Abstract
In comparison to dicot-infecting bacteria, only limited numbers of genome sequences are available for monocot-infecting and in particular cereal-infecting bacteria. Herein we report the characterisation and genome sequence of Xanthomonas translucens isolate DAR61454 pathogenic on wheat and barley. Based on phylogenetic analysis of the ATP synthase beta subunit (atpD) gene, DAR61454 is most closely related to other X. translucens strains and the sugarcane- and banana- infecting Xanthomonas strains, but shares a type III secretion system (T3SS) with X. translucens pv. graminis and more distantly related xanthomonads. Assays with an adenylate cyclase reporter protein demonstrate that DAR61454's T3SS is functional in delivering proteins to wheat cells. X. translucens DAR61454 also encodes two type VI secretion systems with one most closely related to those found in some strains of the rice infecting strain X. oryzae pv. oryzae but not other xanthomonads. Comparative analysis of 18 different Xanthomonas isolates revealed 84 proteins unique to cereal (i.e. rice) infecting isolates and the wheat/barley infecting DAR61454. Genes encoding 60 of these proteins are found in gene clusters in the X. translucens DAR61454 genome, suggesting cereal-specific pathogenicity islands. However, none of the cereal pathogen specific proteins were homologous to known Xanthomonas spp. effectors. Comparative analysis outside of the bacterial kingdom revealed a nucleoside triphosphate pyrophosphohydrolase encoding gene in DAR61454 also present in other bacteria as well as a number of pathogenic Fusarium species, suggesting that this gene may have been transmitted horizontally from bacteria to the Fusarium lineage of pathogenic fungi. This example further highlights the importance of horizontal gene acquisition from bacteria in the evolution of fungi.
Introduction
Bacterial pathogens are of serious economic importance in many crop plants. In monocotyledonous plant hosts, Xanthomonas pathogens cause important diseases such as rice blight and sugarcane leaf scald caused by X. oryzae pv. oryzae and X. albilineans, respectively. Although bacterial diseases of the important cereals wheat and barley are economically less important than fungal diseases, recent genome analyses of fungal pathogens have indicated bacteria, and particularly plant-associated bacteria, are potential sources of new virulence genes contributing to the evolution of fungal virulence [1]. For example, a comparative analysis of the genome of the cereal crown rot pathogen Fusarium pseudograminearum identified 17 genes exclusively found in cereal pathogens and possibly acquired from bacteria by horizontal transfer. One of these genes encoding an amidohydroylase was shown to be required for pathogen virulence on both wheat and barley [2]. Similarly, Klosterman et al. [3] showed that a glucosyltransferase important for virulence in the wilt causing Verticillium fungi likely originated from bacteria. So far, horizontally acquired genes with highly uneven distributions across fungal species but close protein matches from bacterial species have primarily been discovered through the analysis of sequenced fungal genomes. Similarly, examining the genomes of phytopathogenic bacteria for genes that appear preferentially in other pathogens specialised to particular hosts could reveal new insights into the evolution of pathogenicity and/or virulence. Application of this approach to bacteria pathogenic on cereals requires the availability of sequenced genomes from these pathogens and through this and other efforts we have sought to expand the available genomes [4].
Bacterial pathogens and non-pathogens are increasingly utilised as a vehicle to deliver heterologous proteins into plant cells to dissect functions of pathogen proteins with potential roles in virulence [5]. So far, such protein delivery systems have primarily been exploited in dicotyledonous model hosts such as Nicotiana benthamiana and Arabidopsis thaliana [6]. Development of similar systems in monocots using appropriate bacterial pathogens would also be highly desirable to analyse effector function. Previous systems utilizing non-pathogenic strains of Pseudomonas have been described [7] and we recently developed a similar system with improved phenotypic penetrance in cereal in searching for resistance/avirulence gene interactions [8]. The delivery of effectors using pathogenic strains that cause cell death will also be useful to identify effectors that contribute to virulence functions such as suppression of cell death.
We report herein the genome sequence of a X. translucens isolate pathogenic on wheat and barley. We have used the X. translucens genome sequence to search for bacterial genes that may have been acquired by fungal plant pathogens. We have also demonstrated the ability of this pathogen to deliver heterologous proteins into wheat cells. An improved knowledge of the genome and effector composition produced by plant pathogenic bacteria may provide a useful tool for functional genomics research on effector proteins acting on cereal hosts.
Results and Discussion
Xanthomonas translucens DAR61454 is a wheat and barley pathogen
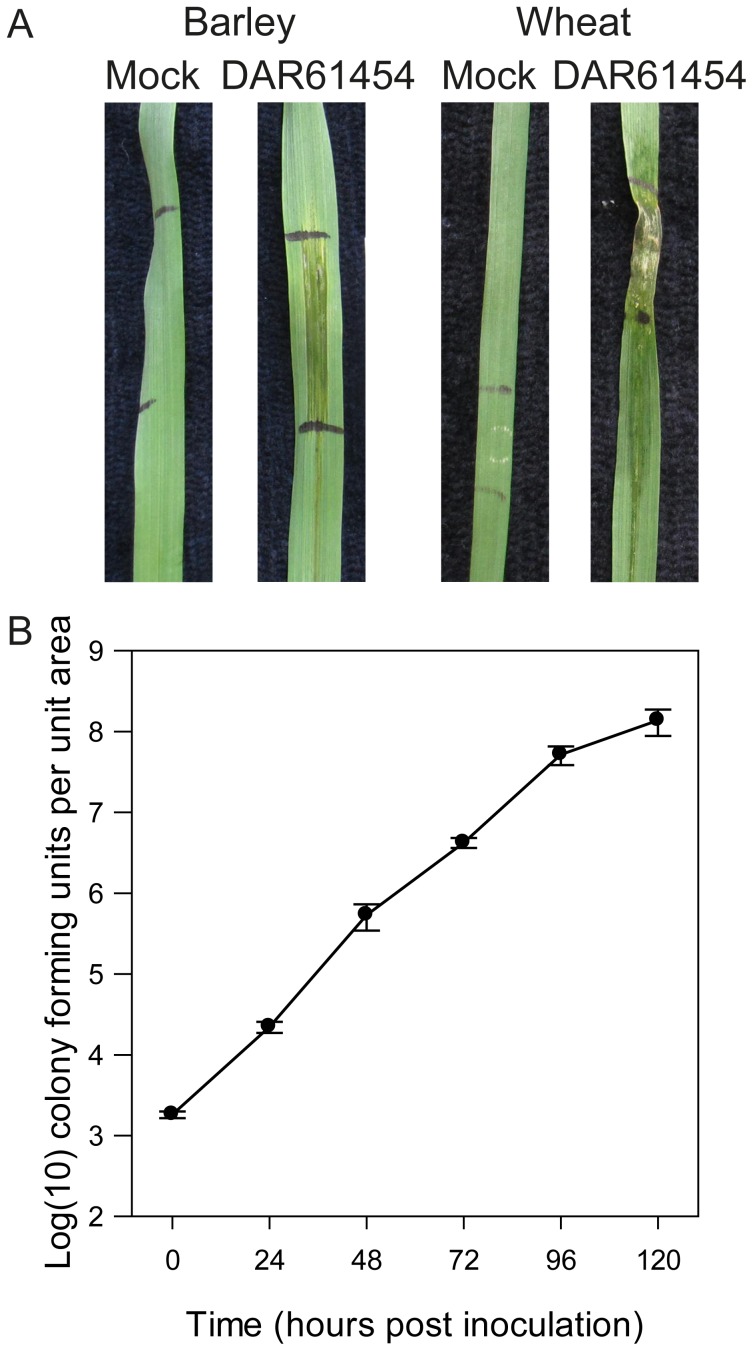
Despite the demonstrated importance of bacterial species as potential donors of virulence genes in fungal cereal pathogens [reviewed in 1], currently limited genome sequence information is available for cereal infecting bacteria and this makes searching for new such genes difficult. In this study, we first aimed to identify an Australian bacterial strain capable of infecting cereal hosts, given that the importation of plant pathogens into Australia is restricted by quarantine laws. Xanthomonas strain DAR61454 identified as X. translucens was isolated from wheat showing symptoms of black chaff disease at a research station in Tamworth, New South Wales, Australia in 1988. To confirm the pathogenicity of this isolate on cereals, we generated a rifampicin resistant derivative of DAR61454 and infiltrated this strain into wheat and barley leaves. Lesions in the infiltrated zone were observed after inoculations in both species (Figure 1A). These lesions expanded over time and became water-soaked and necrotic, consistent with descriptions of X. translucens pathogenicity on wheat [9]. In addition, bacterial concentrations increased exponentially over time in infiltrated barley leaves, consistent with a virulent interaction on this species (Figure 1B). The pathogenicity of DAR61454 was also tested in over 100 wheat accessions which all produced water-soaked lesions following inoculation (data not shown). Together, these experiments indicate that DAR61454 is highly pathogenic on both wheat and barley.
Figure 1. Virulence of Xanthomonas translucens DAR61454 towards wheat and barley.
(A) Exemplar photographs of virulence of X. translucens DAR61454 on wheat and barley (six days post inoculation). Black marks on the leaves highlight the extremities of the bacterial infiltration (B) Bacterial growth over time during infection of barley. No rifampicin resistant bacteria were recovered from mock inoculated plants carried out at the same time. Each time-point is average of four biological replicates with error bars representing the standard error of the mean. One unit area is two leaf disc punches.
Genome assembly and annotation
The DAR61454 genome was sequenced using 100 bp paired end Illumina reads. The total assembly size was 4.5 Mb assembled in 217 scaffolds (split into 404 contigs for annotation). The average coverage depth was 1585 fold. The N75 (number of scaffolds for which 75% of the genome is contained) was 68. The genome was annotated using the NCBI Prokaryote Genome Automatic Annotation Pipeline (PGAAP). The locus tag prefix assigned to this isolate is A989. The genome was predicted to encode 3847 proteins. The sequence data from this whole genome shotgun project has been deposited to DDBJ/EMBL/GenBank under the accession AMXY00000000. The version described in this paper is the first version, AMXY01000000. The raw sequence reads have been deposited into the NCBI sequence read archive under accession SRS502548.
Comparative genome analyses within Xanthomonas spp
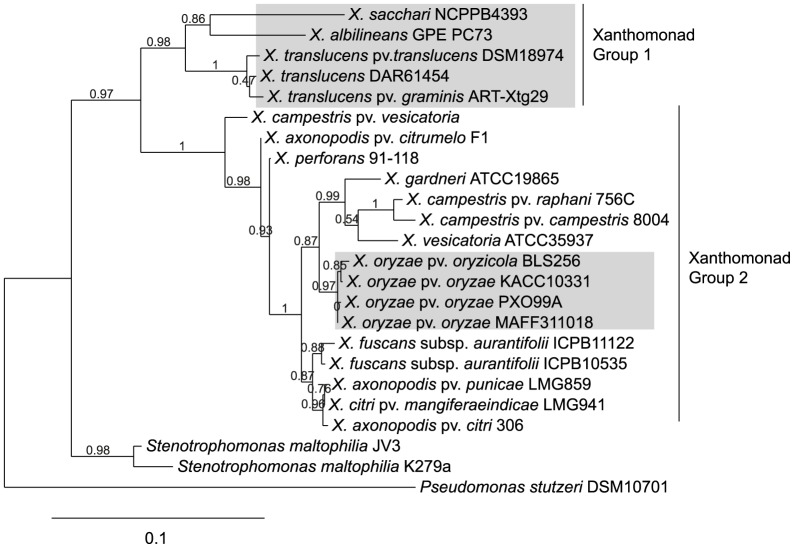
We used the ATP synthase beta subunit encoding gene (atpD) sequence to analyse phylogenetic relationships between DAR61454 and other xanthomonads. This gene has previously been used to infer phylogenetic relationships within xanthomonads [10]. As expected, the best nucleotide matches of DAR61454 atpD were from other X. translucens strains with pathovar specifications of either translucens or graminis. These related sequences have been very recently deposited into GenBank; however, a detailed description of one of these X. translucens genomes was only published whilst this manuscript was under review [11]. The Xanthomonas genus consists of two distinct phylogenetic groups, with most of the extant genomes from isolates in Group 2 [10]. Genome sequences of Group 1 xanthomonads are currently limited to three recently sequenced banana-associated isolates [10], the sugarcane pathogen X. albilineans [12] and the new X. translucens submissions to GenBank. Phylogenetic analysis using atpD suggested DAR61454 belongs to the Group 1 strains of the genus (Figure 2). The gyrB gene, which encodes the B subunit of the DNA gyrase, provides additional resolution for intraspecific identification of xanthomonads [13]. Therefore, additional phylogenetic analysis was carried out using gyrB and, as expected, DAR61454 grouped with other X. translucens pv. translucens strains. However, the branch support for separating the X. translucens pv. translucens group from the single strain of X. translucens pv. graminis was weak (Figure S1). The host infection phenotypes together with phylogenetic analyses of these two genes strongly support the classification of DAR61454 as X. translucens pv. translucens.
Figure 2. Placement of Xanthomonas translucens isolate DAR61454 in the Xanthomonas genus using phylogenetic analysis of the atpD gene nucleotide sequence.
Phylogenetic analysis was carried out at www.phylogeny.fr as detailed in the methods. Numbers of branches represent branch support based on approximate likelihood ratio testing. Monocot infecting or associated xanthomonads are shaded in grey. All other xanthomonads are associated with dicot species.
Orthologous proteins in Xanthomonas spp
To further analyse the DAR61454 genome, 18 sequenced xanthomonad genomes including DAR61454 were compared using orthoMCL cluster analysis [14]. The genome sequences used in this analysis were obtained from www.xanthomonas.org, which has gene annotations associated with the GenBank entries (Table 1). OrthoMCL groups proteins into putative orthologous groups based on sequence similarity. A total of 6590 different groups of proteins were identified in the 18 genomes analysed. Of these, 912 groups of proteins were unique to monocot infecting (sugarcane, rice and wheat), while 727 were specific to cereal infecting (rice and wheat) isolates. The accessions for these 727 cereal specific groups of proteins have been included in File S1. These included 84 proteins (in 74 groups) from DAR61454. Sixty of these 84 proteins were present in clusters of two or more genes in the DAR61454 genome, suggestive of gene transfers of contiguous segments or selective retention of physically co-located genes between cereal infecting xanthomonads, despite the relatively distant overall evolutionary relationship between DAR61454 and the rice infecting Xanthomonas spp. Two large gene clusters, one encoding a type VI secretion system (T6SS, see below) and the other one encoding a number of cytochrome P450 monooxygenases and two polyprenyl transferases (A989_04693-A989_04733), were identified in protein groups specific to cereal infecting isolates. The latter cluster encodes enzymes with closest matches in rhizobial species with the predicted gene functions, suggestive of a secondary metabolite synthetic pathway. One of the most important features of the virulence arsenal of the sugarcane pathogen X. albilineans is the production of the polyketide phytotoxin albicidin [15], but DAR61454 does not encode the genes required for the production of this toxin.
Table 1. Xanthomonas spp. genomes used in comparative analysis.
| Isolate abbreviation | Species | Host(s) | GenBank Accession(s) |
| A989 | Xanthomonas translucens. DAR61454 | wheat and barley | AMXY01000000 |
| PXO | Xanthomonas oryzae pv. oryzae PXO99A | rice | CP000967 |
| XAC | Xanthomonas axonopodis pv. citri str. 306 | citrus | AE008923, AE008924, AE008925 |
| XACM | Xanthomonas axonopodis pv. citrumelo F1 | citrus | CP002914 |
| XALC | Xanthomonas albilineans GPE PC73 | sugarcane | FP565176 |
| XAPC | Xanthomonas axonopodis pv. punicae str. LMG 859 | pomegranate | CAGJ01000001 |
| XAUB | Xanthomonas fuscans subsp. aurantifolii str. ICPB 11122 | citrus | ACPX00000000 |
| XAUC | Xanthomonas fuscans subsp. aurantifolii str. ICPB 10535 | citrus | ACPY00000000 |
| XCXX | Xanthomonas campestris pv. campestris str. 8004 | cauliflower | CP000050 |
| XCR | Xanthomonas campestris pv. raphani 756C | Brassica oleracea var capitata | CP002789 |
| XCV | Xanthomonas campestris pv. vesicatoria str. 85–10 | pepper | AM039948, AM039949, AM039950, AM039951, AM039952 |
| XGA | Xanthomonas gardneri ATCC 19865 | tomato and pepper | AEQX01000000 |
| XMIN | Xanthomonas citri pv. mangiferaeindicae LMG 941 | mango | CAHO01000001 |
| XOC | Xanthomonas oryzae pv. oryzicola BLS256 | rice | CP003057 |
| XOOB | Xanthomonas oryzae pv. oryzae MAFF 311018 | rice | AP008229 |
| XOO | Xanthomonas oryzae pv. oryzae KACC10331 | rice | AE013598 |
| XPE | Xanthomonas perforans 91–118 | tomato | AEQW01000000 |
| XVE | Xanthomonas vesicatoria ATCC 35937 | tomato and pepper | AEQV00000000 |
Secretion systems
Gram negative bacteria can have up to six different types of specialized secretion systems to deal with the unique dual membrane system found in these organisms [16]. Direct roles in virulence towards plants have been described for the T1SS, T2SS, T3SS, T4SS and T6SS [17]–[20]. Using the X. oryzae pv. oryzae T1SS protein sequences (PXO_04478 (RaxA), PXO_04477 (RaxB) and PXO_02621 (RaxC)) as a query, we identified orthologous sequences in the DAR61454 genome (A989_01015, A989_01020 and A989_15402). The T2SS is encoded by 12 or more genes [21]. X. axonopodis pv. citri, X. campestris pv. campestris and X. citri contain two T2SS [22], [23] whereas X. oryzae pv. oryzae encodes one [24]. DAR61454 contains one gene cluster for this system with a typical organization with xpsE through xpsN encoded in one cluster (A989_04551-A989_04506) with xpsD (A989_017673) and xpsO (A989_03190) encoded at other genomic locations. Apart from the separate location of xpsD, the cluster is arranged the same as X. campestris pv. campestris [21].
There does not appear to be a T4SS in DAR61454. X. axonopodis pv. citri encodes two T4SSs, one each on the chromosome and plasmid [25] and X. campestris pv. campestris encodes one T4SS cluster [22]. BLASTp searches using these sequences against DAR61454 failed to identify orthologous proteins, with the exception of a single virD4 homologue encoded by A989_18933.
Our analyses revealed that, in contrast to X. albilineans, which contains a Salmonella SPI-1 type III secretion system (T3SS), and the banana-associated strains that lack a T3SS [10], DAR61454 encodes a Hrp-type T3SS cluster (A989_10802-A989_10942) that most closely resembles that found in the recently sequenced X. translucens pv. graminis genome and those of Group 2 xanthomonads [11]. The organization of the cluster is almost identical to that described X. translucens pv. graminis [11] with the exception of an additional hypothetical protein (A989_10862) encoded between the genes for hpaP and hrcV. Given the central importance of T3 effectors in bacterial virulence in both plants and animals, mutation of the T3SS in xanthomonads typically results in a loss of virulence and this was recently demonstrated for X. translucens pv. graminis [8]; so it can be expected the DAR61454 T3SS is also important for virulence. The identification of the conserved Hrp-T3SS in the DAR61454 genome suggested this strain could be used as a vehicle to deliver heterologous proteins from other pathogens of cereals to understand their roles in virulence. To demonstrate the potential utility of this strain, a X. campestris AvrBs2:calmodulin-dependent adenylate cyclase (Cya) fusion reporter system was used [26], [27]. This reporter system relies on the presence of calmodulin exclusively in eukaryotic cells, meaning that the Cya protein is inactive unless present inside a eukaryotic cell. Thus, the accumulation of cyclic adenosine monophosphate (cAMP) above control levels is dependent on delivery of the calmodulin-dependent-Cya to the host cells. Using this system, we observed significant cAMP accumulation, normalized to total protein content, in wheat leaves 8 and 24 hours after inoculation with X. translucens DAR61454 carrying an AvrBs2T3SS∶Cya expressing plasmid (expression driven by AvrBs2 promoter) in which Cya was translationally fused to the first 100 amino acids of AvrBs2 including its T3SS signal (Figure 3). As a control, a vector containing the same promoter driving Cya expression but lacking the AvrBs2 T3SS signal sequence was used to demonstrate the dependence of the observed Cya activity on the AvrBs2 T3SS. Only low levels of cAMP were detected in leaves infiltrated with this construct, approximately 100-fold lower than that observed with AvrBs2T3SS∶Cya construct at 24 hours (Figure 3). The levels of cAMP observed using DAR61454 to deliver Cya are similar to those observed using this type of reporter system in other bacteria-plant interactions [8], [28], [29]. This result clearly demonstrates the ability of DAR61454 to deliver the reporter protein into wheat cells.
Figure 3. Use of Xanthomonas translucens DAR61454 to deliver a heterologous reporter protein into wheat cells via the Type III secretion system (T3SS).
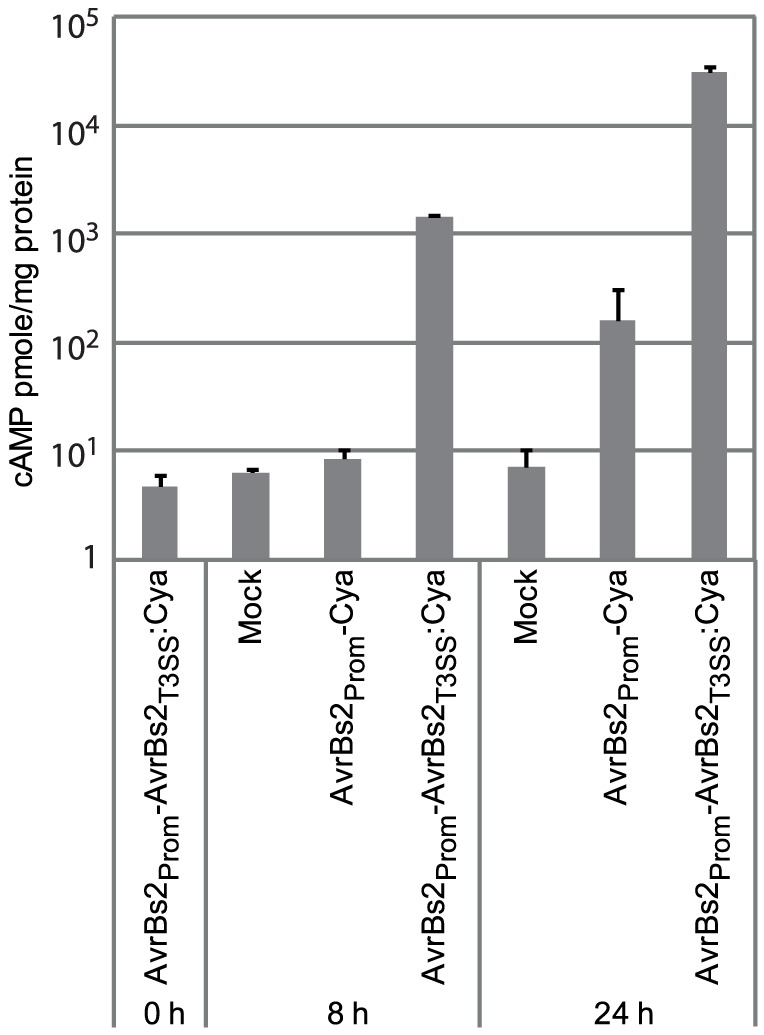
The uniquely eukaryote-functioning calmodulin dependent adenylate cyclase (Cya) reporter was translationally fused to AvrBs2 N-terminus leader in a bacterial expression vector (AvrBs2Prom-AvrBs2T3SS∶Cya). cAMP detection is indicative of delivery of the Cya protein into host cells which was measured by an enzyme immunoassay. The mock treatment used MgCl2 without bacteria. A construct lacking the AvrBs2 T3SS signal but with the AvrBs2 promoter and Cya sequence (AvrBs2Prom-Cya) was used as a control. Some background Cya activity could be observed at the later time point with this construct. The data is average of three biological replicates and error bars represent the standard error of the mean.
DAR61454 also encodes two type VI secretion system (T6SS) gene clusters which both contain nearly all the conserved elements of these clusters (Figure 4) as defined by Boyer et al. [30]. The T6SS was originally identified as a secretion system in the mammalian infecting pathogens Pseudomonas aeruginosa and Vibrio cholerae where it is important for virulence [31], [32]. The T6SS is also used for the secretion of proteins that alter the ability of rhizobia to form functional nodules in a host specific manner [33]. T6SSs are typically encoded by 15–20 genes with 13 of these present at most T6SS loci in different species [30]. One T6SS cluster (A989_06153-A989_06243; Cluster 1) in DAR61454 appears to be found in most Xanthomonads. In contrast, 10 of the 12 genes in the other cluster (A989_05968-A989_06028; Cluster 2) (Figure 4) were identified as cereal specific using the OrthoMCL clustering analysis. Outside of these matches, the next best hits for each of the proteins of the DAR61454 T6SS are to those from a Stenotrophomonas sp., which is a bacterial genus often found in soil and rhizophere samples [34]. These secretion system clusters identified in DAR61454 are likely to be involved in the plant-associated lifestyle of this isolate. Exactly how two separate T6SS that presumably recognise and secrete similar proteins can function in a single bacterium remains obscure.
Figure 4. Type VI secretion system (T6SS) gene clusters in DAR61454. The upper cluster shows the prototypical T6SS from Pseudomonas aeruginosa.
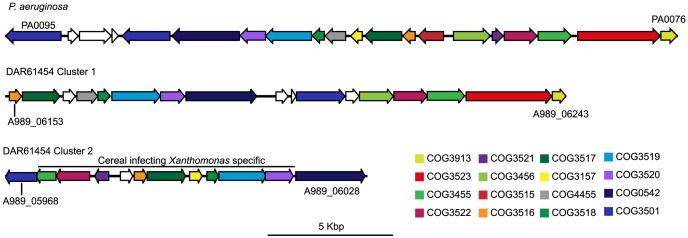
DAR61454 Cluster 1 represents the T6SS cluster found in most Xanthomonas spp. genomes and Cluster 2 appears to be specific to DAR61454 and rice infecting isolates. Orthologous genes are color coded according to their cluster of orthologous groups of proteins (COG) classification in the annotated genomes. Color coding is an approximate of that used by Boyer et al. [30]. Gene numbers at the extremities of the clusters are shown but other numbers are omitted for ease of viewing. In DAR61454 Cluster 1 genes A989_06178 and A989_06183 (COG3519) and Cluster2 genes A989_05968 and A989_05973 (COG3501) were on either side of scaffold gaps and have been fused in this image as they appear to be parts of the same genes.
Ef2fector homologues
BLASTp analysis of the DAR61454 genome revealed the presence of type III effector homologues from 22 different families of Xanthomonas effectors, as classified at www.xanthomonas.org. These DAR61454 effector homologues are listed in Table 2. None of these known effectors are exclusively present in the cereal pathogens. However, it is likely that different effector combinations and/or specific allelic variants of these effectors together with other virulence factors would determine host specificity, as has been suggested for both X. axonopodis [35] and Pseudomonas syringae isolates [36], rather than individual effectors providing the ability to invade a particular host. For five classes of effectors (XopF1, F2, L, X and AE), multiple homologues are present in the DAR61454 genome. DAR61454 is also likely to encode TAL (transcription activator-like) type effectors based on BLASTp analysis. However, DAR61454 genomic regions containing matches to these genes were found on six high coverage contigs which appear to be poorly assembled due to the inherent repetitive nature of TAL-encoding genes [37]. Extraction of the reads from these contigs and assembly against TAL effector coding sequences from X. oryzae pv. oryzae as a reference suggested that there was at least 15 Kbp of sequence in DAR61454 at a coverage equivalent to 1585-fold in these sequence reads. With Xanthomonas TAL effector encoding sequences typically in the range of 3–5 Kbp, this could indicate that there are at least 3–5 TAL effectors in DAR61454. However, longer sequence reads are needed to resolve these.
Table 2. Known effector homologues in Xanthomonas translucens DAR61454.
| A989 protein | Xop Nomenclature | % identity | E-value |
| A989_10180 | AvrBs2 | 58.82 | 0 |
| A989_12395 | XopB | 53.3 | 6e−169 |
| A989_02940 | XopC2 | 63.56 | 5e−161 |
| A989_10570 | XopF1 | 57.37 | 1e−169 |
| A989_10802 | XopF1 | 33.51 | 8e−63 |
| A989_10570 | XopF2 | 76.83 | 0 |
| A989_10802 | XopF2 | 34.98 | 5e−72 |
| A989_02970 | XopJ5 | 88.94 | 2e−116 |
| A989_13784 | XopK | 63.75 | 0 |
| A989_06893 | XopL | 40.06 | 5e−113 |
| A989_16433 | XopL | 30.63 | 2e−31 |
| A989_07258 | XopN | 58.66 | 0 |
| A989_02550 | XopAK | 63.1 | 1e−67 |
| A989_08274 | XopP | 64.98 | 0 |
| A989_00475 | XopP | 39.85 | 1e−92 |
| A989_14199 | XopQ | 47.89 | 9e−95 |
| A989_09773 | XopR | 44.68 | 7e−29 |
| A989_17343 | XopV | 52.34 | 1e−64 |
| A989_02945 | XopX | 60.93 | 0 |
| A989_00070 | XopX | 57.86 | 2e−178 |
| A989_00075 | XopX | 53.49 | 5e−178 |
| A989_04281 | XopZ1 | 57.65 | 0 |
| A989_03235 | XopAA | 80.2 | 0 |
| A989_04481 | XopAD | 67.27 | 0 |
| A989_06893 | XopAE | 32.41 | 1e−26 |
| A989_16433 | XopAE | 31.48 | 7e−10 |
| A989_08851 | XopAF | 33.1 | 5e−19 |
| A989_07073 | XopAH | 32.29 | 2e−33 |
Proteins in this strain have the prefix A989_ supplied by GenBank followed by a unique number.
Evidence for a bacterial to fungal gene transfer
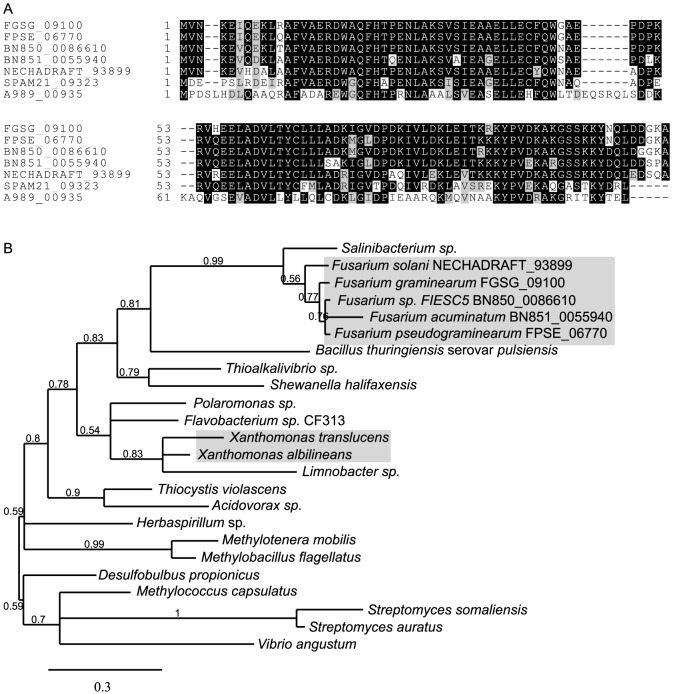
Lateral or horizontal gene transfer is one possible route by which fungal species can acquire new functions to facilitate niche colonisation or gain a competitive advantage. In our recent analysis of the genome of the wheat pathogen F. pseudograminearum, a number of bacterial to fungal gene transfers were identified with strong phylogenetic support [2]. These analyses prompted the sequencing of bacterial pathogens of wheat and barley, such as DAR61454 described here, as possible sources of such horizontally-derived virulence genes. BLASTp analysis of the grass pathogen-specific set of proteins against NCBI's non-redundant (nr) protein database restricted to fungal entries identified a protein encoding a nucleoside triphosphate pyrophosphohydrolase (NTPH) shared between DAR61454 (A989_00935) and the sugarcane pathogen X. albilineans. Interestingly, this gene also had homologues in plant infecting Fusarium species (F. graminearum, F. pseudograminearum, F. solani f. sp. pisi, F. acuminatum and species five of the F. incarnatum-equiseti species complex (F. sp. FIESC5)). These matches to A989_00935 from these five related fungi were the only ones found in the fungal databases at NCBI and whilst only 45% identical, the matches to fungal sequences were throughout the entire length of the proteins at an e-value of 2×10−25. Furthermore, when the Fusarium sequences were used as a query in BLASTp searches, the best hit was to a Salinibacterium sp. with 67% sequence identity and e-value of 2×10−48. The similarity between each of these sequences is shown in a multiple alignment in Figure 5A. Phylogenetic analysis along with the extremely limited distribution of this sequence in fungi supports the view that these three Fusarium spp. acquired this gene from bacteria, but xanthomonads do not appear to be the likely donor genus (Figure 5B). Inspection of the genomic regions of these homologues showed synteny between the A989_00935 homologues and flanking genes in F. graminearum and F. pseudograminearum [2], [38] is conserved. Given these lineages separated ∼1.7 million years ago, the transfer into these species most likely predates this divergence. Synteny is not maintained between the F. pseudograminearum/F. graminearum region and F. solani f. sp. pisi, F. acuminatum or F. sp. FIESC5 regions [39], [40]. NTPHs of this class are involved in the regulation of a cell death pathway encoded by a toxin-antitoxin system in bacteria and form part of a polycistronic RNA with the toxin and antitoxin proteins [41]. However, based on the absence of the other components of this toxin-antioxin system in the genomic region of A989_00935 and the homologue from Salinibacterium sp., these bacterial and fungal homologues may relate to the NTPH enzymatic activity but not in the regulation of toxin-antitoxin systems. Thus a role for NTPHs in bacterial or fungal pathogenesis on plants currently remains obscure.
Figure 5. Sequence similarities between nucleoside triphosphate pyrophosphohydrolases (NTPHs) in bacteria and fungi.
(A) Multiple sequence alignment of the only three fungal NTPHs in GenBank with those of two bacteria. FGSG_09100, FPSE_06770, BN850_0056610, BN851_0055940 and NECHADRAFT_93899 are sequences from Fusarium graminearum, F. pseudograminearum, F. sp. FIESC5, F. acuminatum and F. solani f. sp. pisi, respectively SPAM21_09323 and A989_00935 are sequences from Salinibacterium sp. strain PAMC21357 and Xanthomonas translucens DAR61454 respectively. (B) Phylogenetic analysis of the NTPH protein sequence found in Fusarium spp. and bacteria. The Fusarium and Xanthomonas clades are shaded with all other sequences from bacteria. Analysis was conducted at phylogeny.fr as detailed in the methods. Numbers on each branch indicate support for that branching based on approximate likelihood ratio tests.
In summary, the DAR61454 genome sequenced and initially characterised in this work will be an important resource for comparative analyses of pathogenicity in bacterial and fungal cereal pathogens. In addition the demonstrated ability of DAR61454 as a tool to deliver proteins to the cytoplasm of cereal cells using the type III secretion system can be exploited for molecular pathology studies in a number of plant pathogen interactions. The T6SS found in DAR61454 may also be useful for the delivery of proteins extracellularly, as at least in some bacteria, substrates of this secretion system can be found in culture filtrates [31], although the signature sequences involved in type VI secretion systems are yet to be fully characterised. Finally, the identification of yet another phylogenetically supported horizontal gene acquisition by fungal plant pathogens adds to the growing body of evidence that the acquisition of new genes from bacteria has been a useful evolutionary strategy in fungi. Further sampling of plant associated bacteria via genome sequencing and metagenomics is clearly warranted to fully understand the pervasiveness of this phenomenon in the evolution of virulence in fungal pathogens.
Experimental Procedures
Xanthomonas strain DAR61454, identified as X. translucens, was isolated in the Tamworth region of New South Wales, Australia in March 1988 from wheat and is available from the Australian Collection of Plant Pathogenic Bacteria in the New South Wales Department of Primary Industries. X. translucens DAR61454 was cultured using King B media at 28°C. A rifampicin resistant strain of X. translucens DAR61454 was generated by plating cells from a liquid culture on a King B plate containing a gradient of rifampicin [42] and a single colony picked from the region containing high levels of the antibiotic.
Pathogenicity of X. translucens DAR61454
Pathogenicity of X. translucens DAR61454 was established using two different assays. For visual symptom assessment, a three day culture was diluted to an optical density at 600 nm of 0.05 in 10 mM MgCl2 and approximately 200 µL of this suspension was infiltrated using a 1 mL needleless syringe in to the adaxial side of the second leaf of 10 day old wheat or barley seedlings until the infiltrated liquid started escaping out of stomata. Typically the leaf zone that received inoculum was a 1-2 centimetres either side of the infiltration point. Plants were grown in a glasshouse maintained at 24/16°C and 60/90% relative humidity day/night without any supplementary lighting. For the assessment of bacterial growth over time, a 100-fold dilution of an OD600 = 0.05 suspension of culture of a rifampicin resistant derivative of DAR61454 was made in 10 mM MgCl2 and infiltrated in the same manner as above. The region infiltrated was marked with a permanent pen. Two 7 mm-diameter leaf disks were harvested, one above and one below the infiltration site but both within the infiltrated zones, from each leaf and transferred to a 2 mL eppendorf tube with a single 3 mm ball bearing. The material was ground in a Retsch mill for 30 s at 30 Hz. 400 µL of 10 mM MgCl2 was added to the tube before it was briefly vortexed. A dilution series from each tube was made across six orders of magnitude and 10 µL of these were plated onto King B media containing rifampicin at 50 mg L−1. Bacteria were allowed to grow for two nights at 28°C prior to counting colonies. Each time point consisted of four biological replicates.
Genome sequencing
DNA for genome sequencing was prepared from bacteria grown in liquid culture using a QIAgen Blood and Tissue kit according to the manufacturer's instructions. Illumina library preparation and sequencing was performed by the Australian Genome Research Facility, Melbourne, Australia using one fifth of a HiSeq2000 lane for the sequence generation. Following quality filtering (low quality threshold of 0.05, maximum number of ambiguities set at 2, the removal of 2 terminal nucleotides and a minimum length of 50 bases) of paired end Illumina reads, the genome was assembled from 7.4×107 reads using CLC Genomics Workbench version 5.1 with the scaffolding option selected (minimum contig size 200, minimum and maximum paired end distances set at 316 and 666 bp, respectively. Low coverage contigs (<500-fold with all but four of these being below 100 bp) were removed from the final assembly. As the assembly and scaffolding occurred in one operation in the CLC Genomics Workbench environment, to allow submission to the NCBI Prokaryotic Genome Annotation Pipeline (PGAAP), the 217 scaffolds were split into 404 contigs using custom Perl scripts developed in house (available upon request). Contig relationships were maintained in the GenBank submission by inclusion of a Golden Path (agp) file. No additional curation of the PGAAP annotation was undertaken.
Phylogenetic analysis
The atpD gene was used to infer the taxonomic position of X. translucens DAR61454 based on the analysis performed by Studholme et al. [10]. To provide resolution at the pathovar level, the gyrB gene sequence was used including sequences from the strains described by Parkinson et al. (NCBI population set accession 151936706) [13]. Analysis was performed on nucleotide sequences for these genes at www.phylogeny.fr [43] using a MUSCLE alignment [44], GBLOCKS curation of the nucleotide alignment [45], the PhyML phylogeny package [46] and the HKY85 nucleotide substitution model with approximate likelihood-ratio test for branch support [47]. Tree rendering was performed with TreeDyn.
Phylogenetic analysis of the nucleoside triphosphate pyrophosphohydrolase found in Fusarium spp. and bacteria was performed as described above but on protein sequences manually selected from BLASTp queries at NCBI to represent taxonomically diverse hits to the sequence. The Whelan and Goldman (WAG) evolutionary model for protein sequences was used. Tree rendering was performed with TreeDyn.
Determining protein orthologous relationships
OrthoMCL [14] was implemented on a desktop computer running Ubuntu 12.04 operating system with 16 Gb of RAM. Protein sequences from each of the isolates were extracted from the GenBank entry using custom Perl scripts. Locus tags identifiers were modified for those GenBank entries that did not conform to the three or four character prefixes required by OrthoMCL using custom Perl scripts. OrthoMCL was run as described in the distribution manual using BLAST+ version 2.2.27 with an expectation cut off set at <10−10. Groups of proteins specific to particular pathogen sets were extracted using the grep function in unix.
Delivery of heterologous proteins to host cells by Xanthomonas strain DAR61454
The Cya reporter construct pVSPnPro+AA1-100-AvrBs2∶Cya was obtained from Brian Staskawicz at the University of California, Berkeley, USA. This construct had the AvrBs2 promoter, the first 300 nucleotides of AvrBs2 coding region [48] and the Cya domain from cyclosin gene of Bordetella pertussis [26] cloned as an AvrBs2T3SS∶Cya translational fusion cassette in the broad host range vector pVSP61 [49]. As a control, a vector with the AvrBs2 promoter and Cya but lacking the AvrBs2 leader sequence (AvrBs2Prom-Cya) was used. This construct was cloned by amplification of the AvrBs2 promoter and Cya coding sequence to create BamHI-NcoI and NcoI-EcoRI fragments, respectively, for cloning via a triple fragment ligation into an EcoRI-BamHI cut vector. These constructs were introduced into X. translucens strain DAR61454 (rifampicin marked) by electroporation. The recipient strain DAR61454 was grown at 33°C while making electro-competent cells and during initial selection of transformants to overcome interference from endogenous restriction modification system present in this strain.
Strain DAR61454 containing the AvrBs2T3SS∶Cya expression vector (AvrBs2Prom-AvrBs2T3SS∶Cya) or a control vector without the AvrBs2 leader sequence (AvrBs2Prom-Cya), was grown at 30°C for 36 hrs, cells harvested and resuspended in 10 mM MgCl2 to a OD of 0.4 and infiltrated into leaf blades of three-leaf stage wheat seedlings using needleless syringe. Leaf disc sampling, cAMP extractions and protein measurements were performed as described previously [27] with some modifications. The cAMP levels in the extracted samples were measured with a cAMP Enzyme Immunoassay kit (Cayman Chemical Company) according to manufacturer's instructions and expressed as pmole of cAMP per mg of total protein. Total protein was measured using the Quick Start Bradford Dye Reagent as per the manufacturer's instructions (BioRad).
Supporting Information
Placement of Xanthomonas translucens isolate DAR61454 in the Xanthomonas genus using phylogenetic analysis of the gyrB gene nucleotide sequence. DAR61454 groups with other X. translucens pv. translucens isolates although the support for differentiation from X. translucens pv. graminis is weak. Phylogenetic analysis was carried out at www.phylogeny.fr as detailed in the methods. Numbers of branches represent branch support based on approximate likelihood ratio testing.
(EPS)
Contains the accessions for the groups of proteins specifically found in cereal infecting xanthomonads via the OrthoMCL analysis. Each row of the file has a unique group identifier followed by the individual protein identifiers for entries in that group. Where possible the GenBank locus identifier for that protein has been used. For locus identifiers that did not fit the naming convention required by OrthoMCL the locus identifier prefix was adjusted and can be found in Table 1.
(TXT)
Acknowledgments
The authors wish to thank Drs Brian Staskawicz and Douglas Dahlbeck (University of California, Berkeley) for providing the AvrBs2T3SS∶Cya reporter construct; Eric Cother (New South Wales Department of Primary Industries, Australia) for providing the Xanthomonas strain DAR61454 and Anca Rusu (CSIRO Plant Industry, Brisbane) for excellent technical assistance with pathology assays.
Accession number: AMXY00000000
Funding Statement
This work was supported through access to facilities managed by Bioplatforms Australia and funded by the Australian Government National Collaborative Research Infrastructure Strategy and Education Investment Fund Super Science Initiative (www.bioplatforms.com.au/special-initiatives/agriculture/wheat-datasets) and through funding from the Two Blades Foundation. DMG was partially supported by the Grains Research and Development Corporation (GRDC), an Australian Commonwealth Government Authority. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gardiner DM, Kazan K, Manners JM (2013) Cross-kingdom gene transfer facilitates the evolution of virulence in fungal pathogens. Plant Sci 210: 151–158. [DOI] [PubMed] [Google Scholar]
- 2. Gardiner DM, McDonald MC, Covarelli L, Solomon PS, Rusu AG, et al. (2012) Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog 8: e1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, et al. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog 7: e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gardiner DM, Stiller J, Covarelli L, Lindeberg M, Shivas RG, et al. (2013) Genome sequences of Pseudomonas spp. isolated from cereal crops. genomeA 1: e00209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sohn KH, Lei R, Nemri A, Jones JDG (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana . Plant Cell 19: 4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kleemann J, Rincon-Rivera LJ, Takahara H, Neumann U, van Themaat EVL, et al. (2012) Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum . PLoS Pathog 8: e1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yin C, Hulbert S (2011) Prospects for functional analysis of effectors from cereal rust fungi. Euphytica 179: 57–67. [Google Scholar]
- 8.Upadhyaya NM, Mago R, Staskawicz BJ, Ayliffe M, Ellis J, et al.. (2013) A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol Plant-Microbe Interact In press. [DOI] [PubMed]
- 9.Duveiller E, Bragard C, Maraite H (1997) Bacterial leaf streak and black chaff caused by Xanthomonas translucens. In: Duveiller E, Fucikovsky L, Rudolph K, editors. The bacterial diseases of wheat: concepts and methods of disease management. Mexico, D.F: CIMMYT. pp. 25–47.
- 10. Studholme DJ, Wasukira A, Paszkiewicz K, Aritua V, Thwaites R, et al. (2011) Draft genome sequences of Xanthomonas sacchari and two banana-associated Xanthomonads reveal insights into the Xanthomonas group 1 clade. Genes 2: 1050–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wichmann F, Vorhölter F-J, Hersemann L, Widmer F, Blom J, et al. (2013) The noncanonical type III secretion system of Xanthomonas translucens pv. graminis is essential for forage grass infection. Mol Plant Pathol 14: 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pieretti I, Royer M, Barbe V, Carrere S, Koebnik R, et al. (2009) The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10: 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parkinson N, Cowie C, Heeney J, Stead D (2009) Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int J Syst Evol Microbiol 59: 264–274. [DOI] [PubMed] [Google Scholar]
- 14. Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Birch RG (2001) Xanthomonas albilineans and the antipathogenesis approach to disease control. Mol Plant Pathol 2: 1–11. [DOI] [PubMed] [Google Scholar]
- 16. Tseng T-T, Tyler B, Setubal J (2009) Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Records AR (2011) The Type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant-Microbe Interact 24: 751–757. [DOI] [PubMed] [Google Scholar]
- 18. Yan Q, Wang N (2011) High-throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol Plant-Microbe Interact 25: 69–84. [DOI] [PubMed] [Google Scholar]
- 19. Büttner D, Bonas U (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34: 107–133. [DOI] [PubMed] [Google Scholar]
- 20. da Silva FG, Shen Y, Dardick C, Burdman S, Yadav RC, et al. (2004) Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol Plant-Microbe Interact 17: 593–601. [DOI] [PubMed] [Google Scholar]
- 21. Sandkvist M (2001) Type II Secretion and Pathogenesis. Infect Immun 69: 3523–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. da Silva ACR, Ferro JA, Reinach FC, Farah CS, Furlan LR, et al. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417: 459–463. [DOI] [PubMed] [Google Scholar]
- 23. Brunings AM, Gabriel DW (2003) Xanthomonas citri: breaking the surface. Mol Plant Pathol 4: 141–157. [DOI] [PubMed] [Google Scholar]
- 24. Lee BM, Park YJ, Park DS, Kang HW, Kim JG, et al. (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res 33: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alegria MC, Souza DP, Andrade MO, Docena C, Khater L, et al. (2005) Identification of new protein-protein interactions involving the products of the chromosome- and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri . J Bacteriol 187: 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sory MP, Cornelis GR (1994) Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol 14: 583–594. [DOI] [PubMed] [Google Scholar]
- 27. Casper-Lindley C, Dahlbeck D, Clark ET, Staskawicz BJ (2002) Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc Natl Acad Sci USA 99: 8336–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol 186: 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schechter LM, Guenther J, Olcay EA, Jang S, Krishnan HB (2010) Translocation of NopP by Sinorhizobium fredii USDA257 into Vigna unguiculata root nodules. Appl Environ Microbiol 76: 3758–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, et al. (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103: 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, et al. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312: 1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bladergroen MR, Badelt K, Spaink HP (2003) Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant-Microbe Interact 16: 53–64. [DOI] [PubMed] [Google Scholar]
- 34. Hayward AC, Fegan N, Fegan M, Stirling GR (2010) Stenotrophomonas and Lysobacter: ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J Appl Microbiol 108: 756–770. [DOI] [PubMed] [Google Scholar]
- 35. Hajri A, Brin C, Hunault G, Lardeux F, Lemaire C, et al. (2009) A repertoire for repertoire hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas . PLoS ONE 4: e6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, et al. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7: e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48: 419–436. [DOI] [PubMed] [Google Scholar]
- 38. Cuomo CA, Güldener U, Xu J-R, Trail F, Turgeon BG, et al. (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317: 1400–1402. [DOI] [PubMed] [Google Scholar]
- 39. Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, et al. (2009) The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet 5: e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moolhuijzen PM, Manners JM, Wilcox SA, Bellgard MI, Gardiner DM (2013) Genome sequences of six wheat-infecting Fusarium species isolates. genomeA 1: e00670–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gross M, Marianovsky I, Glaser G (2006) MazG – a regulator of programmed cell death in Escherichia coli. Mol Microbiol 59: 590–601. [DOI] [PubMed] [Google Scholar]
- 42. Bryson V, Szybalski W (1952) Microbial selection. Science 116: 45–51. [PubMed] [Google Scholar]
- 43. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Edgar R (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 46. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 47. Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol 55: 539–552. [DOI] [PubMed] [Google Scholar]
- 48. Minsavage GV, Dahlebeck D, Whalen MC, Kearney B, Bonas U, et al. (1990) Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria – pepper interactions. Mol Plant-Microbe Interact 3: 41–47. [Google Scholar]
- 49. Loper JE, Lindow SE (1987) Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringe pv. syringae on bean leaf surfaces. Phytopathology 77: 1449–1454. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Placement of Xanthomonas translucens isolate DAR61454 in the Xanthomonas genus using phylogenetic analysis of the gyrB gene nucleotide sequence. DAR61454 groups with other X. translucens pv. translucens isolates although the support for differentiation from X. translucens pv. graminis is weak. Phylogenetic analysis was carried out at www.phylogeny.fr as detailed in the methods. Numbers of branches represent branch support based on approximate likelihood ratio testing.
(EPS)
Contains the accessions for the groups of proteins specifically found in cereal infecting xanthomonads via the OrthoMCL analysis. Each row of the file has a unique group identifier followed by the individual protein identifiers for entries in that group. Where possible the GenBank locus identifier for that protein has been used. For locus identifiers that did not fit the naming convention required by OrthoMCL the locus identifier prefix was adjusted and can be found in Table 1.
(TXT)