc family cytokines. CD132 is expressed as a transmembrane glycoprotein on CD4+ and CD8+ T cells, B cells, NK cells, monocytes/macrophages, neutrophils, and granulocytes, but non-lymphocytes, such as keratinocytes and human gingival fibroblasts, also express CD132.
Structures of CD132 molecules with γc family cytokines have also been formulated [1]. CD132 is involved in either spontaneous or GH-induced cell cycle progression and regulates lymphocyte development and proliferation [2], [3]. Chicken CD132 (chCD132), the only CD132 molecule identified in domestic fowl, is transcribed in the spleen, thymus, and bursa of Fabricius (BF). Recently, we determined the structure of the chCD132 functional domain bound to chicken interleukin (chIL)-2 [4].
Infectious bursal disease virus (IBDV) is a member of the Birnaviridae family, and mainly replicates in the bursa of Fabricius (BF) of chickens. Replication of IBDV in the bursa is accompanied by an influx of T cells. The marked influx of T cells into the infected bursa indicates that cell-mediated immunity plays important roles in the clearance of virus particles. The T cells in the bursa of chickens infected by virus are activated, with up-regulated expression of a number of cytokine genes, such as IL-1b, IL-6, IFN-g, IL-2 and chIFN-γ. The change in the level of cytokine expression is closely associated with organizational destruction, inflammation and apoptosis. Further, extrabursal replication and persistence of the virus in vivo may determine the extent to which the cellular immune systems gets stimulated. IBDV induces an immunosuppressive response in chickens, which manifests as a necrosis of B lymphoid cells in the BF, a decrease in macrophages, a chIFN-γ over production by T lymphocytes and a subsequent reduction in the ability to respond to secondary infections. Recently, Liu et al [5] reported that IFN-γ, IL-2, IL-12P40, IL-4, IL-5, IL-10 and IL-13 are strongly induced during IBDV infection. However, the action of ILs and their cellular receptors in IBDV pathogenesis remains unknown. Here, we generated the mAb to chicken CD132 (chCD132) and analyzed the dynamic profiles of CD132 and γc family cytokines (γc cytokines) in IBDV-infected chickens. Our data confirmed that IBDV infection influences on the expression of CD132 and γc cytokines.
Results
Generation and Identification of the mAb to chCD132
The sequencing results showed that the chCD132 cDNA sequence (GenBank Accession NO. D1852357) is 1047 bp in length and encodes a 327 amino acid (aa) polypeptide after truncation of the 21 aa signal peptide at the N-terminus. The chCD132 cDNA was cloned into the pET28a plasmid. The resulting plasmid was transformed into the E. coli strain BL21 (DE3) for chCD132 expression, and rchCD132 (recombinant chCD132) with a 6×-His tag was optimally expressed in E. coli as insoluble inclusions after induction by 0.5 mM IPTG. As shown in Fig. 1A, the molecular weight of rchCD132 was approximately 28 kDa, according to SDS-PAGE results. Subsequently, rchCD132 protein was further purified by using a nickel column under denaturing conditions. SPF BALB/c mice were immunized subcutaneously with purified rchCD132, and 6 hybridoma cell lines secreting anti-chCD132 antibodies were established by the clone technique of limiting dilution. Western blot assays demonstrated that all 6 anti-chCD132 mAbs bound strongly to the rchCD132 protein expressed in E. coli (Fig. 1B); however, one (mAb C10) of the 6 mAbs exhibited the binding affinity similar to chCD132 protein expressed on the con A-stimulated SMC (Fig. 1C), indicating that the C10 mAb binds to cellular chCD132 located on the surface of SMC.
Figure 1. Identification of anti-chCD132 mAb bound to cellular CD132 on the SMC surface.
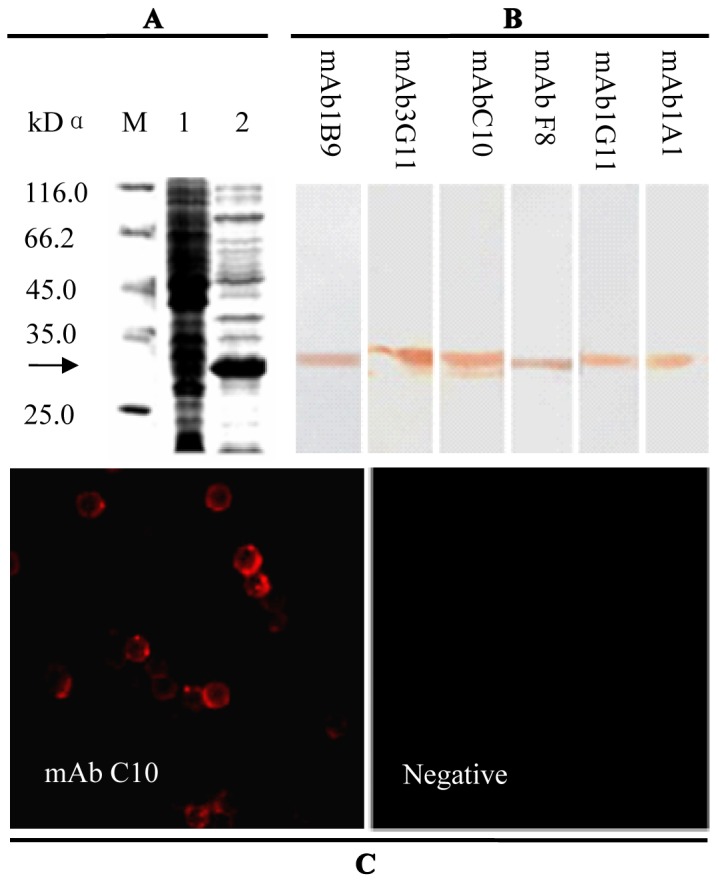
(A) SDS-PAGE analysis of Escherichia coli-expressed chCD132; M, molecular weight marker; lane 1, bacterial lysates of E. coli BL21 (DE3) transformed with pET28a; lane 2, bacterial lysates containing rchCD132. (B) Western blot analysis of rchCD132 recognized by 6 anti-chCD132 mAbs. (C) anti-chCD132 mAb C10 recognized by chCD132 expressed on the SMC surface using indirect immunofluorescencestaining (×10).
Transcription and Expression of Gene chCD132 in IBDV-infected CEF
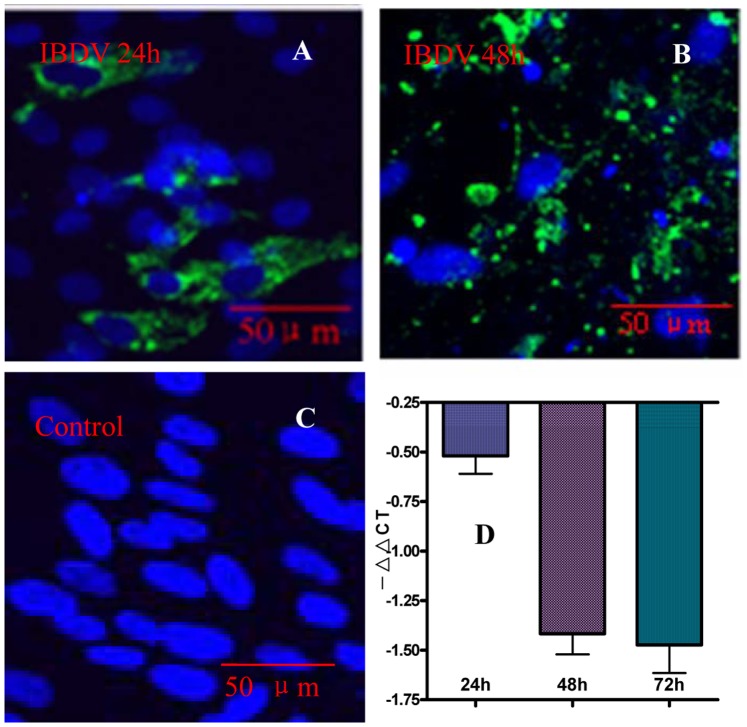
The chCD132 expressed in IBDV-infected CEF was examined. As shown in Fig. 2A, 2B and 2C, chCD132 was not detected by the anti-chCD132 mAb C10 in the IBDV-inoculated and mock-infected CEF monolayer at 24 hpi and 48 hpi. These data demonstrate that chCD132 expression is not detected at a detectable protein level in uninfected and IBDV-infected CEF. To further analyze chCD132 changes on the transcriptional level, the chCD132 transcript of the CEF monolayer with and without IBDV infection were analyzed at 24, 48, and 72 hpi by qRT-PCR. Data in Fig. 2D shows that during virus infection, compared with the mock-infected CEF monolayer, the γc mRNA level was persistently downregulated in the IBDV-infected CEF (p<0.05), indicating that γc mRNA transcription was inhibited during IBDV infection.
Figure 2. The mRNA abundance and protein expression of chCD132 on an IBDV-infected CEF monolayer.
CEFs were infected with IBDV a 100 TCID dose of the eNB virus. (A)–(C) Double-stained immunofluorescence images with anti-chCD132 mAb (red) and chicken serum (green) to IBDV under laser confocal microscopy. (A), (B) and (C) There is no chCD132 (red) expression in IBDV- and mock-infected CEF. (D) Transcription kinetics of chCD132 analyzed by qRT-PCR. Samples were normalized with the β-actin gene as a control and uninfected CEF at each time point as a reference. Each experiment was conducted in triplicate. Values are expressed as −ΔΔCT ± SD.
In vivo Inhibition of chCD132 Expression during IBDV Infection
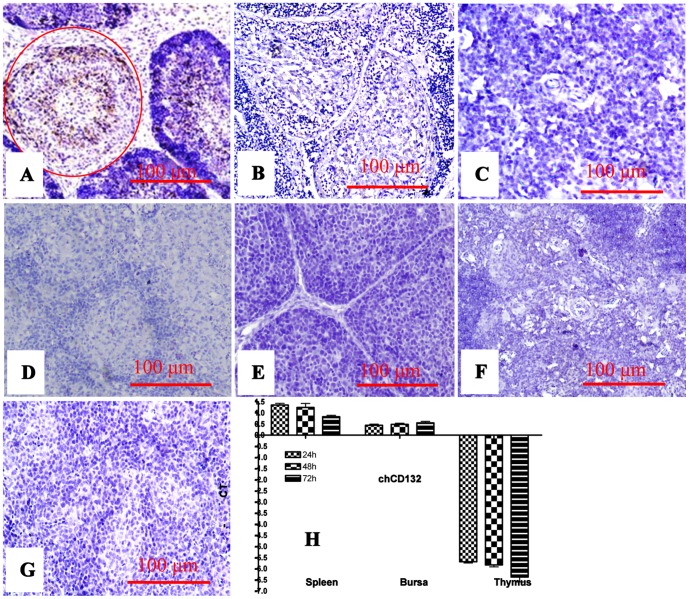
The chCD132 was analysed in bursa of chickens infected with virulent IBDV. As shown in Fig. 3A–G, an immunohistochemical assay demonstrated that chCD132 protein is not detected with mouse anti-chCD132 mAb at different time points when post-inoculated in the bursa, thymus, and spleen of mock- and IBDV-infected chickens. Correspondingly, when compared to the mock-infected chickens, up-regulation of chCD132 was not significant (p>0.05) in spleen and bursa of IBDV-infected chickens (Fig. 3H). However, down-regulation of chCD132 mRNA transcription within thymus of IBDV-infected chickens was down regulated. These data indicate that the change of chCD132 transcription within the bursa is insignificant during IBDV infection and that chCD132 transcript is gradually decreased in the spleen and inhibited in thymus.
Figure 3. Expression and transcription of chCD132 mRNAs in SPF chickens infected with IBDV.
(A)–(G) represents immunohistochemical staining of bursa, thymus, and spleen of mock- and IBDV-infected chickens. (A) IBDV antigens (brown) in bursa lymph follicles (red, round); (B) Bursa lymph follicles of IBDV-infected chicken unrecognized by anti-chCD132 mAb; (C) Thymus of IBDV-infected chicken unrecognized by anti-chCD132 mAb; (D) ChCD132 antigens in spleen of IBDV-infected chicken unrecognized by anti-chCD132 mAb; (E)–(G) Bursa, thymus and spleen of mock-infected chickens un-reactive with anti-chCD132 mAb; (H) chCD132 mRNA transcription in bursa, thymus, and spleen of chicken infected with IBDV. Samples were normalized with the β-actin gene as a negative control, and uninfected SPF chicken as a reference. All samples were assayed in triplicate and the values are expressed as −ΔΔCT ± SD.
Transcriptional Kinetics of γc Cytokines in IBDV-infected Chickens
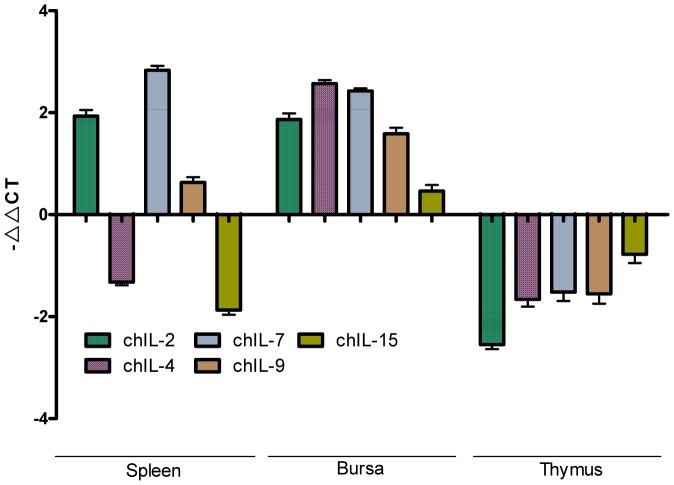
Data shown in Fig. 4 demonstrates that IBDV infection has opposite effects on the transcription of thymic and bursal γc cytokines in chickens, that is, the transcription of all 5 γc cytokines is downregulated in thymus and upregulated in bursa. However, in the spleen of IBDV-infected chickens, the transcription of chIL-4 and chIL-15 is downregulated, and the transcription of chIL-2, chIL-7 and chIL-9 is upregulated in comparison to mock infected chickens. This indicates that the balance of γc cytokines changed in spleen of IBDV-infected chickens.
Figure 4. Kinetics of γc cytokines mRNA expression in chicken immune tissues during infection measured by qRT-PCR.
Samples from spleen, thymus, and bursa were normalized with the β-actin gene as a control. All cDNAs were assayed in triplicate. The values are expressed as −ΔΔCT ± SD.
Kinetics of CD132-phenotypic T Cells of Chickens Infected with the Virulent IBDV
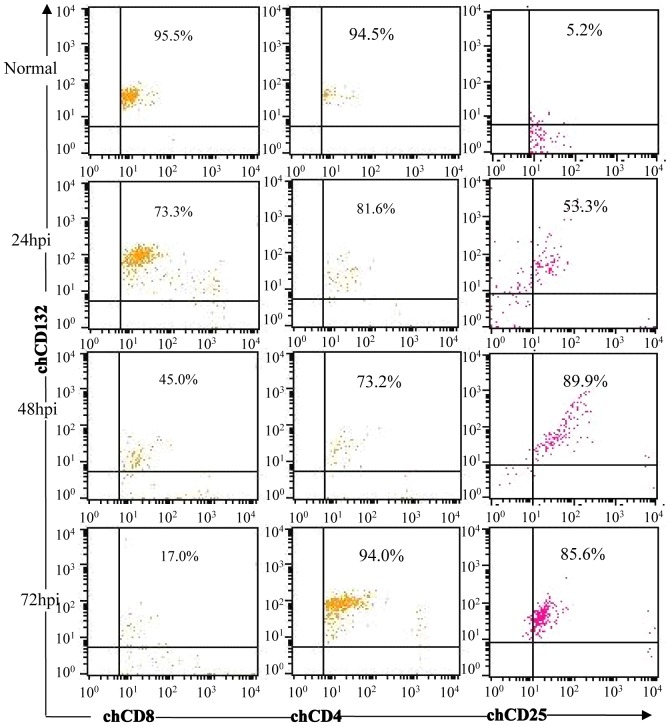
To analyze the role of chCD132+ T lymphocytes, we measured the profiles of CD132-labeled T lymphocytes in the bursa of IBDV-infected chickens by using a FACS assay after all the detected cell samples were stained using the anti-chCD3 mAb. As shown in Fig. 5, from 24 h to 72 h after IBDV infection, the percentage of bursal CD8+CD132+ T lymphocytes decreases persistently, in contrast with CD132+CD25+ T cells in IBDV-infected chickens. However, compared to mock-infected chickens, there are no remarkable changes in CD132+CD4+ T cell frequency (Fig. 5) in bursa of IBDV-infected chickens. These data show that IBDV infection induces downregulation of CD132+CD8+ T cells and upregulation of CD132+CD25+ T cells in bursa of IBDV-infected chickens.
Figure 5. FACS analysis of chCD132-phenotypic T lymphocytes in bursa of IBDV-infected chickens.
Bursacytes were incubated with FITC-labeled mouse anti-chicken CD3 mAb and PE-labeled mouse anti-chicken CD4 or CD8 mAb and APC-labeled mouse anti-chicken CD132 mAbs. Cells were counted on a Cytomics FC500 and analyzed using CELLQUEST software. Mock-infected SPF chickens were used as a negative control. All samples were assayed in triplicate.
Discussion
Avian cytokines, like their mammalian counterparts, are influential in host immune response to pathogenic infection [5]. CD132, a shared cellular receptor for IL-2, -4, -7, -9, -15, and -21, plays important roles in immunoregulatory networks. CD132 genetic mutations eliminate the activity of all CD132-dependent cytokines, and which result in human XSCID [6]. However, little information is available regarding CD132 molecules in domestic fowl except for that the γc chain gene and chCD132 functional domain binding to chIL-2 in chickens were identified [4]. In this study, we first developed mouse mAbs to rchCD132 by using rchCD132 expressed in E. coli as an immunogen. Subsequently, through chCD132 detection of the Con A-stimulated SMC surface, we obtained one hybridoma cell line secreting an anti-chCD132 mAb that binds cellular chCD132. The ability to produce this interaction provides a crucial tool for future functional investigation of chCD132.
Chickens infected with IBDV experience suppression in both humoral and cellular immunity. To date, little is known about the chCD132 molecules that plays important roles in T and NK cell development and activation of γc cytokine signal pathways. In present study, the transcription of intrathymic γc is significantly inhibited even if the intrasplenic and intra-bursal γc transcripts were slightly upregulated in IBDV-infected chickens (Fig. 3H), demonstrating that IBDV infection influences on γc transcription. Previous researches reported that the acute phase of IBD lasts for about 7 to 10 days; within this phase, the thymus undergoes marked atrophy and extensive apoptosis of thymocytes and its lesions are quickly overcome [7]. Therefore, we guessed that the possible explanation could be that γc-labeled cells impaired in thymus resulted in downregulation of γc transcripts and that γc-labeled cells activated in spleen and bursa revealed upregulation of γc transcripts, or that γc-labeled T cells moving from thymus to spleen and bursa induced the downregulation of intrathymic γc transcripts and upregulation of intrasplenic and intra-bursal γc transcripts to resist on IBDV infection. However, an accurate explanation regulating γc transcription needs further investigation during IBDV infection.
The γc family cytokines broadly contribute to lymphocyte proliferation, differentiation and survival [3]. IL-2 promotes proliferation of B cells, contributes to the development of regulatory T cells and regulates the proliferation and apoptosis of activated T cells [8]. IL-4 is required for T helper 2 cell developments and is a potential contributor to impaired CD8+ T-cell function in some anti-viral responses [9]. IL-7 has a central role in T-cell development [10], [11], [12], increased intrathymic IL-7 signaling significantly enhances the maintenance of immature thymocytes. IL-9 induces activation of epithelial cells, B cells, eosinophils, and mast cells [13]. IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation [14], and an essential role in CD8+ T-cell homeostasis [15]. Our experiments show that in IBDV-infected chickens mRNA transcripts of 5 γc family cytokines are upregulated in bursa and inhibited in thymus, however in spleen IL-2, -7, and -9 transcripts were upregulated and the transcripts of IL-4 and -15 were inhibited, indicating that IBDV infection results in disruption of homeostasis of γc family cytokines. On the basis of our experimental results, we considered that the development of thymic T lymphocytes secreting γc family cytokines may reveal a functional disturbance during IBDV infection. Delusively, we can not explain why are the kinetics of splenic γc family cytokines different from those of thymus and bursa γc family cytokines.
Regulation of cell surface γc expression levels is critical for cellular response by γc cytokines [3]. Generally, little was known about CD132-phenotypic T cells with CD4 or CD8 molecules, although some reports have demonstrated that HIV infection increased CD132 expression in all CD4+ T cells [16] and in human CD4+CD8+ thymocytes lost IL-7 signaling [11]. Interestingly, in present study, we found that CD132+CD8+ T cells were decreased in the bursa of IBDV-infected chickens and CD132+CD25+ T cells were upregulated (Fig. 5), that is, the profile of CD132+CD8+ and CD132+CD25+ T cells revealed a reverse dynamic change, and the CD132+CD4+ T cells is an insignificant decrease. Therefore it was reasonable to hypothesize that cellular immunity was relevant to CD132-immunophentypic T cells labelled with CD8 or CD25 molecules during IBDV infection. However, we cannot explain the roles of CD132+CD8+ T cells and CD132+CD25+ T cells during viral infection. Therefore further studies of CD132+ T cells are needed in future.
Materials and Methods
Viruses and Specific Pathogen-free Chickens
The chicken embryo fibroblast (CEF)-adapted IBDV strain NB (107.3 TCID50/0.1 ml, eNB virus), BF-replicated virulent IBDV strain NB (106.15 BLD50/0.1 ml, cNB virus) were identified and stored in our laboratory [17], [18]. Specific pathogen-free (SPF) chickens and embryonated chicken eggs were purchased from Beijing Merial Vital Laboratory Animal Technology Co., Ltd, Beijing, China. The use of all laboratory animals in this study was approved by the scientific ethical committee of Zhejiang University.
Prokaryotic Expression of chCD132 Protein and Monoclonal Antibody to chCD132
Splenic mononuclear cells (SMCs) were prepared as described previously [19]. An SMC culture was grown in RPMI 1640 medium supplemented with 10% NCS and 10 µg/ml con A at a final concentration of 5×106 cells/ml and was incubated at 41°C for 21 h. SMCs were washed 3 times with ice-cold PBS, lysed with Trizol reagent (Invitrogen, Carlsbad, CA), and total cellular RNA was extracted. Nucleotide sequences encoding the chCD132 extracellular domain were amplified from cultured SMC total cellular RNA by RT-PCR and the primer pair: 5′-ATGAATTCGCATCCCCCAGCCCCAAAG-3′ (containing the EcoRI site), and 5′-CGGTCGACTCACGTGTGGATCCAGAATC-3′ (containing the SalI site). PCR products were digested with the enzymes EcoRI and SalI, and inserted into the pET28a expression vector (Novagen, Madison, WI). After sequencing, the recombinant chCD132 (rchCD132) protein was expressed under IPTG induction in the Escherichia coli BL21 (DE3) strain and purified using a nickel column under denaturing conditions following the manufacturer's instructions (Qiagen Inc., Valencia, CA). Finally, the protein was analyzed by SDS-PAGE and western blot with the anti-His mAb (Amersham, USA) as described previously [20]. Preparation of the mAb to chCD132 was performed according to a previously reported protocol [19]. The mAb binding to native chCD132 was identified using Con A-activated SMCs in indirect immunofluorescence assays as stated previously [19]; PE-conjugated goat anti-mouse IgG was used as the secondary antibody.
Virus Inoculation and Sampling
CEFs were prepared from 10-day-old SPF embryonated chicken eggs and maintained in Hank's medium supplemented with 8% NCS, then seeded in 96- or 24-well plates. After formation of a CEF monolayer, the monolayer was inoculated with a 100 TCID50 dose of the eNB virus. The media-treated CEF monolayers were used as negative controls. The inoculated CEF monolayer was incubated at 37°C for 24 h, 48 h, and 72 h according to experimental demands, and 3 samples were collected at each time point. SPF chickens (25 days old) were raised in negative pressure isolators and inoculated intranasally at 0.1 ml/per chicken with a 300 BLD50 dose of the cNB virus. At 48 h post-infection (hpi), blood, spleen, thymus, and bursa from infected chickens were collected for RNA isolation, fluorescence-activated cell sorting (FACS) analysis, and immunohistochemical staining. The mock-infected SPF chickens were used as a negative control. The tissue samples of 3 infected chickens were collected at each time point.
Quantitative Real-time RT-PCR (qRT-PCR)
Primers specific for simultaneous amplification of γc cytokine genes, summarized in Table 1, were designed using the Lasergene sequence analysis software (DNAStar, Inc., Madison, WI) according to available gene information in the GenBank TM library. Total cellular RNA was extracted from the CEF monolayer and chicken tissues by using the Trizol reagent (Invitrogen), and RNA concentrations were detected using a spectrophotometer (260/280 nm). After heating at 65°C for 5 min to denature RNA and inactivate RNases, 1 µg of total RNA was subject to reverse transcription using 200 units of SuperScript III reverse transcriptase (Invitrogen), 40 units of RNaseOUT recombinant RNase inhibitor (Invitrogen), 200 ng of random hexamer primers (TaKaRa), 0.5 mM (each) dNTPs (TaKaRa), 4 µl of 5× First-Strand Buffer (Invitrogen), and 1 µl of 0.1 M DTT (Invitrogen) in a total volume of 20 µl at 25°C for 5 min, and then, incubated at 50°C for 1 h. The reaction was terminated by heating at 70°C for 15 min. qRT-PCR was performed by using 500 Real Time PCR system (ABI) in a total volume of 20 µl containing 1 µl cDNA template, 1× SYBR Premix Ex Taq (Perfect Real Time, TakaRa), and 200 nM of each primer. Melting curves were generated, and quantitative analysis of the data was performed using the realplex 2.0 software in a relative quantification (ddCt) study model (Applied Biosystems). The cellular β-actin gene previously used as the reference gene [21] in the IBDV infection was taken in the present study. Each sample was assayed in triplicate. Parallel mock-infected CEFs or chicken tissues were used as negative controls. Relative expression level changes greater than 2-fold (ddCT ≥1, p≤0.05) were considered significant.
Table 1. Primers for chicken CD132-dependent cytokines and β-actin in the method of Real-time PCR.
| Sequences of primers (5′-3′) | Source | Accession No |
| TCAGGGTGTGATGGTTGGT | β-actin | L08165 |
| TCTGTTGGCTTTGGGGTT | ||
| ATCTTTGGCTGTATTTCGGTAG | chIL-2 | AF502412 |
| CCTGGGTCTCAGTTGGTGTGT | ||
| AGTGCCGCTGATGGAGA | chIL-4 | AJ621249 |
| GGAGCTGACGCATGTTGA | ||
| CATACTCAGCCATGACATCGA | chIL-7 | AJ852017 |
| AGAACCTGTAACGTCCCACA | ||
| CTCTCTTGTGTGCTGCTCTCTG | chIL-9 | DQ294769 |
| CAACATGTGCTTGGTTCCTTC | ||
| ATGCTGGGGATGGCACA | chIL-15 | AF152927 |
| GCACATAGGAAGAAGATGGTTAGT |
Indirect Immunofluorescence Assay
Indirect immunofluorescence assays (IFA) were performed as stated previously [22]. CEFs inoculated with the eNB virus were cultured at 37°C for 24 h, 48 h, and 72 h, respectively. The cells were washed with PBS and fixed with cold acetone/methanol (1∶1) for 20 min at −20°C. After washing, CEFs were respectively incubated with anti-chCD132 mAbs at a 1∶500 dilution, followed by incubation with PE-conjugated goat anti-mouse IgG (1∶1000, Southern Biotechnology Associates Inc., Birmingham, AL) for 1 h at 37°C in a humidified chamber. Subsequently, the stained CEF monolayer was incubated with chicken anti-IBDV serum prepared in our laboratory [23], followed by incubation with FITC-conjugated goat anti-chicken IgG (1∶1000; Sigma Chemical Co., St.Louis, MO). Nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) was performed as described previously [24]. The triple-stained cells were washed 3 times with PBS. Finally, the stained cells were examined under a Zeiss LSM510 laser confocal microscope.
Immunohistochemical Staining
The levels of chCD132 protein in IBDV-infected chicken tissues were measured to understand the kinetics of in vivo protein expression. Immunohistochemical assay (IHC) was performed as previously described [25]. Briefly, the collected immune tissues were fixed in 10% neutral buffered formalin, routinely processed, and embedded in paraffin. Subsequently, the sections were treated with 0.3% H2O2 in PBS to inactivate endogenous peroxidase, washed 3 times in PBS, and digested for 10 min at 37°C with 0.1% trypsin (pH 7.6, 0.1% CaCl2) for antigen retrieval. The mAbs specific for chCD132, chicken anti-IBDV serum was applied and allowed to incubate for 2 h at 37°C. The primary antibody was detected by HRP-conjugated goat anti-mouse IgG or goat anti-chicken IgG secondary antibody (KPL, Gaithersburg, Maryland, USA). Finally, the sections were counterstained with hematoxylin.
Measurement of chCD132 Expression
Anti-chCD132 and anti-chCD25 mAbs were labeled with allophycocyanin (APC) and peridinin-chlorophyll (PerCP) (Donjindo, Japan, and Prozyme, USA, respectively) according to the labeling kit instructions. The bursa of IBDV-infected chickens described above were minced and passed through a stainless steel screen to obtain homogeneous cell suspensions, and bursacytes from IBDV-infected SPF chicken were isolated with Histopaque-1077 (Sigma). To examine CD132 expression in T lymphocytes, bursacytes were stained with an FITC-labeled mouse anti-chicken CD3 mAb (Southern Biotechnology Associates (SBA), Birmingham, AL), PE-labeled mouse anti-chicken CD4 or CD8 mAb (SBA), PerCP-labeled mouse anti-chicken CD25, and APC-labeled mouse anti-chicken CD132 mAb. Each experiment included a control sample with the same combination of antibodies. FACS analysis was performed on a Cytomics FC500 (Beckman Coulter, USA) and analyzed using CELLQUEST software (Beckman).
Statistical Analysis
All data analysis was performed using Microsoft Excel 2007. Student's t-test was used to detect significant differences between infected and control groups. A P-value ≤0.05 was considered significant.
Funding Statement
This work was funded by NSFC (31001075), National Science and Technology Support Program (grant number 2010BAD04B01-10), Key Scientific and Technological Innovation Team of Zhejiang Province (2010R50031), China Agriculture Research System (CARS-41-K11) and Chinese Universities Scientific Fund, Zhejiang A&F University Talent Starting Program (2012FR021, 2012FR061), and Zhejiang Provincial Natural Science Foundation (grant number LQ12C17001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wang X, Lupardus P, Laporte SL, Garcia KC (2009) Structural biology of shared cytokine receptors. Annu Rev Immunol 27: 29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amorosi S, Russo I, Amodio G, Garbi C, Vitiello L, et al. (2009) The cellular amount of the common gamma-chain influences spontaneous or induced cell proliferation. J Immunol 182: 3304–3309. [DOI] [PubMed] [Google Scholar]
- 3. Rochman Y, Spolski R, Leonard WJ (2009) New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol 9: 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gu J, Teng Q, Huang Z, Ruan X, Zhou J (2010) Identification of the functional interleukin-2 binding domain of the chicken common cytokine receptor gamma chain. Dev Comp Immunol 34: 258–263. [DOI] [PubMed] [Google Scholar]
- 5. Liu H, Zhang M, Han H, Yuan J, Li Z (2010) Comparison of the expression of cytokine genes in the bursal tissues of the chickens following challenge with infectious bursal disease viruses of varying virulence. Virol J 7: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leonard WJ (2001) Cytokines and immunodeficiency diseases. Nat Rev Immunol 1: 200–208. [DOI] [PubMed] [Google Scholar]
- 7. Tanimura N, Sharma JM (1998) In-situ apoptosis in chickens infected with infectious bursal disease virus. J Comp Pathol 118: 15–27. [DOI] [PubMed] [Google Scholar]
- 8. D'Souza WN, Lefrancois L (2003) IL-2 is not required for the initiation of CD8 T cell cycling but sustains expansion. J Immunol 171: 5727–5735. [DOI] [PubMed] [Google Scholar]
- 9. Crawley AM, Vranjkovic A, Young C, Angel JB (2010) Interleukin-4 downregulates CD127 expression and activity on human thymocytes and mature CD8+ T cells. Eur J Immunol 40: 1396–1407. [DOI] [PubMed] [Google Scholar]
- 10. Mazzucchelli R, Durum SK (2007) Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol 7: 144–154. [DOI] [PubMed] [Google Scholar]
- 11. Marino JH, Tan C, Taylor AA, Bentley C, Van De Wiele CJ, et al. (2010) Differential IL-7 responses in developing human thymocytes. Hum Immunol 71: 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Surh CD, Sprent J (2008) Homeostasis of naive and memory T cells. Immunity 29: 848–862. [DOI] [PubMed] [Google Scholar]
- 13. Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, et al. (2008) IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat Immunol 9: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Armitage RJ, Macduff BM, Eisenman J, Paxton R, Grabstein KH (1995) IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation. J Immunol 154: 483–490. [PubMed] [Google Scholar]
- 15. Sun JC, Ma A, Lanier LL (2009) Cutting edge: IL-15-independent NK cell response to mouse cytomegalovirus infection. J Immunol 183: 2911–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Juffroy O, Bugault F, Lambotte O, Landires I, Viard JP, et al. (2010) Dual mechanism of impairment of interleukin-7 (IL-7) responses in human immunodeficiency virus infection: decreased IL-7 binding and abnormal activation of the JAK/STAT5 pathway. J Virol 84: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi L, Li H, Ma G, Zhou J, Hong L, et al. (2009) Competitive replication of different genotypes of infectious bursal disease virus on chicken embryo fibroblasts. Virus Genes 39: 46–52. [DOI] [PubMed] [Google Scholar]
- 18. Zhou JY, Shen HG, Chen HX, Tong GZ, Liao M, et al. (2006) Characterization of a highly pathogenic H5N1 influenza virus derived from bar-headed geese in China. J Gen Virol 87: 1823–1833. [DOI] [PubMed] [Google Scholar]
- 19. Teng QY, Zhou JY, Wu JJ, Guo JQ, Shen HG (2006) Characterization of chicken interleukin 2 receptor alpha chain, a homolog to mammalian CD25. FEBS Lett 580: 4274–4281. [DOI] [PubMed] [Google Scholar]
- 20. Zhou JY, Wang JY, Chen JG, Wu JX, Gong H, et al. (2005) Cloning, in vitro expression and bioactivity of duck interleukin-2. Mol Immunol 42: 589–598. [DOI] [PubMed] [Google Scholar]
- 21. Zheng X, Hong L, Shi L, Guo J, Sun Z, et al. (2008) Proteomics analysis of host cells infected with infectious bursal disease virus. Mol Cell Proteomics 7: 612–625. [DOI] [PubMed] [Google Scholar]
- 22. Wu Y, Peng C, Xu L, Zheng X, Liao M, et al. (2012) Proteome dynamics in primary target organ of infectious bursal disease virus. Proteomics 12: 1844–1859. [DOI] [PubMed] [Google Scholar]
- 23. Zheng X, Hong L, Li Y, Guo J, Zhang G, et al. (2006) In vitro expression and monoclonal antibody of RNA-dependent RNA polymerase for infectious bursal disease virus. DNA Cell Biol 25: 646–653. [DOI] [PubMed] [Google Scholar]
- 24. Wu Y, Hong L, Ye J, Huang Z, Zhou J (2009) The VP5 protein of infectious bursal disease virus promotes virion release from infected cells and is not involved in cell death. Arch Virol 154: 1873–1882. [DOI] [PubMed] [Google Scholar]
- 25. Wang Y, Wu X, Li H, Wu Y, Shi L, et al. (2009) Antibody to VP4 protein is an indicator discriminating pathogenic and nonpathogenic IBDV infection. Mol Immunol 46: 1964–1969. [DOI] [PubMed] [Google Scholar]