Abstract
Background
The scalloped (sd) and vestigial (vg) genes function together in Drosophila wing development. Little is known about sd protein (SD) expression during development, or whether sd and vg interact in other developing tissues. To begin to address these questions, we generated an anti-SD antibody.
Results
During embryogenesis, SD is expressed in both central and peripheral nervous systems, and the musculature. SD is also expressed in developing flight appendages. Despite SD expression herein, the peripheral nervous system, musculature, and dorsal limb primordia appeared generally normal in the absence of sd function. SD is also expressed in subsets of ventral nerve cord cells, including neuroblast 1–2 descendants and ventral unpaired median motor neurons (mVUMs). While sd function is not required to specify these neurons, it is necessary for the correct innervation of somatic muscles by the mVUMs. We also show that SD and VG are co-expressed in overlapping and distinctive subsets of cells in embryonic and larval tissues.
Conclusions
We describe the full breadth of SD expression during Drosophila embryogenesis, and identify a requirement for sd function in a subset of motor neurons. This work provides the necessary foundation for functional studies regarding the roles of sd during Drosophila development.
Keywords: transcription factor, vestigial, imaginal disc, central nervous system, peripheral nervous system, muscle
INTRODUCTION
The classical Drosophila scalloped (sd) mutant wing phenotype was first described in 1929 (Gruneberg, 1929), and the gene was subsequently cloned over 60 years later (Campbell et al., 1991). It was established that sd encodes a protein with a TEA family DNA-binding domain (Campbell et al., 1992; Jacquemin and Davidson, 1997; Anbanandam et al., 2006) and binds DNA in a sequence-specific manner (Halder et al., 1998b; Guss et al., 2001). SD acts as a transcription factor with two coactivators, vestigial (VG) and yorkie (YKI), functioning in wing development and the control of cell proliferation. With VG (Williams et al., 1991), SD functions as the selector (Mann and Carroll, 2002) for wing identity, forming a complex that is necessary and sufficient to direct wing development (Kim et al., 1996; Halder et al., 1998b; Simmonds et al., 1998; Halder and Carroll, 2001). SD binds to and directly regulates the wing-specific cis-regulatory elements for genes controlling many aspects of wing morphogenesis (Halder et al., 1998b; Guss et al., 2001; Lunde et al., 2003). With YKI, SD functions as an activator of growth and proliferation, in a complex regulated by the Hippo signaling pathway (reviewed in Bandura and Edgar, 2008).
Despite these extensive investigations of sd function, it has not before been possible to examine the expression of the SD protein. To address this challenge, we made an anti-SD antibody, and used it to characterize the full breadth of SD expression during Drosophila development. In addition to the expected distribution of SD in the wing and haltere imaginal discs, we show that SD is strongly expressed in subsets of neuronal and muscle cells, where it colocalizes with VG. Based on this characterization, we present data that SD functions in neuronal pathfinding to muscle innervation of the ventral unpaired median motor neurons. This work lays the foundation for the systematic study of SD function in the development of the neuromusculature.
RESULTS AND DISCUSSION
Synthesis and specificity of the SD antibody
To define the cellular expression of SD during Drosophila development, we generated an anti-SD antibody. To test the specificity of this probe, its immunoreactivity was assessed in embryos showing loss of expression, or overexpression, of sd (Figure 1). The SD antibody failed to label any tissues in Df(1)Exel6251/Y embryos, which carry an X chromosome deficiency that uncovers the sd locus (Parks et al., 2004; Fig. 1B). Expression was also assessed in embryos carrying the recessive lethal sd3L allele (data not shown). The molecular lesion associated with this allele is a nonsense mutation in the first leucine codon within the Vestigial Binding Domain (Campbell et al., 1991; Srivastava et al., 2004), Very low, diffuse signal was observed in the central nervous systems of sd3L embryos suggesting it is a strong hypomorphic mutation. Conversely, the antibody labeled additional cells in embryos in which patched-GAL4 drove UAS-sd in many cells in every segment (Fig. 1D). Taken together, these data show that our antibody recognizes specifically the SD protein.
Figure 1. Specificity of the anti-SD antibody.
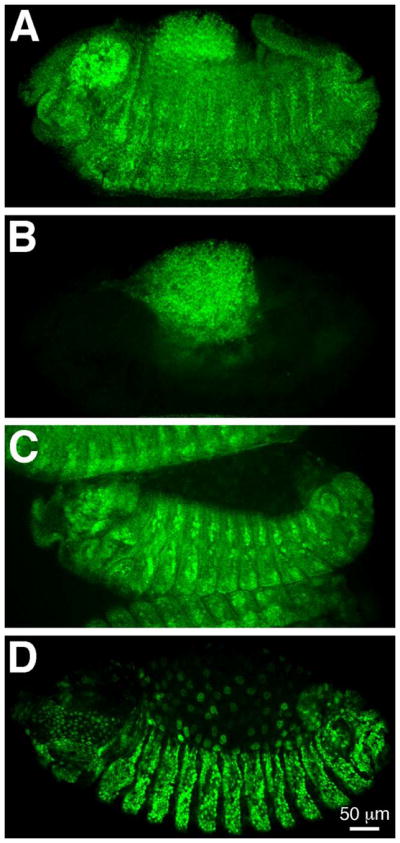
(A, B) Detection of the guinea pig anti-SD antibody in stage 14 embryos of genotypes Df(1)Exel6251 or FM7c act-lacZ/FM7c act-lacZ (A) and Df(1)Exel6251/Y (B). While the anti-SD antibody is detectable in embryos carrying the balancer chromosome (A; βGAL expression not shown), no detection is observed in embryos missing the sd locus (B). (C, D) Detection of the guinea pig anti-SD antibody in stage 12 embryos in which sd is expressed at normal levels (C) or overexpressed (D). The pattern of anti-SD antibody detection in embryos carrying patched-GAL4 is comparable to that observed in wild type embryos (C; compare to Figure 2I), while ectopic expression is detected in embryos that additionally carry UAS-sd (D; note that this image was collected using lower laser settings than panel C). Our rat anti-SD antibody showed the same results in sd loss of expression and overexpression experiments (data not shown). Anterior is to the left in all panels.
SD expression during embryonic development
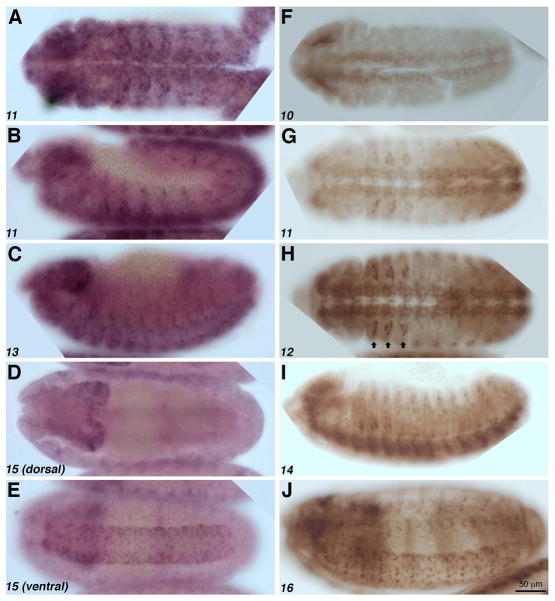
We observe close correlation between the patterns of expression the sd RNA and SD protein, which further supports the specificity of our antibody (Fig. 2). The sd RNA is detectable in the central nervous system (CNS; Fig. 2A, D, E; see also Campbell et al., 1992) and the peripheral nervous system (PNS; Fig. 2B, C). SD protein is expressed in the ventral nerve cord (VNC) detectable beginning at stage 10 (Fig. 2F). The expression in the CNS continues through stages 11 (Fig. 2G), 12 (Fig. 2H), 14 (Fig. 2I), and 16 (Fig. 2J). Expression in the PNS is also observed at stage 14 (Fig. 2I), while expression in the muscles is discernible at stage 16 (Fig 2J). We also observe strong SD expression in adepithelial cells (Bate et al., 1991) located in the between the dorsal and ventral limb primordia (arrows, Fig. 2H; see also Fig. 3P–R).
Figure 2. SD expression during embryogenesis.
(A–E) Whole mount in situ hybridization showing sd RNA distribution (purple) in embryos at stages 11 (A, B), 13 (C), and 15 (D, E). sd is expressed in the central nervous system at stages 11 (A) and 15 (D, E) and is also detectable in the peripheral nervous system of embryos at stages 11 (B) and 13 (C). (F–J). SD protein expression (brown). SD is detectable in the ventral nerve cord at stages 10 (F), 11 (G), 12 (H) and becomes localized to subsets of cells by stage 16 (J). SD is also expressed in the peripheral nervous system at stage 14 (I), the developing somatic mesoderm and cells between the dorsal and ventral limb primordia (G–I; arrows, H). Anterior is to the left in all panels, and stages of development are in the lower left.
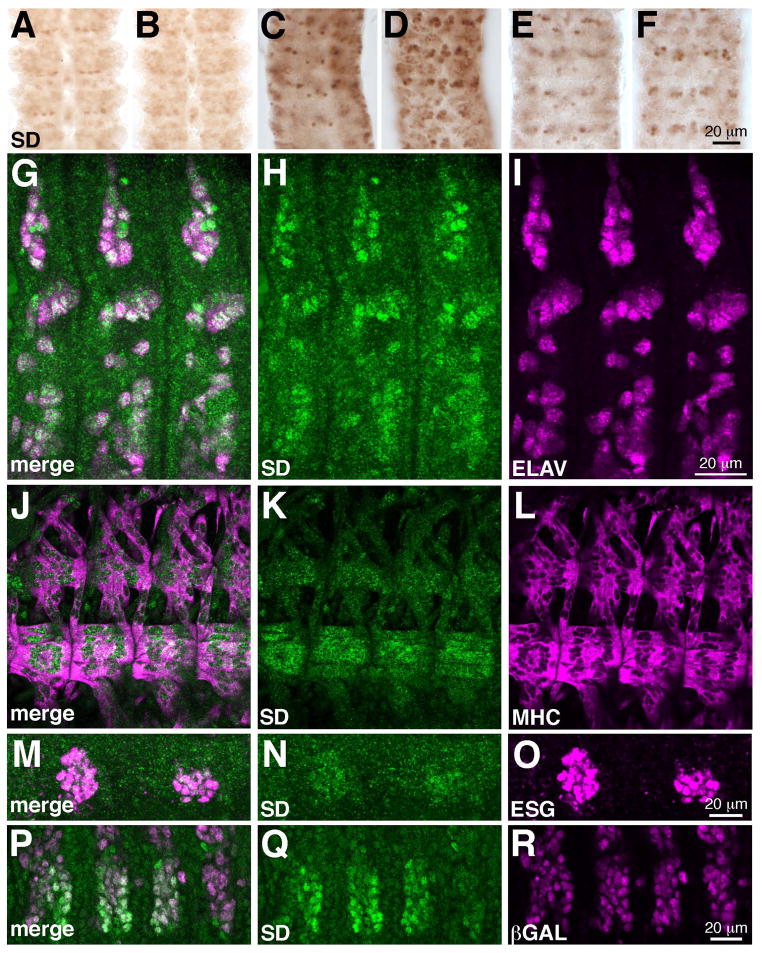
Figure 3. SD expression in the embryonic CNS, PNS, limb primordia, and somatic muscles.
(A–F) Expression of SD expression VNCs dissected from stage 12 (A, B), stage 15 (C, D) and stage 16 (E, F) embryos. For each stage, dorsal (A, C, E) and ventral (B, D, F) focal planes are shown. SD is initially broadly expressed in the VNC, and expression resolves to subsets of cells as development proceeds. (G–I) SD (green) and ELAV (magenta) in three abdominal segments of a stage 12 embryo, showing that most SD expressing cells are sensory neurons. These images are maximum projections of multiple confocal slices. (J–L) Four abdominal segments of a late stage 16 embryo showing SD (green) expression in muscles VL1–4 and LL1, which are marked by MHC (magenta). (M–O) SD (green) is coexpressed with ESG (magenta) in the dorsal limb primordia of the second and third thoracic segments of a stage 17 embryo. These images are maximum projections of multiple confocal slices. (P–O) Three thoracic segments and first abdominal segment of a stage 12 embryo. SD (green) is coexpressed with βGAL driven by twist-GAL4 in the three thoracic segments. Specimens are oriented with anterior up (A–F) or to the left (G–R).
We examined more closely the expression of SD during development (Fig. 3). In the VNC, SD appears to be initially expressed in many cells (Fig. 3A, B), although the expression resolves to higher levels in a subset of cells as embryogenesis proceeds (Fig. 3C–F). To more thoroughly characterize the SD expression in other tissues during embryogenesis, we performed colocalization studies with tissue or cell type specific molecular markers. In the PNS, SD is expressed in most sensory neurons, based on close coexpression with Embryonic Lethal Abnormal Vision (ELAV; (ELAV; Robinow and White, 1991; Fig. 3G–I). This coexpression continues through embryogenesis, although the level of SD expression in cells in the PNS is reduced. SD is also expressed in the somatic bodywall muscles during embryogenesis. At stage 12, SD is detectable in the ventral lateral muscles 1–4 (VLs 1–4, muscle nomenclature per Bate, 1993). This expression continues through stage 16, with higher expression in the muscle nuclei (Fig. 3J–L). By this stage SD is also detectable in ventral acute muscles 1 and 2 (VA1 and 2) and lateral longitudinal muscle 1 (LL1). In late stage 16 embryos, additional dorsal muscles also express SD. Therefore, expression of SD occurs in the most ventral muscles first, then expands to more dorsal muscles as development proceeds. Deng et al. (2009) observed broader sd expression, earlier in development, by employing sd RNA and enhancer trap detection methods, with reduced expression by late stage 16, whereas we observed the most extensive expression this late embryonic stage.
SD is also expressed at low levels in the embryonic wing and haltere primordia, based on colocalization with Esgargot (ESG; Fig. 3M–O). The cells expressing the higher level of SD in the lateral regions of the three thoracic segments are progenitors of the adult flight muscles (Fig. 3P–R). We make this conclusion based on the coexpression of SD and βGAL driven by twist-GAL4, a marker for these adepithelial cells (Bate et al., 1991). Our observation correlates well with those made by Bernard et al. (2003), who showed that VG and sd-lacZ are coexpressed in these cells later in development.
Scalloped expression in the larval imaginal discs
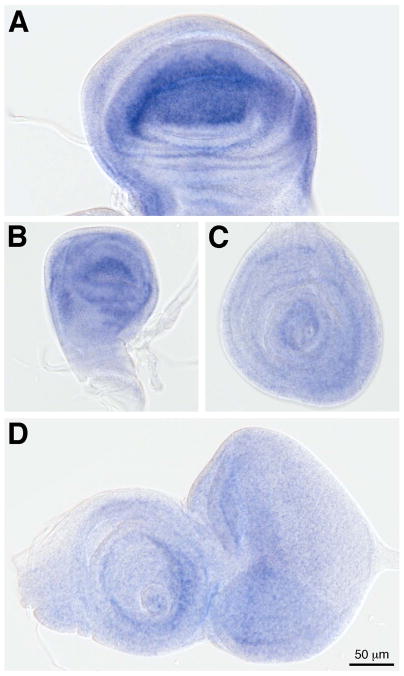
We also examined SD expression in tissues of the wandering third instar larva (Fig. 4). In the wing imaginal disc, SD is expressed in the wing pouch and margin (Fig. 4A), with expression extending into the hinge and notum (Cohen, 1993). SD is expressed in the corresponding structures in the haltere disc (Fig. 4B). The strong expression of SD in the wing disc correlates well with its documented role in wing development. In comparison, SD is expressed at much lower levels in the leg (Fig. 4C) and eye antennal (Fig. 4D) imaginal discs. We also examined the expression of sd RNA in the imaginal discs (Fig. 5). Concordant with the high levels of the protein in the wing and haltere, strong RNA expression is observed in these imaginal discs (Fig. 5A, B). We also observe expression, at lower levels, in the leg and eye antennal discs (Fig. 5C, D). In the leg and antenna, sd appears to be expressed in rings that may correspond to different locations along the proximal-distal axes of these tissues.
Figure 4. SD expression in larval imaginal discs.
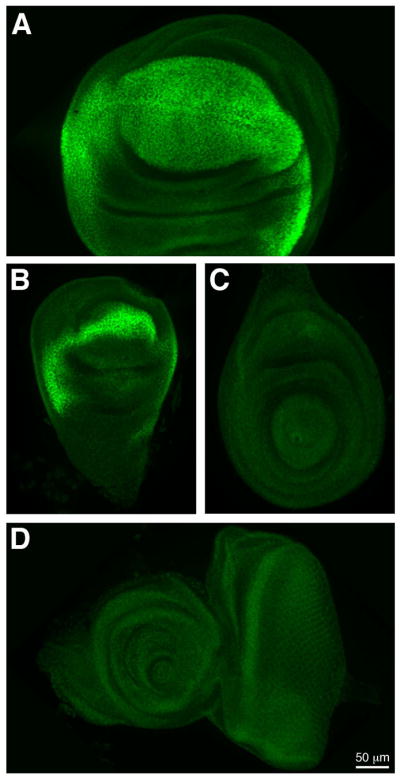
SD (green) is expressed in the wing (A) and haltere (B) imaginal discs, and at low levels in the leg (C) and eye antennal (D) discs.
Figure 5. sd expression in larval imaginal discs.
sd (blue) is expressed in the wing (A) and haltere (B) imaginal discs. It is also expressed at lower levels in the leg (C) and eye antennal (D) discs.
Our observations generally agree with the expression pattern of sd that has been reported using the sd enhancer trap line ETX4 (Campbell et al., 1992), although we do see some differences. For example, we report here what appear to be higher levels of expression of sd and SD in cells in the embryo using direct detection of these gene products versus those levels reflected by ETX4 (Campbell et al., 1992 and our own observations). In addition, Campbell et al. (1992) show that ETX4 drives high expression posterior to the morphogenetic furrow in eye imaginal disc, whereas we observe both RNA and protein at higher levels to the anterior of this landmark. Such discrepancies may be due to subtle differences in regulation, and/or perdurance, of the reporter protein in the enhancer trap line.
Coexpression of Scalloped and Vestigial in developing tissues
Scalloped (SD) and Vestigial (VG) have a well-established role working together in wing development (Halder et al., 1998b; Simmonds et al., 1998). Based on a characterized VG antibody, the identity of many VG-expressing cells have been determined in developing tissues (Williams et al., 1991; Landgraf et al., 2003; Guss et al., 2008). Therefore, using this tool, we next examined the coexpression of SD and VG in both embryos and larvae. This approach allowed us to determine the identity of SD-expressing cells, and provides the necessary foundation to identify tissues in which SD and VG may function together, as well as tissues in which they may function independently. A summary of these data is shown in Figure 6.
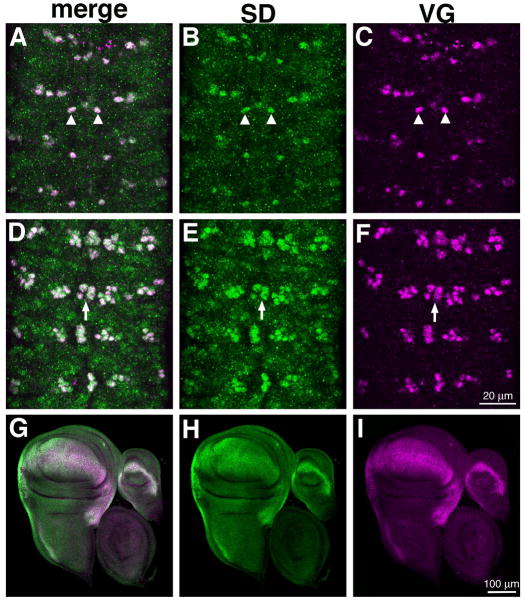
Figure 6. SD and VG coexpression in the ventral nerve cord and imaginal discs.
Dorsal (A–C) and ventral (D–F) focal planes of a stage 16 embryonic VNC showing expression of SD (green) and VG (magenta). These images are maximum projections of multiple confocal slices. SD is expressed at high levels in cells that also express VG, including descendants of NB1-2 (arrowheads, A–C) and the mVUMS (arrows, D–E). (G–I) SD (green) and VG (magenta) expression in the somatic muscles of abdominal segments of a stage 15 embryo. At this stage, both proteins are expressed in VLs 1–4 (arrowheads, G–I), but VG is expressed in dorsal muscles as well (arrowheads, G–I). Specimens are oriented with anterior up (A–F) or to the left (G–I). (J–L) Wing, haltere and leg imaginal discs from a wandering third instar larva. SD (green) and VG (magenta) are coexpressed at high levels in the wing and haltere imaginal discs.
SD is coexpressed with VG in subsets of cells in both the embryo and in the wing and haltere imaginal discs (Fig. 6). As discussed above, SD expression appears to be initially broad in the ventral nerve cord, and to resolve to high expression in a clear subset of cells. Interestingly, these same identified neurons also express VG (Fig. 6A–F). These cells include neuroblast 1–2 (NB1-2) descendants (Fig. 6A–C; referred to as mVg in Landgraf et al., 2003; Guss et al., 2008) and the ventral unpaired median motor neurons (mVUMs; Fig. 6D–F). With respect to the somatic muscles, both SD and VG are expressed at elevated levels in the ventral muscles VLs 1–4 and LL1 (Fig. 6G–I) At this stage, VG is additionally expressed in dorsal acute muscles (DA)1–3 (Landgraf et al., 1999). In the larval imaginal discs, consistent with their shared role as selectors for wing development, both SD and VG are expressed in the wing and haltere imaginal discs (Fig. 6J–K).
Functional requirements for Scalloped in the neuromusculature
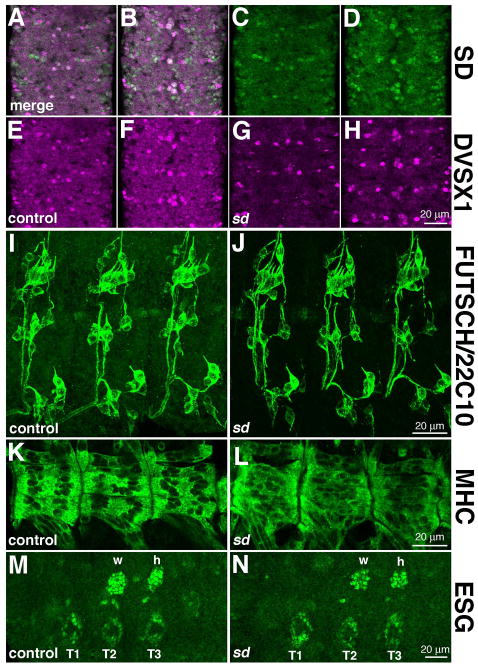
To begin to investigate the function of Scalloped in embryonic tissues, we surveyed general morphological markers the nervous systems and muscle in sd3L (strong hypomorph) and Df(1)Exel6251 (null) embryos (Fig. 7). It appears that sd function is not required for cell fate specification in the CNS, including in the SD-expressing NB1-2 descendants and mVUMs. Based on labeling for DVSX1 (Erclik et al., 2008), which is expressed in the NB1-2 descendants and mVUMs, we found no consistent difference between control and Df(1)Exel6251 null embryos (Fig. 7A–H). Although SD is broadly expressed in the PNS, we did not observe a detectable change in the number/position of neuronal cell bodies or the gross morphology of the sensory neuron axons. This conclusion is based on similar expression of microtubule-associated Futsch/22C10 (Hummel et al., 2000) in control and sd mutant embryos (Fig. 7I, J). Similarly, we observed no detectable change in embryonic muscle patterning or differentiation in the absence of SD function. Based on labeling for Myosin Heavy Chain (MHC), we observed a subtle change in the distribution of muscle nuclei in sd mutant versus control embryos (Fig. 7K, L). We focused on VLs 1–4 and LL1, because we observed these muscles to consistently express SD from stage 12 through later embryogenesis. For this reason, we did not examine the ventral oblique muscles and cardiac cells, reported by Deng et al. (2009) to show defects in sd mutants.
Figure 7. sd function is not required for global patterning of the PNS, CNS, muscles, and limb primordia.
(A–H) sd is not required for cells fate specification of the mVUMs and NB1-2 descendants in the ventral nerve cord. SD (green, A–D) is coexpressed with DVSX1 (magenta, A, B, E–H) in cells in the ventral nerve cord, including the mVUMs and NB1-2 descendants. The distribution of these cells is normal in Df(1)Exel6251 embryos, which are null for sd. (I, J) Antibody 22C10 was used to assess the distribution of cell bodies and axons in control (I) and sd3L embryos (J); these images are maximum projections of multiple confocal slices. We observed no reproducible difference between the wild type and mutant, showing that sd is not required for general PNS patterning. (K, L) Muscle structure was assessed using myosin heavy chain (MHC) in control (K) and Df(1)Exel6251 (L) embryos. Muscle structure is generally normally in sd mutant embryos, although altered distribution of nuclei was observed. (M, N) Distribution of ESG expressing cells in the dorsal and ventral limb primordia of control (M) and Df(1)Exel6251 (N) embryos. The normal distribution of these cells in sd mutant embryos confirms the observation that sd function is not required for the establishment of the limb primordia or the dorsal migration of the flight appendage cells (marked w, h) from the leg primordia (marked T1, T2, T3). Specimens are oriented with anterior up (A–H) or to the left (I–N).
Finally, we observe that sd function is not required for the establishment of the flight appendage primordia. This conclusion is based on the same distribution of ESG expressing cells in the dorsal and ventral flight appendage primordia in the thoracic segments of sd mutant and control embryos (Fig. 7M, N), and is consistent with the observation that the wing disc forms in sd mutants (Williams et al., 1993). We conclude that SD is either not required for the specification of these tissues, or that its effects are subtler or address different aspects of differentiation than are accessible with the markers we used.
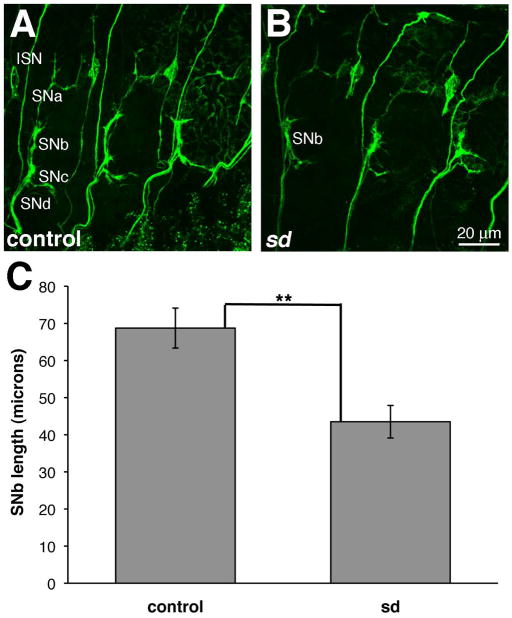
The fact that sd is not required for the cell fate specification in the neuromusculature led us to ask whether it may function in a later differentiation stage. Because the mVUM gene expression profile (Wheeler et al., 2006) and target muscles (Landgraf et al., 1997) have been characterized, we tested for sd phenotypes in this select cell population. Collectively, the three mVUM neurons innervate dorsal oblique muscles 3–5 (DO 3–5) via the intersegemental nerve (ISN), lateral transverse muscles 1 and 2 (LT1 and 2), via segmental nerve a (SNa), and ventral oblique muscles 1–6 (VO 1–6) via SNb and SNd (Landgraf et al., 1997). More recent accounts (e.g., Ruiz-Canada and Budnik, 2006) refer to only the ventral muscles VO 1–5 as being innervated by mVUMs, so we have focused on these muscles, and the structures of SNb and SNd. The monoclonal antibody ID4 for Fasciclin II (FasII; Van Vactor et al., 1993) was used to label all the motorneuron axons in control and sd mutant embryos. We observed disruptions of the segmental nerve branches in the absence of sd (Fig. 8). Because SNd could not consistently be identified and it was difficult to identify the stereotyped branches of SNb (Halpern et al., 1991), we focused on SNb length, from its projection from ISN to its distal tip. The length of SNb is shorter in sd mutant embryos than in controls (mean=43.52, s.e.m.=4.38 versus mean=68.73, s.e.m.=5.38, p=0.0034). This result indicates that sd function is required for the normal innervation of the ventral muscles.
Figure 8. sd is required for the innervation ventral muscles.
Embryos of genotypes (A) w1118 and (B) sd3L showing expression of FASII in three posterior abdominal segments of late stage 16 embryos. ISN and SNa-d are labeled in a representative control segment (A) and SNb is labeled in a segment in the sd mutant (B). These images, which are maximum projections of multiple confocal sections, are oriented anterior to the left. (C) SNb length is shorter in the sd mutant than the control. Error bars represent the s.e.m, and p<0.01.
SUMMARY
Here we use a new antibody probe to characterize the expression of the TEA domain transcription factor SD during Drosophila development. Although this protein is known for its role in wing development and growth control, SD is expressed in many tissue types during embryogenesis, including the central and peripheral nervous systems, and the muscles. We report a new role for sd, in the innervation of the ventral muscles. We also examined the coexpression of SD with VG, its cofactor in wing development. These proteins are distributed in similar but not identical expression patterns, presenting the possibilities that they may fine tune each other’s expression, function together outside the wing, and/or function independently.
EXPERIMENTAL PROCEDURES
Drosophila stocks
The genotype w1118 was used as wild type. Df(1)Exel6251 is a deficiency that uncovers the sd locus (FlyBase), and sd3L is a recessive lethal allele (Campbell et al., 1991; Srivastava et al., 2004). Both mutations were maintained over FM7c act-lacZ (Bloomington Drosophila Stock Center) and male embryos carrying the mutation were identified by the absence of beta-galatosidase (βGAL) expression. Additional stocks used were patched-GAL4, twist-GAL4, UAS-lacZ.nls (all from Bloomington Drosophila Stock Center) and UAS-sd (Sean Carroll).
SD antibody generation
Both known SD isoforms span from N-terminal MVDSK through C-terminal RLIKE (FlyBase R5.8, 2008). This 375 amino acid SD sequence was subjected to the Jameson-Wolf antigenic index (Lasergene, Madison, WI). PCR primers were designed to generate a form of SD containing 369 amino acids, and had the following sequences, including restriction enzyme recognition sequences (bold) and end sequences to facilitate enzyme activity (underline):
SD_7_28: 5′ CTACATATGGATAGCAAAAACCTGGATGTCG 3′
SD_1113_1092: 5′ CTAAAGCTTACGGTATATGTGATGGGTGGTG 3′
Samples containing as template the sd open reading frame and the thermostable polymerase PfuTurbo were subjected to 30 cycles of amplification using an annealing temperature of 65°C. The PCR products of triplicate reactions were processed using the QIAQuick PCR purification protocol. The products were then digested with NdeI and HindIII for cloning. Following digestion, the products were again processed using the QIAQuick PCR purification protocol. The insert was cloned in-frame via NdeI and HindIII sites in the pET29a expression vector. The construct was transformed into competent BL21 cells, with expression induced by addition of IPTG to 1 mM to bacterial culture with OD600 approximately 0.5, followed by an additional 2.5 hours growth. The 46 kDa protein was recovered as described by Williams et al. (1995). Inclusion bodies recovered by treatment with lysozyme followed by recovery with a French press were solubilized in 10% SDS, then dialyzed with phosphate buffered saline (PBS). The protein at ~2 mg/ml concentration was injected by Pocono Rabbit Farm and Laboratory, Inc. (Canadensis, PA) into 3 rats and 3 rabbits. The final exsanguinations were affinity purified against the SD protein.
Whole mount in situ hybridization
Detection of RNA in Drosophila embryos was performed as described by Small (2000), and in third instar larval imaginal discs as described by Mark Sturtevant and Ethan Bier (1997). A DNA construct containing the sd open reading frame cloned into pBluescript (Anj Hudson and Sean Carroll) was used as template to synthesize T3-primed sense or T7-primed antisense RNA probes using digoxigenin-labeled RNA synthesis materials from Roche. No signal was observed in embryos or imaginal tissues processed with the sense RNA probes (data not shown).
Immunocytochemistry
Embryos from overnight collections were washed with 0.4% NaCl/0.1% Triton X-100, dechorionated by incubating in bleach for 2.5 minutes, and rinsed with dH20. Embryos were then fixed by agitating for 20 minutes in equal volumes heptane and fix buffer comprised of 0.1 M PIPES, 2 mM EGTA, 1 mM MgSO4, and 4% formaldehyde. Following fixation, embryos were devitellinized and washed with methanol. For immunolabeling, embryos were rehydrated into PBS with 0.1% Triton X-100 and 1% bovine serum albumin, which was used for all antibody incubations and washes. Wandering third instar larval imaginal discs were dissected, fixed, blocked, and processed with antibodies as described by Halder et al (1998a).
Antibodies
SD was detected using either guinea pig anti-SD (1:500 or 1:1000 in embryos, 1:1000 in larvae) or rat anti-SD (1:5 in embryos). Guinea pig anti-DVSX1 (1:750; Erclik et al., 2008) was provided by Howard Lipshitz. Rabbit anti-VG (1:25 in embryos, 1:100 in larvae; Williams et al., 1991) was the gift of Sean Carroll and in embryos was detected with biotinyl tyramide amplification as described in Guss et al (2008). Rat anti-ESG (1:500; Fuse et al., 1994) was provided by Shigeo Hayashi. Rabbit anti MHC (1:500) was provided by Dan Kiehart via Emma Rushton. Rat anti-ELAV (7E8A10, 1:200), mouse anti FUTSCH (22C10; 1:100), mouse anti-FASII (ID4, 1:20) were developed by Gerald Rubin, Seymour Benzer, and Corey Goodman, respectively, and obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. βGAL was detected using antibodies raised in either rabbit (Invitrogen) or mouse (Promega) and diluted 1:1000. Primary antibodies were detected by either Alexa Fluor-conjugated secondary antibodies (1:300, Molecular Probes) or biotin-conjugated secondary antibodies (1:400, Jackson) using the ABC kit (Vector Laboratories) and DAB Substrate Kit (Pierce). All antibodies were routinely preadsorbed against fixed embryos before use. Samples were incubated overnight in primary antibodies at 4°C, and two hours at room temperature in secondary antibodies. For imaging, embryos were suspended and mounted in VECTASHIELD® (Vector Laboratories) or 70% glycerol.
Image collection and presentation
Fluorescent specimens were imaged using Zeiss LSM 510 META and Olympus Fluoview 500 inverted laser scanning confocal microscopes. Data were collected using sequential scans to avoid artifacts from overlapping emission spectra, with image slices of 0.5 μM. Except as noted, each figure panel shows a single confocal section. Non-fluorescent specimens were collected using Zeiss Axioplan and Nikon E800 compound microscopes equipped for differential interference contrast illumination. Images were merged, cropped and adjusted for brightness, contrast and levels using Adobe Photoshop versions 10.0.1 and 13.0.1. Except as noted, images of specimens being directly compared (e.g., wild type versus mutant, wing versus leg imaginal discs) were collected using the same microscope settings, and subjected to the same image adjustments.
Characterization of SNb length
To assess the length of segmental nerve projections, maximum projections of Z-series were collected of FASII expression in posterior abdominal segments of late stage 16 embryos. The length of SNb was measured from the branch from the ISN to the most distal tip of SNb using MetaMorph Software. In sd3L embryos, eight segments in three animals were measured, while in controls, seven segments in three animals were measured. The data was analyzed using the Student’s t-test with a two-tailed distribution assuming unequal variance. The standard error of the mean (s.e.m) is reported.
Bullet points.
Scalloped (SD) protein expression is characterized for the first time using an anti-SD antibody.
SD is expressed in the central and peripheral nervous systems, the musculature and the flight appendage primordia of the Drosophila embryo, and in the larval imaginal discs.
sd function is required for the correct innervation of the somatic muscles by the ventral unpaired median motor neurons.
Acknowledgments
We thank Anj Hudson for the DNA construct containing the sd open reading frame, John Bell, Sean Carroll, Shigeo Hayashi, Howard Lipshitz, and Emma Rushton for antibodies and/or fly stocks, Yi Zhu and Beth Wilson for advice and assistance during the SD antibody synthesis, Cheryl Gatto for advice on statistical analysis of SNb length, Erik Boczko for translating the first sd reference from the original German, and past and present members of the Skeath and Broadie labs for hospitality during research visits and sabbatical stays. As undergraduates at Dickinson College, Chelsea Gurvis, Evan Hennessy, Aaron Kopp, Kelly LaRue, Brandon Lee, Liz Norris, and Rachel Yonker assisted in preliminary characterization of the sd RNA or protein expression in wild type or sd mutant embryos, and Elise Fiala and Matthew Weddig processed the samples shown in Figure 3, panels P-R. Several monoclonal antibodies used in the course of this work were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. We also thank the Bloomington Drosophila Stock Center for providing fly stocks. This work was supported by NSF 0744261 to J.B.S. and K.A.G., and NIH RO1 MH096832 to K.B.
References
- Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandura JL, Edgar BA. Yorkie and Scalloped: partners in growth activation. Dev Cell. 2008;14:315–316. doi: 10.1016/j.devcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Bate M. The mesoderm and its derivatives. In: Martinez Arias A, Bate M, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Bate M, Rushton E, Currie DA. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- Bernard F, Lalouette A, Gullaud M, Jeantet AY, Cossard R, Zider A, Ferveur JF, Silber J. Control of apterous by vestigial drives indirect flight muscle development in Drosophila. Dev Biol. 2003;260:391–403. doi: 10.1016/s0012-1606(03)00255-0. [DOI] [PubMed] [Google Scholar]
- Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- Campbell SD, Duttaroy A, Katzen AL, Chovnick A. Cloning and characterization of the scalloped region of Drosophila melanogaster. Genetics. 1991;127:367–380. doi: 10.1093/genetics/127.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM. Imaginal disc development. In: Martinez-Arias A, Bate M, editors. The Development of Drosophila melanogaster. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Deng H, Hughes SC, Bell JB, Simmonds AJ. Alternative requirements for Vestigial, Scalloped, and Dmef2 during muscle differentiation in Drosophila melanogaster. Mol Biol Cell. 2009;20:256–269. doi: 10.1091/mbc.E08-03-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erclik T, Hartenstein V, Lipshitz HD, McInnes RR. Conserved role of the Vsx genes supports a monophyletic origin for bilaterian visual systems. Curr Biol. 2008;18:1278–1287. doi: 10.1016/j.cub.2008.07.076. [DOI] [PubMed] [Google Scholar]
- Fuse N, Hirose S, Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 1994;8:2270–2281. doi: 10.1101/gad.8.19.2270. [DOI] [PubMed] [Google Scholar]
- Gruneberg VH. Ein Beitrag zur Kenntnis der Rontgenmutationen des X-Chromosoms von Drosophila melanogaster. Biol Zentbl. 1929;49:680–694. [Google Scholar]
- Guss KA, Mistry H, Skeath JB. Vestigial expression in the Drosophila embryonic central nervous system. Dev Dyn. 2008;237:2483–2489. doi: 10.1002/dvdy.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss KA, Nelson CE, Hudson A, Kraus ME, Carroll SB. Control of a genetic regulatory network by a selector gene. Science. 2001;292:1164–1167. doi: 10.1126/science.1058312. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998a;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Halder G, Carroll SB. Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development. 2001;128:3295–3305. doi: 10.1242/dev.128.17.3295. [DOI] [PubMed] [Google Scholar]
- Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Carroll SB. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in response to signaling proteins. Genes and Development. 1998b;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern ME, Chiba A, Johansen J, Keshishian H. Growth cone behavior underlying the development of stereotypic synaptic connections in Drosophila embryos. J Neurosci. 1991;11:3227–3238. doi: 10.1523/JNEUROSCI.11-10-03227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, Klambt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Davidson I. The role of the TEF transcription factors in cardiogenesis and other developmental processes. TCM. 1997;7:192–197. doi: 10.1016/S1050-1738(97)00052-2. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Baylies M, Bate M. Muscle founder cells regulate defasciculation and targeting of motor axons in the Drosophila embryo. Curr Biol. 1999;9:589–592. doi: 10.1016/s0960-9822(99)80262-0. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Bossing T, Technau GM, Bate M. The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J Neurosci. 1997;17:9642–9655. doi: 10.1523/JNEUROSCI.17-24-09642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf M, Sanchez-Soriano N, Technau GM, Urban J, Prokop A. Charting the Drosophila neuropile: a strategy for the standardised characterisation of genetically amenable neurites. Dev Biol. 2003;260:207–225. doi: 10.1016/s0012-1606(03)00215-x. [DOI] [PubMed] [Google Scholar]
- Lunde K, Trimble JL, Guichard A, Guss KA, Nauber U, Bier E. Activation of the knirps locus links patterning of morphogenesis of the second wing vein in Drosophila. Development. 2003;130:235–248. doi: 10.1242/dev.00207. [DOI] [PubMed] [Google Scholar]
- Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Budnik V. Introduction on the use of the Drosophila embryonic/larval neuromuscular junction as a model system to study synapse development and function, and a brief summary of pathfinding and target recognition. Int Rev Neurobiol. 2006;75:1–31. doi: 10.1016/S0074-7742(06)75001-2. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Liu X, Soanes KH, Krause HM, Irvine KD, Bell JB. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes and Development. 1998;12:3815–3820. doi: 10.1101/gad.12.24.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S. In vivo analysis of lacZ fusion genes in transgenic Drosophila melanogaster. Methods Enzymol. 2000;326:146–159. doi: 10.1016/s0076-6879(00)26052-7. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Simmonds AJ, Garg A, Fossheim L, Campbell SD, Bell JB. Molecular and functional analysis of scalloped recessive lethal alleles in Drosophila melanogaster. Genetics. 2004;166:1833–1843. doi: 10.1534/genetics.166.4.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant M, Bier E. In situ protocol for imaginal discs and pupal wings. 1997 Retrieved October 24, 2012, from superfly.ucsd.edu/bierlab/research/protocols/imagdisc.html.
- Van Vactor D, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Kearney JB, Guardiola AR, Crews ST. Single-cell mapping of neural and glial gene expression in the developing Drosophila CNS midline cells. Dev Biol. 2006;294:509–524. doi: 10.1016/j.ydbio.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Bell J, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes and Development. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Carroll SB. Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development. 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- Williams JA, Langeland JA, Thalley BS, Skeath JB, Carroll S. Expression of foreign protiens in E. coli using plasmid vectors and purification of specific polyclonal antibodies. New York: IRL Press; 1995. [Google Scholar]